
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CYP4F3-PTMS (FusionGDB2 ID:21097) |
Fusion Gene Summary for CYP4F3-PTMS |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CYP4F3-PTMS | Fusion gene ID: 21097 | Hgene | Tgene | Gene symbol | CYP4F3 | PTMS | Gene ID | 4051 | 5763 |
| Gene name | cytochrome P450 family 4 subfamily F member 3 | parathymosin | |
| Synonyms | CPF3|CYP4F|CYPIVF3|LTB4H | ParaT | |
| Cytomap | 19p13.12 | 12p13.31 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cytochrome P450 4F320-HETE synthase20-hydroxyeicosatetraenoic acid synthasecytochrome P-450cytochrome P450, family 4, subfamily F, polypeptide 3cytochrome P450, subfamily IVF, polypeptide 3 (leukotriene B4 omega hydroxylase)cytochrome P450-LTB-omega | parathymosin | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q08477 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000221307, ENST00000585846, ENST00000586182, ENST00000591058, ENST00000588886, | ENST00000538057, ENST00000309083, ENST00000389462, | |
| Fusion gene scores | * DoF score | 1 X 1 X 1=1 | 16 X 10 X 10=1600 |
| # samples | 1 | 16 | |
| ** MAII score | log2(1/1*10)=3.32192809488736 | log2(16/1600*10)=-3.32192809488736 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CYP4F3 [Title/Abstract] AND PTMS [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CYP4F3(15752423)-PTMS(6878769), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across CYP4F3 (5'-gene) Fusion gene breakpoints across CYP4F3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
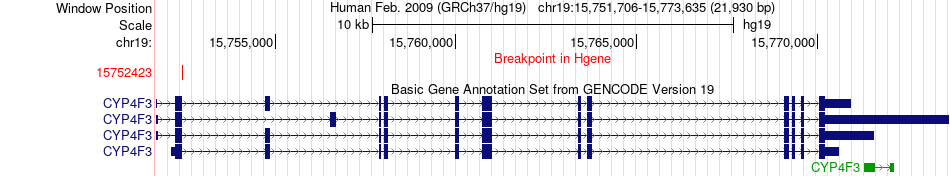 |
 Fusion gene breakpoints across PTMS (3'-gene) Fusion gene breakpoints across PTMS (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
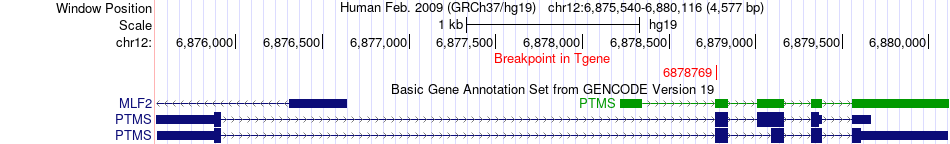 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LIHC | TCGA-EP-A2KB-01A | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + |
Top |
Fusion Gene ORF analysis for CYP4F3-PTMS |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000221307 | ENST00000538057 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + |
| 5CDS-3UTR | ENST00000585846 | ENST00000538057 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + |
| 5CDS-3UTR | ENST00000586182 | ENST00000538057 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + |
| 5CDS-3UTR | ENST00000591058 | ENST00000538057 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + |
| In-frame | ENST00000221307 | ENST00000309083 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + |
| In-frame | ENST00000221307 | ENST00000389462 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + |
| In-frame | ENST00000585846 | ENST00000309083 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + |
| In-frame | ENST00000585846 | ENST00000389462 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + |
| In-frame | ENST00000586182 | ENST00000309083 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + |
| In-frame | ENST00000586182 | ENST00000389462 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + |
| In-frame | ENST00000591058 | ENST00000309083 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + |
| In-frame | ENST00000591058 | ENST00000389462 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + |
| intron-3CDS | ENST00000588886 | ENST00000309083 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + |
| intron-3CDS | ENST00000588886 | ENST00000389462 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + |
| intron-3UTR | ENST00000588886 | ENST00000538057 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000586182 | CYP4F3 | chr19 | 15752423 | + | ENST00000389462 | PTMS | chr12 | 6878769 | + | 629 | 232 | 564 | 1 | 188 |
| ENST00000586182 | CYP4F3 | chr19 | 15752423 | + | ENST00000309083 | PTMS | chr12 | 6878769 | + | 997 | 232 | 567 | 1 | 189 |
| ENST00000591058 | CYP4F3 | chr19 | 15752423 | + | ENST00000389462 | PTMS | chr12 | 6878769 | + | 642 | 245 | 577 | 2 | 192 |
| ENST00000591058 | CYP4F3 | chr19 | 15752423 | + | ENST00000309083 | PTMS | chr12 | 6878769 | + | 1010 | 245 | 580 | 2 | 193 |
| ENST00000221307 | CYP4F3 | chr19 | 15752423 | + | ENST00000389462 | PTMS | chr12 | 6878769 | + | 642 | 245 | 577 | 2 | 192 |
| ENST00000221307 | CYP4F3 | chr19 | 15752423 | + | ENST00000309083 | PTMS | chr12 | 6878769 | + | 1010 | 245 | 580 | 2 | 193 |
| ENST00000585846 | CYP4F3 | chr19 | 15752423 | + | ENST00000389462 | PTMS | chr12 | 6878769 | + | 702 | 305 | 637 | 83 | 184 |
| ENST00000585846 | CYP4F3 | chr19 | 15752423 | + | ENST00000309083 | PTMS | chr12 | 6878769 | + | 1070 | 305 | 858 | 292 | 188 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000586182 | ENST00000389462 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + | 0.025140723 | 0.97485936 |
