
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CUL4B-DAPK2 (FusionGDB2 ID:20689) |
Fusion Gene Summary for CUL4B-DAPK2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CUL4B-DAPK2 | Fusion gene ID: 20689 | Hgene | Tgene | Gene symbol | CUL4B | DAPK2 | Gene ID | 8450 | 23604 |
| Gene name | cullin 4B | death associated protein kinase 2 | |
| Synonyms | CUL-4B|MRXHF2|MRXS15|MRXSC|SFM2 | DRP-1|DRP1 | |
| Cytomap | Xq24 | 15q22.31 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cullin-4B | death-associated protein kinase 2DAP kinase 2DAP-kinase-related protein 1 beta isoform | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | Q13620 | Q9UIK4 | |
| Ensembl transtripts involved in fusion gene | ENST00000336592, ENST00000371322, ENST00000404115, ENST00000486604, | ENST00000558482, ENST00000261891, ENST00000457488, ENST00000558069, | |
| Fusion gene scores | * DoF score | 5 X 6 X 4=120 | 8 X 9 X 8=576 |
| # samples | 6 | 13 | |
| ** MAII score | log2(6/120*10)=-1 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(13/576*10)=-2.14755718841386 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CUL4B [Title/Abstract] AND DAPK2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CUL4B(119691779)-DAPK2(64275953), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | CUL4B-DAPK2 seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. CUL4B-DAPK2 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. CUL4B-DAPK2 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. CUL4B-DAPK2 seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. CUL4B-DAPK2 seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CUL4B | GO:0010498 | proteasomal protein catabolic process | 25970626 |
| Hgene | CUL4B | GO:0035518 | histone H2A monoubiquitination | 22334663 |
| Hgene | CUL4B | GO:0070914 | UV-damage excision repair | 22334663 |
| Tgene | DAPK2 | GO:0006468 | protein phosphorylation | 10376525 |
| Tgene | DAPK2 | GO:0035556 | intracellular signal transduction | 10376525 |
 Fusion gene breakpoints across CUL4B (5'-gene) Fusion gene breakpoints across CUL4B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
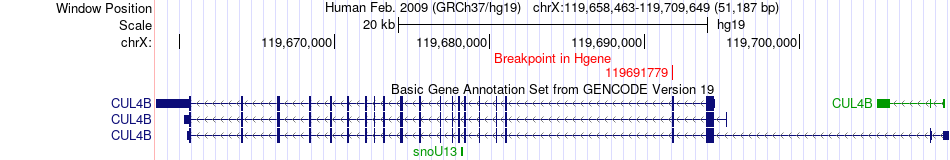 |
 Fusion gene breakpoints across DAPK2 (3'-gene) Fusion gene breakpoints across DAPK2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
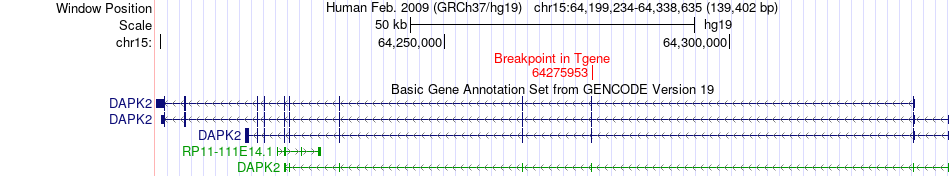 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LIHC | TCGA-DD-A11C-01A | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - |
Top |
Fusion Gene ORF analysis for CUL4B-DAPK2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000336592 | ENST00000558482 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - |
| 5CDS-5UTR | ENST00000371322 | ENST00000558482 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - |
| 5CDS-5UTR | ENST00000404115 | ENST00000558482 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - |
| Frame-shift | ENST00000336592 | ENST00000261891 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - |
| Frame-shift | ENST00000336592 | ENST00000457488 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - |
| Frame-shift | ENST00000336592 | ENST00000558069 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - |
| Frame-shift | ENST00000371322 | ENST00000261891 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - |
| Frame-shift | ENST00000371322 | ENST00000457488 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - |
| Frame-shift | ENST00000371322 | ENST00000558069 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - |
| In-frame | ENST00000404115 | ENST00000261891 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - |
| In-frame | ENST00000404115 | ENST00000457488 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - |
| In-frame | ENST00000404115 | ENST00000558069 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - |
| intron-3CDS | ENST00000486604 | ENST00000261891 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - |
| intron-3CDS | ENST00000486604 | ENST00000457488 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - |
| intron-3CDS | ENST00000486604 | ENST00000558069 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - |
| intron-5UTR | ENST00000486604 | ENST00000558482 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000404115 | CUL4B | chrX | 119691779 | - | ENST00000261891 | DAPK2 | chr15 | 64275953 | - | 3633 | 1128 | 1267 | 2148 | 293 |
| ENST00000404115 | CUL4B | chrX | 119691779 | - | ENST00000457488 | DAPK2 | chr15 | 64275953 | - | 2746 | 1128 | 1267 | 2148 | 293 |
| ENST00000404115 | CUL4B | chrX | 119691779 | - | ENST00000558069 | DAPK2 | chr15 | 64275953 | - | 2503 | 1128 | 1267 | 2502 | 412 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000404115 | ENST00000261891 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - | 0.0118952 | 0.9881048 |
| ENST00000404115 | ENST00000457488 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - | 0.013315667 | 0.9866844 |
| ENST00000404115 | ENST00000558069 | CUL4B | chrX | 119691779 | - | DAPK2 | chr15 | 64275953 | - | 0.004364174 | 0.9956358 |
Top |
Fusion Genomic Features for CUL4B-DAPK2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