| ENST00000586182 | ENST00000309083 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + | 0.003724387 | 0.99627566 |
| ENST00000591058 | ENST00000389462 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + | 0.026985902 | 0.9730142 |
| ENST00000591058 | ENST00000309083 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + | 0.003055219 | 0.9969447 |
| ENST00000221307 | ENST00000389462 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + | 0.026985902 | 0.9730142 |
| ENST00000221307 | ENST00000309083 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + | 0.003055219 | 0.9969447 |
| ENST00000585846 | ENST00000389462 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + | 0.03468168 | 0.96531826 |
| ENST00000585846 | ENST00000309083 | CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878769 | + | 0.003566206 | 0.99643373 |
Top |
Fusion Genomic Features for CYP4F3-PTMS |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878768 | + | 5.96E-06 | 0.99999404 |
| CYP4F3 | chr19 | 15752423 | + | PTMS | chr12 | 6878768 | + | 5.96E-06 | 0.99999404 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
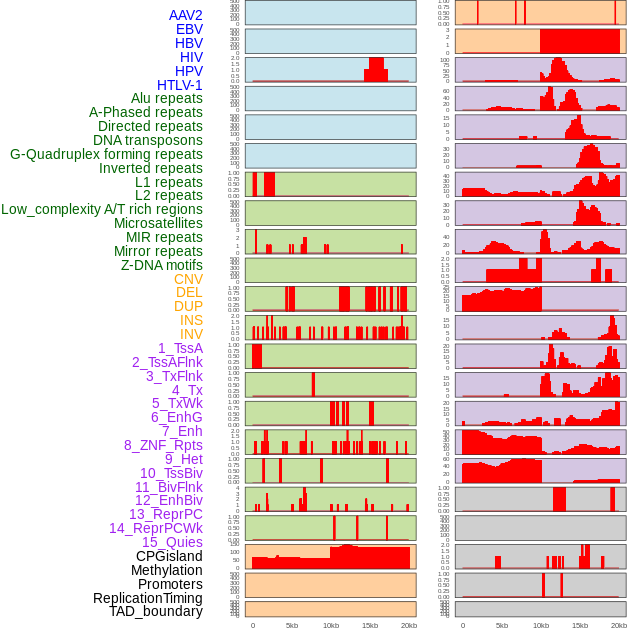 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
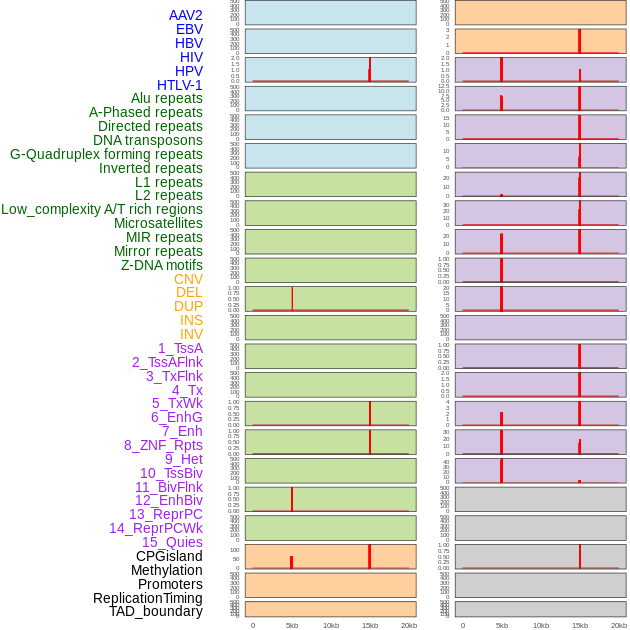 |
Top |
Fusion Protein Features for CYP4F3-PTMS |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:15752423/chr12:6878769) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CYP4F3 | . |
| FUNCTION: A cytochrome P450 monooxygenase involved in the metabolism of various endogenous substrates, including fatty acids and their oxygenated derivatives (oxylipins) (PubMed:8486631, PubMed:9675028, PubMed:11461919, PubMed:15145985, PubMed:16547005, PubMed:16820285, PubMed:18182499, PubMed:18065749, PubMed:18577768). Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (CPR; NADPH-ferrihemoprotein reductase) (PubMed:9675028). May play a role in inactivation of proinflammatory and anti-inflammatory oxylipins during the resolution of inflammation (PubMed:8486631, PubMed:9675028, PubMed:11461919, PubMed:15145985, PubMed:15364545, PubMed:16547005, PubMed:16820285, PubMed:18182499, PubMed:18065749, PubMed:18577768). {ECO:0000269|PubMed:11461919, ECO:0000269|PubMed:15145985, ECO:0000269|PubMed:15364545, ECO:0000269|PubMed:16547005, ECO:0000269|PubMed:16820285, ECO:0000269|PubMed:18065749, ECO:0000269|PubMed:18182499, ECO:0000269|PubMed:18577768, ECO:0000269|PubMed:8486631, ECO:0000269|PubMed:9675028}.; FUNCTION: [Isoform CYP4F3A]: Catalyzes predominantly the oxidation of the terminal carbon (omega-oxidation) of oxylipins in myeloid cells, displaying higher affinity for arachidonate metabolite leukotriene B4 (LTB4) (PubMed:8486631, PubMed:9675028, PubMed:11461919, PubMed:15364545). Inactivates LTB4 via three successive oxidative transformations to 20-hydroxy-LTB4, then to 20-oxo-LTB4 and to 20-carboxy-LTB4 (PubMed:9675028). Has omega-hydroxylase activity toward long-chain fatty acid epoxides with preference for 8,9-epoxy-(5Z,11Z,14Z)-eicosatrienoate (EET) and 9,10-epoxyoctadecanoate (PubMed:15145985). Omega-hydroxylates monohydroxy polyunsaturated fatty acids (PUFAs), including hydroxyeicosatetraenoates (HETEs) and hydroxyeicosapentaenoates (HEPEs), to dihydroxy compounds (PubMed:15364545, PubMed:9675028). Contributes to the degradation of saturated very long-chain fatty acids (VLCFAs) such as docosanoic acid, by catalyzing successive omega-oxidations to the corresponding dicarboxylic acid, thereby initiating chain shortening (PubMed:18182499). Has low hydroxylase activity toward PUFAs (PubMed:18577768, PubMed:11461919). {ECO:0000269|PubMed:11461919, ECO:0000269|PubMed:15145985, ECO:0000269|PubMed:15364545, ECO:0000269|PubMed:18182499, ECO:0000269|PubMed:18577768, ECO:0000269|PubMed:8486631, ECO:0000269|PubMed:9675028}.; FUNCTION: [Isoform CYP4F3B]: Catalyzes predominantly the oxidation of the terminal carbon (omega-oxidation) of polyunsaturated fatty acids (PUFAs) (PubMed:11461919, PubMed:16820285, PubMed:18577768). Participates in the conversion of arachidonic acid to 20-hydroxyeicosatetraenoic acid (20-HETE), a signaling molecule acting both as vasoconstrictive and natriuretic with overall effect on arterial blood pressure (PubMed:11461919, PubMed:16820285, PubMed:18577768). Has high omega-hydroxylase activity toward other PUFAs, including eicosatrienoic acid (ETA), eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) (PubMed:16820285, PubMed:18577768). Can also catalyze