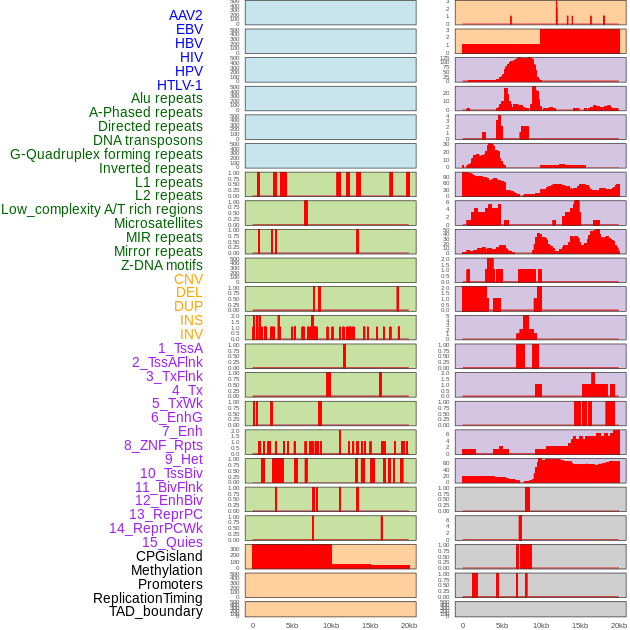 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for CUL4B-DAPK2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chrX:119691779/chr15:64275953) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CUL4B | DAPK2 |
| FUNCTION: Core component of multiple cullin-RING-based E3 ubiquitin-protein ligase complexes which mediate the ubiquitination and subsequent proteasomal degradation of target proteins. The functional specificity of the E3 ubiquitin-protein ligase complex depends on the variable substrate recognition subunit. CUL4B may act within the complex as a scaffold protein, contributing to catalysis through positioning of the substrate and the ubiquitin-conjugating enzyme. Plays a role as part of the E3 ubiquitin-protein ligase complex in polyubiquitination of CDT1, histone H2A, histone H3 and histone H4 in response to radiation-induced DNA damage. Targeted to UV damaged chromatin by DDB2 and may be important for DNA repair and DNA replication. Required for ubiquitination of cyclin E, and consequently, normal G1 cell cycle progression. Regulates the mammalian target-of-rapamycin (mTOR) pathway involved in control of cell growth, size and metabolism. Specific CUL4B regulation of the mTORC1-mediated pathway is dependent upon 26S proteasome function and requires interaction between CUL4B and MLST8. With CUL4A, contributes to ribosome biogenesis (PubMed:26711351). {ECO:0000269|PubMed:14578910, ECO:0000269|PubMed:16322693, ECO:0000269|PubMed:16678110, ECO:0000269|PubMed:18235224, ECO:0000269|PubMed:18593899, ECO:0000269|PubMed:19801544, ECO:0000269|PubMed:22118460, ECO:0000269|PubMed:26711351}. | FUNCTION: Calcium/calmodulin-dependent serine/threonine kinase involved in multiple cellular signaling pathways that trigger cell survival, apoptosis, and autophagy. Regulates both type I apoptotic and type II autophagic cell death signals, depending on the cellular setting. The former is caspase-dependent, while the latter is caspase-independent and is characterized by the accumulation of autophagic vesicles. Acts as a mediator of anoikis and a suppressor of beta-catenin-dependent anchorage-independent growth of malignant epithelial cells. May play a role in granulocytic maturation (PubMed:17347302). Regulates granulocytic motility by controlling cell spreading and polarization (PubMed:24163421). {ECO:0000269|PubMed:17347302, ECO:0000269|PubMed:24163421, ECO:0000269|PubMed:26047703}.; FUNCTION: Isoform 2 is not regulated by calmodulin. It can phosphorylate MYL9. It can induce membrane blebbing and autophagic cell death. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CUL4B | chrX:119691779 | chr15:64275953 | ENST00000371322 | - | 2 | 20 | 3_193 | 224 | 896.0 | Compositional bias | Note=Ser-rich |
| Hgene | CUL4B | chrX:119691779 | chr15:64275953 | ENST00000404115 | - | 4 | 22 | 3_193 | 242 | 914.0 | Compositional bias | Note=Ser-rich |
| Tgene | DAPK2 | chrX:119691779 | chr15:64275953 | ENST00000261891 | 0 | 11 | 29_37 | 30 | 371.0 | Nucleotide binding | ATP | |
| Tgene | DAPK2 | chrX:119691779 | chr15:64275953 | ENST00000457488 | 1 | 12 | 29_37 | 30 | 371.0 | Nucleotide binding | ATP | |
| Tgene | DAPK2 | chrX:119691779 | chr15:64275953 | ENST00000558069 | 1 | 10 | 29_37 | 30 | 489.0 | Nucleotide binding | ATP | |
| Tgene | DAPK2 | chrX:119691779 | chr15:64275953 | ENST00000261891 | 0 | 11 | 277_344 | 30 | 371.0 | Region | Note=Calmodulin-binding | |
| Tgene | DAPK2 | chrX:119691779 | chr15:64275953 | ENST00000261891 | 0 | 11 | 292_301 | 30 | 371.0 | Region | Autoinhibitory domain | |
| Tgene | DAPK2 | chrX:119691779 | chr15:64275953 | ENST00000457488 | 1 | 12 | 277_344 | 30 | 371.0 | Region | Note=Calmodulin-binding | |
| Tgene | DAPK2 | chrX:119691779 | chr15:64275953 | ENST00000457488 | 1 | 12 | 292_301 | 30 | 371.0 | Region | Autoinhibitory domain | |
| Tgene | DAPK2 | chrX:119691779 | chr15:64275953 | ENST00000558069 | 1 | 10 | 277_344 | 30 | 489.0 | Region | Note=Calmodulin-binding | |
| Tgene | DAPK2 | chrX:119691779 | chr15:64275953 | ENST00000558069 | 1 | 10 | 292_301 | 30 | 489.0 | Region | Autoinhibitory domain |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | DAPK2 | chrX:119691779 | chr15:64275953 | ENST00000261891 | 0 | 11 | 23_285 | 30 | 371.0 | Domain | Protein kinase | |
| Tgene | DAPK2 | chrX:119691779 | chr15:64275953 | ENST00000457488 | 1 | 12 | 23_285 | 30 | 371.0 | Domain | Protein kinase | |
| Tgene | DAPK2 | chrX:119691779 | chr15:64275953 | ENST00000558069 | 1 | 10 | 23_285 | 30 | 489.0 | Domain | Protein kinase |
Top |
Fusion Gene Sequence for CUL4B-DAPK2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >20689_20689_1_CUL4B-DAPK2_CUL4B_chrX_119691779_ENST00000404115_DAPK2_chr15_64275953_ENST00000261891_length(transcript)=3633nt_BP=1128nt AGTTCAGTTTAGAAACTGCCAGTCTTGGTATAGAACAACCTAGGCTTATCTGTTTAAGTCCCATAGGTAGTATTATTAGAGAATAACCCA GGCTTTGGGAACTGTATAACTCTGAGATCGTTTGAAATTTTCGTTTTGTTTTTCTTTTGGCTCCAACCAGGCCTCTGTTGTTAGACTAAT GGGGTTTCCCCTGCAGAGATTGGTCAGCCCTGCAGTCTATCATCCCGGAAGGTGTTAAATACACATGAAAGCCAGCCTCGGCGGGCAGAA CCTAGCCCGCCGAAAGGGCGGGCATTGCTAAAGTGTTTTCAAAAGGTCTTCCCAATCCTCTGCTTTAGACAGCTCGGACCTTAGATAGAA