the oxidation of the penultimate carbon (omega-1 oxidation) of PUFAs with lower efficiency (PubMed:18577768). Contributes to the degradation of saturated very long-chain fatty acids (VLCFAs) such as docosanoic acid and hexacosanoic acid, by catalyzing successive omega-oxidations to the corresponding dicarboxylic acids, thereby initiating chain shortening (PubMed:16547005, PubMed:18182499). Omega-hydroxylates long-chain 3-hydroxy fatty acids, likely initiating the oxidative conversion to the corresponding 3-hydroxydicarboxylic fatty acids (PubMed:18065749). Has omega-hydroxylase activity toward long-chain fatty acid epoxides with preference for 8,9-epoxy-(5Z,11Z,14Z)-eicosatrienoate (EET) and 9,10-epoxyoctadecanoate (PubMed:15145985). {ECO:0000269|PubMed:11461919, ECO:0000269|PubMed:15145985, ECO:0000269|PubMed:16547005, ECO:0000269|PubMed:16820285, ECO:0000269|PubMed:18065749, ECO:0000269|PubMed:18182499, ECO:0000269|PubMed:18577768}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CYP4F3 | chr19:15752423 | chr12:6878769 | ENST00000221307 | + | 2 | 13 | 11_31 | 66 | 521.0 | Transmembrane | Helical |
| Hgene | CYP4F3 | chr19:15752423 | chr12:6878769 | ENST00000585846 | + | 1 | 12 | 11_31 | 66 | 521.0 | Transmembrane | Helical |
| Hgene | CYP4F3 | chr19:15752423 | chr12:6878769 | ENST00000586182 | + | 2 | 13 | 11_31 | 66 | 521.0 | Transmembrane | Helical |
| Hgene | CYP4F3 | chr19:15752423 | chr12:6878769 | ENST00000591058 | + | 2 | 13 | 11_31 | 66 | 521.0 | Transmembrane | Helical |
| Tgene | PTMS | chr19:15752423 | chr12:6878769 | ENST00000309083 | 0 | 5 | 36_90 | 15 | 103.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Gene Sequence for CYP4F3-PTMS |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >21097_21097_1_CYP4F3-PTMS_CYP4F3_chr19_15752423_ENST00000221307_PTMS_chr12_6878769_ENST00000309083_length(transcript)=1010nt_BP=245nt AGAAGGGGAGAGGAGGTTGTGTGGGACAAGGTGCTCCTGACAGAAGGATGCCACAGCTGAGCCTGTCCTCGCTGGGCCTTTGGCCAATGG CAGCATCCCCGTGGCTGCTCCTGCTGCTGGTTGGGGCCTCCTGGCTCCTGGCCCGCATCCTGGCCTGGACCTATACCTTCTATGACAACT GCTGCCGCCTCCGGTGTTTCCCGCAACCCCCGAAACGGAATTGGTTCTTGGGTCACCTGGGCCTGGACCTGAAGGAGAAGAAGGAGAAGG TGGAGGAGAAGGCAAGCCGGAAAGAGCGAAAGAAAGAAGTGGTGGAGGAGGAGGAGAACGGGGCTGAGGAGGAAGAAGAAGAAACTGCCG AGGATGGAGAGGAGGAAGATGAAGGGGAAGAAGAAGATGAGGAAGAAGAAGAAGAGGATGATGAAGGGCCCGCGCTGAAGAGAGCTGCCG