GAACACAACGGATCCTCCAGTGACTGACGACCCAAAGGACGGATGATGTCACAGTCATCTGGATCAGGAGATGGGAATGATGATGAGGCT ACTACCTCTAAAGACGGTGGTTTTTCTTCCCCCAGTCCCTCAGCTGCTGCTGCTGCTCAGGAGGTCAGATCTGCCACTGATGGTAATACC AGCACCACTCCGCCCACCTCTGCCAAGAAGAGAAAGTTAAACAGCAGCAGCAGTAGCAGCAGTAACAGTAGTAACGAGAGAGAAGACTTT GATTCCACCTCTTCCTCCTCTTCCACTCCTCCTTTACAACCCAGGGATTCGGCATCCCCTTCAACCTCGTCCTTCTGCCTGGGGGTTTCA GTGGCTGCTTCCAGCCACGTACCGATACAGAAGAAGCTGCGTTTTGAAGACACCCTGGAGTTTGTAGGGTTTGATGCGAAGATGGCTGAG GAATCCTCCTCCTCCTCCTCCTCATCTTCACCAACTGCTGCAACATCTCAGCAGCAGCAACTTAAAAATAAGAGTATATTAATCTCTTCT GTGGCTTCGGTGCATCATGCAAACGGCCTAGCCAAATCTTCTACCACCGTCTCTAGCTTTGCTAACAGCAAACCTGGCTCTGCTAAGAAG TTAGTGATCAAGAACTTTAAAGATAAGCCTAAATTACCAGAAAACTACACAGATGAAACCTGGCAAAAACTGAAAGAAGCAGTGGAAGCT ATTCAGAATAGTACTTCAATTAAGTACAATTTAGAAGAACTCTACCAGTGGCCAGTTTGCCATCGTGAAGAAGTGCCGGGAGAAGAGCAC GGGGCTTGAGTATGCAGCCAAGTTCATCAAGAAGCGGCAGAGCCGGGCGAGCCGGCGCGGTGTGAGCCGGGAGGAGATCGAGCGGGAGGT GAGCATCCTGCGGCAGGTGCTGCACCACAATGTCATCACGCTGCACGACGTCTATGAGAACCGCACCGACGTGGTGCTCATCCTTGAGCT AGTGTCTGGAGGAGAGCTCTTCGATTTCCTGGCCCAGAAGGAGTCACTGAGTGAGGAGGAGGCCACCAGCTTCATTAAGCAGATCCTGGA TGGGGTGAACTACCTTCACACAAAGAAAATTGCTCACTTTGATCTCAAGCCAGAAAACATTATGTTGTTAGACAAGAATATTCCCATTCC ACACATCAAGCTGATTGACTTTGGTCTGGCTCACGAAATAGAAGATGGAGTTGAATTTAAGAATATTTTTGGGACGCCGGAATTTGTTGC TCCAGAAATTGTGAACTACGAGCCCCTGGGTCTGGAGGCTGACATGTGGAGCATAGGCGTCATCACCTACATCCTCTTAAGTGGAGCATC CCCTTTCCTGGGAGACACGAAGCAGGAAACACTGGCAAATATCACAGCAGTGAGTTACGACTTTGATGAGGAATTCTTCAGCCAGACGAG CGAGCTGGCCAAGGACTTTATTCGGAAGCTTCTGGTTAAAGAGACCCGGAAACGGCTCACAATCCAAGAGGCTCTCAGACACCCCTGGAT CACGCCGGTGGACAACCAGCAAGCCATGGTGCGCAGGGAGTCTGTGGTCAATCTGGAGAACTTCAGGAAGCAGTATGTCCGCAGGCGGTG GAAGCTTTCCTTCAGCATCGTGTCCCTGTGCAACCACCTCACCCGCTCGCTGATGAAGAAGGTGCACCTGAGGCCGGATGAGGACCTGAG GAACTGTGAGAGTGACACTGAGGAGGACATCGCCAGGAGGAAAGCCCTCCACCCACGGAGGAGGAGCAGCACCTCCTAACTGGCCTGACC TGCAGTGGCCGCCAGGGAGGTCTGGGCCCAGCGGGGCTCCCTTCTGTGCAGACTTTTGGACCCAGCTCAGCACCAGCACCCGGGCGTCCT GAGCACTTTGCAAGAGAGATGGGCCCAAGGAATTCAGAAGAGCTTGCAGGCAAGCCAGGAGACCCTGGGAGCTGTGGCTGTCTTCTGTGG AGGAGGCTCCAGCATTCCCAAAGCTCTTAATTCTCCATAAAATGGGCTTTCCTCTGTCTGCCATCCTCAGAGTCTGGGGTGGGAGTGTGG ACTTAGGAAAACAATATAAAGGACATCCTCATCATCACGGGGTGAAGGTCAGACTAAGGCAGCCTTCTTCACAGGCTGAGGGGGTTCAGA ACCAGCCTGGCCAAAAATTACACCAGAGAGACAGAGTCCTCCCCATTGGGAACAGGGTGATTGAGGAAAGTGAACCTTGGGTGTGAGGGA CCAATCCTGTGACCTCCCAGAACCATGGAAGCCAGGACGTCAGGCTGACCAACACCTCAGACCTTCTGAAGCAGCCCATTGCTGGCCCGC CATGTTGTAATTTTGCTCATTTTTATTAAACTTCTGGTTTACCTGATGCTTGGCTTCTTTTAGGGCTACCCCCATCTCATTTCCTTTAGC CCGTGTGCCTGTAACTCTGAGGGGGGGCACCCAGTGGGGTGCTGAGTGGGCAGAATCTCAGAAGGTCCTCCTGAACCGTCCGCGCAGGCC TGCAGTGGGCCTGCCTCCTCCTTGCTTCCCTAACAGGAAGGTGTCCAGTTCAAGAGAACCCACCCAGAGACTGGGAGTGGTGGCTCACGC CTATAATCCCTGCGCTTTGGCAGTCCGAGGCAGGGGAATTGCTTGAACTCAGGAGTTGGAGACCAGCCTGGGCAACATGGCAAAACGCAG TCTGTACAAAAAATACAAAAAATTAGCCAGGTGTAGGGGTAGGCACCTGGCATCCCAGCTACTCCAGGGGCTGAGGTGACAGCATTGCTT AAGCCCAGAAGGTCGAGGCTGCAGTGAGCTGAGATCACGCCACTGCACTCCAGTCTGGGTGACAGAGAGAGACCATATCCAAAAAAAAAA AAAGTTGCCAGAGACGAGTATGCCCATGCTCCCTCTACCTCACTGCCACCACTCCTGCTGTTAGGAGCTGAGTGTGTCTCCCTAAAATTT CTATGTTGAAGTCTTAACCCTTGGTACCACAGAATATCACTGTATTTGGAGATGGGGTCTTTAGAAAGGCACTTAAATTAAAATGAGCTC ACTGATATGGGCCCCGATGCAATATAATTGGTGTCCTTATAAGAAGGGGAGGTTAGGACACGCAGGAAAGACCACATGAAGGCCCAGGAG TGGGAGGGGGAATAGCCATCGACAAACTAAGGGGGCCTCAGAGGAAACCAACCCTGCTGACACCTCAATCTTAGACTCTGGCCTCAAAAA >20689_20689_1_CUL4B-DAPK2_CUL4B_chrX_119691779_ENST00000404115_DAPK2_chr15_64275953_ENST00000261891_length(amino acids)=293AA_BP= MRQVLHHNVITLHDVYENRTDVVLILELVSGGELFDFLAQKESLSEEEATSFIKQILDGVNYLHTKKIAHFDLKPENIMLLDKNIPIPHI KLIDFGLAHEIEDGVEFKNIFGTPEFVAPEIVNYEPLGLEADMWSIGVITYILLSGASPFLGDTKQETLANITAVSYDFDEEFFSQTSEL AKDFIRKLLVKETRKRLTIQEALRHPWITPVDNQQAMVRRESVVNLENFRKQYVRRRWKLSFSIVSLCNHLTRSLMKKVHLRPDEDLRNC -------------------------------------------------------------- >20689_20689_2_CUL4B-DAPK2_CUL4B_chrX_119691779_ENST00000404115_DAPK2_chr15_64275953_ENST00000457488_length(transcript)=2746nt_BP=1128nt AGTTCAGTTTAGAAACTGCCAGTCTTGGTATAGAACAACCTAGGCTTATCTGTTTAAGTCCCATAGGTAGTATTATTAGAGAATAACCCA GGCTTTGGGAACTGTATAACTCTGAGATCGTTTGAAATTTTCGTTTTGTTTTTCTTTTGGCTCCAACCAGGCCTCTGTTGTTAGACTAAT GGGGTTTCCCCTGCAGAGATTGGTCAGCCCTGCAGTCTATCATCCCGGAAGGTGTTAAATACACATGAAAGCCAGCCTCGGCGGGCAGAA CCTAGCCCGCCGAAAGGGCGGGCATTGCTAAAGTGTTTTCAAAAGGTCTTCCCAATCCTCTGCTTTAGACAGCTCGGACCTTAGATAGAA GAACACAACGGATCCTCCAGTGACTGACGACCCAAAGGACGGATGATGTCACAGTCATCTGGATCAGGAGATGGGAATGATGATGAGGCT ACTACCTCTAAAGACGGTGGTTTTTCTTCCCCCAGTCCCTCAGCTGCTGCTGCTGCTCAGGAGGTCAGATCTGCCACTGATGGTAATACC AGCACCACTCCGCCCACCTCTGCCAAGAAGAGAAAGTTAAACAGCAGCAGCAGTAGCAGCAGTAACAGTAGTAACGAGAGAGAAGACTTT GATTCCACCTCTTCCTCCTCTTCCACTCCTCCTTTACAACCCAGGGATTCGGCATCCCCTTCAACCTCGTCCTTCTGCCTGGGGGTTTCA GTGGCTGCTTCCAGCCACGTACCGATACAGAAGAAGCTGCGTTTTGAAGACACCCTGGAGTTTGTAGGGTTTGATGCGAAGATGGCTGAG GAATCCTCCTCCTCCTCCTCCTCATCTTCACCAACTGCTGCAACATCTCAGCAGCAGCAACTTAAAAATAAGAGTATATTAATCTCTTCT GTGGCTTCGGTGCATCATGCAAACGGCCTAGCCAAATCTTCTACCACCGTCTCTAGCTTTGCTAACAGCAAACCTGGCTCTGCTAAGAAG TTAGTGATCAAGAACTTTAAAGATAAGCCTAAATTACCAGAAAACTACACAGATGAAACCTGGCAAAAACTGAAAGAAGCAGTGGAAGCT ATTCAGAATAGTACTTCAATTAAGTACAATTTAGAAGAACTCTACCAGTGGCCAGTTTGCCATCGTGAAGAAGTGCCGGGAGAAGAGCAC GGGGCTTGAGTATGCAGCCAAGTTCATCAAGAAGCGGCAGAGCCGGGCGAGCCGGCGCGGTGTGAGCCGGGAGGAGATCGAGCGGGAGGT GAGCATCCTGCGGCAGGTGCTGCACCACAATGTCATCACGCTGCACGACGTCTATGAGAACCGCACCGACGTGGTGCTCATCCTTGAGCT