AAGAGGAGGATGAAGCGGATCCCAAACGGCAGAAGACAGAAAATGGGGCATCGGCGTGAGCCCCTGCCAACAGGCTGGGGTTGGGAGGCC TCTCTGGGCCTGGAGGTGGGGGTGGGGGCAGCCAAGTCCAGCCACTCTTCACCTGGCTCCCTGCTCTGGGCCCTGCACCAGAGCTGCCAC CCTCTTCTTTCTCCCCAGCCTTCTCATTTCCGCCTCTCCAGACACTGCGCCCTCCACCCTCACTCTGCCATTGTTCCACCTCCTGACCTG CTCCATCTGAGCTCTCCAGCTGGCCCCCAATTGCTCCTCTCTCTCTTTGCTCTCTTTCTCCCTCCCCTACCAGCCTCATTCTTCCTCCGG TAGCCTCTCCCACCTAACCTCTGCATCCCCCAGCCTCATGTCCTGCCCCATCCCTATCCTGCCTGATCCCTGGATCTCCCTCAGATCCCC TCTTCTCAGACAGCGCCAGGCCGGGGTGGGGCCGGGGTTGGGGCCGAGCCCCACAGCTGCCCCCCTCCCCTCCCTTTTTGTATAATTTAA >21097_21097_1_CYP4F3-PTMS_CYP4F3_chr19_15752423_ENST00000221307_PTMS_chr12_6878769_ENST00000309083_length(amino acids)=193AA_BP=1 MDLAAPTPTSRPREASQPQPVGRGSRRCPIFCLLPFGIRFILLFGSSLQRGPFIILFFFFLIFFFPFIFLLSILGSFFFFLLSPVLLLLH HFFLSLFPACLLLHLLLLLLQVQAQVTQEPIPFRGLRETPEAAAVVIEGIGPGQDAGQEPGGPNQQQEQPRGCCHWPKAQRGQAQLWHPS -------------------------------------------------------------- >21097_21097_2_CYP4F3-PTMS_CYP4F3_chr19_15752423_ENST00000221307_PTMS_chr12_6878769_ENST00000389462_length(transcript)=642nt_BP=245nt AGAAGGGGAGAGGAGGTTGTGTGGGACAAGGTGCTCCTGACAGAAGGATGCCACAGCTGAGCCTGTCCTCGCTGGGCCTTTGGCCAATGG CAGCATCCCCGTGGCTGCTCCTGCTGCTGGTTGGGGCCTCCTGGCTCCTGGCCCGCATCCTGGCCTGGACCTATACCTTCTATGACAACT GCTGCCGCCTCCGGTGTTTCCCGCAACCCCCGAAACGGAATTGGTTCTTGGGTCACCTGGGCCTGGACCTGAAGGAGAAGAAGGAGAAGG TGGAGGAGAAGGCAAGCCGGAAAGAGCGAAAGAAAGAAGTGGTGGAGGCTCCAGTCCCATTGGTGGTGATGGTGGGCAAGGCATGCAGCC AGCCTGAGGCTACTCCCTCTCCTGGTCCCCACAGGAGGAGGAGAACGGGGCTGAGGAGGAAGAAGAAGAAACTGCCGAGGATGGAGAGGA GGAAGATGAAGGGGAAGAAGAAGATGAGGAAGAAGAAGAAGAGGATGATGAAGGGCCCGCGCTGAAGAGAGCTGCCGAAGAGGAGGATGA AGCGGATCCCAAACGGCAGAAGACAGAAAATGGGGCATCGGCGTGAGCCCCTGCCAACAGGCTGGGGTTGGGAGGCCTCTCTGGGCCTGG >21097_21097_2_CYP4F3-PTMS_CYP4F3_chr19_15752423_ENST00000221307_PTMS_chr12_6878769_ENST00000389462_length(amino acids)=192AA_BP=0 MPHFLSSAVWDPLHPPLRQLSSARALHHPLLLLPHLLLPLHLPPLHPRQFLLLPPQPRSPPPVGTRRGSSLRLAACLAHHHHQWDWSLHH FFLSLFPACLLLHLLLLLLQVQAQVTQEPIPFRGLRETPEAAAVVIEGIGPGQDAGQEPGGPNQQQEQPRGCCHWPKAQRGQAQLWHPSV -------------------------------------------------------------- >21097_21097_3_CYP4F3-PTMS_CYP4F3_chr19_15752423_ENST00000585846_PTMS_chr12_6878769_ENST00000309083_length(transcript)=1070nt_BP=305nt TCTATCTCCAGCACTGCCCGTCTCTGCCTCTCTCCACTGTCCCTGGAGCTCCCTGGGCCCTTCCCTGGGCCTCAGGACCTCACTCACCAC CCCATCTGCCCTGCAGGATGCCACAGCTGAGCCTGTCCTCGCTGGGCCTTTGGCCAATGGCAGCATCCCCGTGGCTGCTCCTGCTGCTGG TTGGGGCCTCCTGGCTCCTGGCCCGCATCCTGGCCTGGACCTATACCTTCTATGACAACTGCTGCCGCCTCCGGTGTTTCCCGCAACCCC CGAAACGGAATTGGTTCTTGGGTCACCTGGGCCTGGACCTGAAGGAGAAGAAGGAGAAGGTGGAGGAGAAGGCAAGCCGGAAAGAGCGAA AGAAAGAAGTGGTGGAGGAGGAGGAGAACGGGGCTGAGGAGGAAGAAGAAGAAACTGCCGAGGATGGAGAGGAGGAAGATGAAGGGGAAG AAGAAGATGAGGAAGAAGAAGAAGAGGATGATGAAGGGCCCGCGCTGAAGAGAGCTGCCGAAGAGGAGGATGAAGCGGATCCCAAACGGC AGAAGACAGAAAATGGGGCATCGGCGTGAGCCCCTGCCAACAGGCTGGGGTTGGGAGGCCTCTCTGGGCCTGGAGGTGGGGGTGGGGGCA GCCAAGTCCAGCCACTCTTCACCTGGCTCCCTGCTCTGGGCCCTGCACCAGAGCTGCCACCCTCTTCTTTCTCCCCAGCCTTCTCATTTC CGCCTCTCCAGACACTGCGCCCTCCACCCTCACTCTGCCATTGTTCCACCTCCTGACCTGCTCCATCTGAGCTCTCCAGCTGGCCCCCAA TTGCTCCTCTCTCTCTTTGCTCTCTTTCTCCCTCCCCTACCAGCCTCATTCTTCCTCCGGTAGCCTCTCCCACCTAACCTCTGCATCCCC CAGCCTCATGTCCTGCCCCATCCCTATCCTGCCTGATCCCTGGATCTCCCTCAGATCCCCTCTTCTCAGACAGCGCCAGGCCGGGGTGGG >21097_21097_3_CYP4F3-PTMS_CYP4F3_chr19_15752423_ENST00000585846_PTMS_chr12_6878769_ENST00000309083_length(amino acids)=188AA_BP=0 MRLVGEGEREQRERGAIGGQLESSDGAGQEVEQWQSEGGGRSVWRGGNEKAGEKEEGGSSGAGPRAGSQVKSGWTWLPPPPPPGPERPPN PSLLAGAHADAPFSVFCRLGSASSSSSAALFSAGPSSSSSSSSSSSSSPSSSSSPSSAVSSSSSSAPFSSSSTTSFFRSFRLAFSSTFSF -------------------------------------------------------------- >21097_21097_4_CYP4F3-PTMS_CYP4F3_chr19_15752423_ENST00000585846_PTMS_chr12_6878769_ENST00000389462_length(transcript)=702nt_BP=305nt TCTATCTCCAGCACTGCCCGTCTCTGCCTCTCTCCACTGTCCCTGGAGCTCCCTGGGCCCTTCCCTGGGCCTCAGGACCTCACTCACCAC CCCATCTGCCCTGCAGGATGCCACAGCTGAGCCTGTCCTCGCTGGGCCTTTGGCCAATGGCAGCATCCCCGTGGCTGCTCCTGCTGCTGG TTGGGGCCTCCTGGCTCCTGGCCCGCATCCTGGCCTGGACCTATACCTTCTATGACAACTGCTGCCGCCTCCGGTGTTTCCCGCAACCCC CGAAACGGAATTGGTTCTTGGGTCACCTGGGCCTGGACCTGAAGGAGAAGAAGGAGAAGGTGGAGGAGAAGGCAAGCCGGAAAGAGCGAA AGAAAGAAGTGGTGGAGGCTCCAGTCCCATTGGTGGTGATGGTGGGCAAGGCATGCAGCCAGCCTGAGGCTACTCCCTCTCCTGGTCCCC ACAGGAGGAGGAGAACGGGGCTGAGGAGGAAGAAGAAGAAACTGCCGAGGATGGAGAGGAGGAAGATGAAGGGGAAGAAGAAGATGAGGA AGAAGAAGAAGAGGATGATGAAGGGCCCGCGCTGAAGAGAGCTGCCGAAGAGGAGGATGAAGCGGATCCCAAACGGCAGAAGACAGAAAA >21097_21097_4_CYP4F3-PTMS_CYP4F3_chr19_15752423_ENST00000585846_PTMS_chr12_6878769_ENST00000389462_length(amino acids)=184AA_BP=0 MPHFLSSAVWDPLHPPLRQLSSARALHHPLLLLPHLLLPLHLPPLHPRQFLLLPPQPRSPPPVGTRRGSSLRLAACLAHHHHQWDWSLHH