AGTGTCTGGAGGAGAGCTCTTCGATTTCCTGGCCCAGAAGGAGTCACTGAGTGAGGAGGAGGCCACCAGCTTCATTAAGCAGATCCTGGA TGGGGTGAACTACCTTCACACAAAGAAAATTGCTCACTTTGATCTCAAGCCAGAAAACATTATGTTGTTAGACAAGAATATTCCCATTCC ACACATCAAGCTGATTGACTTTGGTCTGGCTCACGAAATAGAAGATGGAGTTGAATTTAAGAATATTTTTGGGACGCCGGAATTTGTTGC TCCAGAAATTGTGAACTACGAGCCCCTGGGTCTGGAGGCTGACATGTGGAGCATAGGCGTCATCACCTACATCCTCTTAAGTGGAGCATC CCCTTTCCTGGGAGACACGAAGCAGGAAACACTGGCAAATATCACAGCAGTGAGTTACGACTTTGATGAGGAATTCTTCAGCCAGACGAG CGAGCTGGCCAAGGACTTTATTCGGAAGCTTCTGGTTAAAGAGACCCGGAAACGGCTCACAATCCAAGAGGCTCTCAGACACCCCTGGAT CACGCCGGTGGACAACCAGCAAGCCATGGTGCGCAGGGAGTCTGTGGTCAATCTGGAGAACTTCAGGAAGCAGTATGTCCGCAGGCGGTG GAAGCTTTCCTTCAGCATCGTGTCCCTGTGCAACCACCTCACCCGCTCGCTGATGAAGAAGGTGCACCTGAGGCCGGATGAGGACCTGAG GAACTGTGAGAGTGACACTGAGGAGGACATCGCCAGGAGGAAAGCCCTCCACCCACGGAGGAGGAGCAGCACCTCCTAACTGGCCTGACC TGCAGTGGCCGCCAGGGAGGTCTGGGCCCAGCGGGGCTCCCTTCTGTGCAGACTTTTGGACCCAGCTCAGCACCAGCACCCGGGCGTCCT GAGCACTTTGCAAGAGAGATGGGCCCAAGGAATTCAGAAGAGCTTGCAGGCAAGCCAGGAGACCCTGGGAGCTGTGGCTGTCTTCTGTGG AGGAGGCTCCAGCATTCCCAAAGCTCTTAATTCTCCATAAAATGGGCTTTCCTCTGTCTGCCATCCTCAGAGTCTGGGGTGGGAGTGTGG ACTTAGGAAAACAATATAAAGGACATCCTCATCATCACGGGGTGAAGGTCAGACTAAGGCAGCCTTCTTCACAGGCTGAGGGGGTTCAGA ACCAGCCTGGCCAAAAATTACACCAGAGAGACAGAGTCCTCCCCATTGGGAACAGGGTGATTGAGGAAAGTGAACCTTGGGTGTGAGGGA CCAATCCTGTGACCTCCCAGAACCATGGAAGCCAGGACGTCAGGCTGACCAACACCTCAGACCTTCTGAAGCAGCCCATTGCTGGCCCGC >20689_20689_2_CUL4B-DAPK2_CUL4B_chrX_119691779_ENST00000404115_DAPK2_chr15_64275953_ENST00000457488_length(amino acids)=293AA_BP= MRQVLHHNVITLHDVYENRTDVVLILELVSGGELFDFLAQKESLSEEEATSFIKQILDGVNYLHTKKIAHFDLKPENIMLLDKNIPIPHI KLIDFGLAHEIEDGVEFKNIFGTPEFVAPEIVNYEPLGLEADMWSIGVITYILLSGASPFLGDTKQETLANITAVSYDFDEEFFSQTSEL AKDFIRKLLVKETRKRLTIQEALRHPWITPVDNQQAMVRRESVVNLENFRKQYVRRRWKLSFSIVSLCNHLTRSLMKKVHLRPDEDLRNC -------------------------------------------------------------- >20689_20689_3_CUL4B-DAPK2_CUL4B_chrX_119691779_ENST00000404115_DAPK2_chr15_64275953_ENST00000558069_length(transcript)=2503nt_BP=1128nt AGTTCAGTTTAGAAACTGCCAGTCTTGGTATAGAACAACCTAGGCTTATCTGTTTAAGTCCCATAGGTAGTATTATTAGAGAATAACCCA GGCTTTGGGAACTGTATAACTCTGAGATCGTTTGAAATTTTCGTTTTGTTTTTCTTTTGGCTCCAACCAGGCCTCTGTTGTTAGACTAAT GGGGTTTCCCCTGCAGAGATTGGTCAGCCCTGCAGTCTATCATCCCGGAAGGTGTTAAATACACATGAAAGCCAGCCTCGGCGGGCAGAA CCTAGCCCGCCGAAAGGGCGGGCATTGCTAAAGTGTTTTCAAAAGGTCTTCCCAATCCTCTGCTTTAGACAGCTCGGACCTTAGATAGAA GAACACAACGGATCCTCCAGTGACTGACGACCCAAAGGACGGATGATGTCACAGTCATCTGGATCAGGAGATGGGAATGATGATGAGGCT ACTACCTCTAAAGACGGTGGTTTTTCTTCCCCCAGTCCCTCAGCTGCTGCTGCTGCTCAGGAGGTCAGATCTGCCACTGATGGTAATACC AGCACCACTCCGCCCACCTCTGCCAAGAAGAGAAAGTTAAACAGCAGCAGCAGTAGCAGCAGTAACAGTAGTAACGAGAGAGAAGACTTT GATTCCACCTCTTCCTCCTCTTCCACTCCTCCTTTACAACCCAGGGATTCGGCATCCCCTTCAACCTCGTCCTTCTGCCTGGGGGTTTCA GTGGCTGCTTCCAGCCACGTACCGATACAGAAGAAGCTGCGTTTTGAAGACACCCTGGAGTTTGTAGGGTTTGATGCGAAGATGGCTGAG GAATCCTCCTCCTCCTCCTCCTCATCTTCACCAACTGCTGCAACATCTCAGCAGCAGCAACTTAAAAATAAGAGTATATTAATCTCTTCT GTGGCTTCGGTGCATCATGCAAACGGCCTAGCCAAATCTTCTACCACCGTCTCTAGCTTTGCTAACAGCAAACCTGGCTCTGCTAAGAAG TTAGTGATCAAGAACTTTAAAGATAAGCCTAAATTACCAGAAAACTACACAGATGAAACCTGGCAAAAACTGAAAGAAGCAGTGGAAGCT ATTCAGAATAGTACTTCAATTAAGTACAATTTAGAAGAACTCTACCAGTGGCCAGTTTGCCATCGTGAAGAAGTGCCGGGAGAAGAGCAC GGGGCTTGAGTATGCAGCCAAGTTCATCAAGAAGCGGCAGAGCCGGGCGAGCCGGCGCGGTGTGAGCCGGGAGGAGATCGAGCGGGAGGT GAGCATCCTGCGGCAGGTGCTGCACCACAATGTCATCACGCTGCACGACGTCTATGAGAACCGCACCGACGTGGTGCTCATCCTTGAGCT AGTGTCTGGAGGAGAGCTCTTCGATTTCCTGGCCCAGAAGGAGTCACTGAGTGAGGAGGAGGCCACCAGCTTCATTAAGCAGATCCTGGA TGGGGTGAACTACCTTCACACAAAGAAAATTGCTCACTTTGATCTCAAGCCAGAAAACATTATGTTGTTAGACAAGAATATTCCCATTCC ACACATCAAGCTGATTGACTTTGGTCTGGCTCACGAAATAGAAGATGGAGTTGAATTTAAGAATATTTTTGGGACGCCGGAATTTGTTGC TCCAGAAATTGTGAACTACGAGCCCCTGGGTCTGGAGGCTGACATGTGGAGCATAGGCGTCATCACCTACATCCTCTTAAGTGGAGCATC CCCTTTCCTGGGAGACACGAAGCAGGAAACACTGGCAAATATCACAGCAGTGAGTTACGACTTTGATGAGGAATTCTTCAGCCAGACGAG CGAGCTGGCCAAGGACTTTATTCGGAAGCTTCTGGTTAAAGAGACCCGGAAACGGCTCACAATCCAAGAGGCTCTCAGACACCCCTGGAT CACGTCAAAAGGAGAAGGCAGAGCCCCTGAACAGCGGAAGACAGAGCCCACCCAGCTGAAGACCAAGCACCTGAGAGAGTACACTCTCAA ATGCCACTCAAGCATGCCTCCCAACAACTCCTATGTCAACTTCGAGCGTTTTGCCTGTGTGGTGGAAGACGTGGCTCGGGTCGACCTGGG ATGCCGTGCCCTGGTGGAGGCCCACGATACCATCCAGGACGATGTGGAGGCCCTGGTCTCCATCTTCAATGAGAAGGAAGCTTGGTACCG AGAGGAGAATGAGAGCGCCCGGCATGACCTGTCCCAGCTCAGGTATGAGTTCCGCAAGGTGGAGTCATTGAAGAAGCTGCTGCGAGAAGA CATCCAGGCCACGGGATGTAGCCTCGGAAGCATGGCCAGGAAACTGGACCATCTGCAAGCCCAGTTTGAGATTCTGAGGCAGGAGCTCTC CGCAGACCTGCAGTGGATTCAGGAGCTGGTGGGCAGCTTCCAGCTGGAGAGTGGGAGCTCTGAGGGCCTGGGCTCCACATTCTACCAGGA >20689_20689_3_CUL4B-DAPK2_CUL4B_chrX_119691779_ENST00000404115_DAPK2_chr15_64275953_ENST00000558069_length(amino acids)=412AA_BP= MRQVLHHNVITLHDVYENRTDVVLILELVSGGELFDFLAQKESLSEEEATSFIKQILDGVNYLHTKKIAHFDLKPENIMLLDKNIPIPHI KLIDFGLAHEIEDGVEFKNIFGTPEFVAPEIVNYEPLGLEADMWSIGVITYILLSGASPFLGDTKQETLANITAVSYDFDEEFFSQTSEL AKDFIRKLLVKETRKRLTIQEALRHPWITSKGEGRAPEQRKTEPTQLKTKHLREYTLKCHSSMPPNNSYVNFERFACVVEDVARVDLGCR ALVEAHDTIQDDVEALVSIFNEKEAWYREENESARHDLSQLRYEFRKVESLKKLLREDIQATGCSLGSMARKLDHLQAQFEILRQELSAD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CUL4B-DAPK2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CUL4B-DAPK2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CUL4B-DAPK2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