FFLSLFPACLLLHLLLLLLQVQAQVTQEPIPFRGLRETPEAAAVVIEGIGPGQDAGQEPGGPNQQQEQPRGCCHWPKAQRGQAQLWHPAG -------------------------------------------------------------- >21097_21097_5_CYP4F3-PTMS_CYP4F3_chr19_15752423_ENST00000586182_PTMS_chr12_6878769_ENST00000309083_length(transcript)=997nt_BP=232nt AGAAGAAGGGGAGAGGAGGTTGTGTGGGACAAGGATGCCACAGCTGAGCCTGTCCTCGCTGGGCCTTTGGCCAATGGCAGCATCCCCGTG GCTGCTCCTGCTGCTGGTTGGGGCCTCCTGGCTCCTGGCCCGCATCCTGGCCTGGACCTATACCTTCTATGACAACTGCTGCCGCCTCCG GTGTTTCCCGCAACCCCCGAAACGGAATTGGTTCTTGGGTCACCTGGGCCTGGACCTGAAGGAGAAGAAGGAGAAGGTGGAGGAGAAGGC AAGCCGGAAAGAGCGAAAGAAAGAAGTGGTGGAGGAGGAGGAGAACGGGGCTGAGGAGGAAGAAGAAGAAACTGCCGAGGATGGAGAGGA GGAAGATGAAGGGGAAGAAGAAGATGAGGAAGAAGAAGAAGAGGATGATGAAGGGCCCGCGCTGAAGAGAGCTGCCGAAGAGGAGGATGA AGCGGATCCCAAACGGCAGAAGACAGAAAATGGGGCATCGGCGTGAGCCCCTGCCAACAGGCTGGGGTTGGGAGGCCTCTCTGGGCCTGG AGGTGGGGGTGGGGGCAGCCAAGTCCAGCCACTCTTCACCTGGCTCCCTGCTCTGGGCCCTGCACCAGAGCTGCCACCCTCTTCTTTCTC CCCAGCCTTCTCATTTCCGCCTCTCCAGACACTGCGCCCTCCACCCTCACTCTGCCATTGTTCCACCTCCTGACCTGCTCCATCTGAGCT CTCCAGCTGGCCCCCAATTGCTCCTCTCTCTCTTTGCTCTCTTTCTCCCTCCCCTACCAGCCTCATTCTTCCTCCGGTAGCCTCTCCCAC CTAACCTCTGCATCCCCCAGCCTCATGTCCTGCCCCATCCCTATCCTGCCTGATCCCTGGATCTCCCTCAGATCCCCTCTTCTCAGACAG CGCCAGGCCGGGGTGGGGCCGGGGTTGGGGCCGAGCCCCACAGCTGCCCCCCTCCCCTCCCTTTTTGTATAATTTAATAAAGAAATGGTC >21097_21097_5_CYP4F3-PTMS_CYP4F3_chr19_15752423_ENST00000586182_PTMS_chr12_6878769_ENST00000309083_length(amino acids)=189AA_BP=1 MDLAAPTPTSRPREASQPQPVGRGSRRCPIFCLLPFGIRFILLFGSSLQRGPFIILFFFFLIFFFPFIFLLSILGSFFFFLLSPVLLLLH HFFLSLFPACLLLHLLLLLLQVQAQVTQEPIPFRGLRETPEAAAVVIEGIGPGQDAGQEPGGPNQQQEQPRGCCHWPKAQRGQAQLWHPC -------------------------------------------------------------- >21097_21097_6_CYP4F3-PTMS_CYP4F3_chr19_15752423_ENST00000586182_PTMS_chr12_6878769_ENST00000389462_length(transcript)=629nt_BP=232nt AGAAGAAGGGGAGAGGAGGTTGTGTGGGACAAGGATGCCACAGCTGAGCCTGTCCTCGCTGGGCCTTTGGCCAATGGCAGCATCCCCGTG GCTGCTCCTGCTGCTGGTTGGGGCCTCCTGGCTCCTGGCCCGCATCCTGGCCTGGACCTATACCTTCTATGACAACTGCTGCCGCCTCCG GTGTTTCCCGCAACCCCCGAAACGGAATTGGTTCTTGGGTCACCTGGGCCTGGACCTGAAGGAGAAGAAGGAGAAGGTGGAGGAGAAGGC AAGCCGGAAAGAGCGAAAGAAAGAAGTGGTGGAGGCTCCAGTCCCATTGGTGGTGATGGTGGGCAAGGCATGCAGCCAGCCTGAGGCTAC TCCCTCTCCTGGTCCCCACAGGAGGAGGAGAACGGGGCTGAGGAGGAAGAAGAAGAAACTGCCGAGGATGGAGAGGAGGAAGATGAAGGG GAAGAAGAAGATGAGGAAGAAGAAGAAGAGGATGATGAAGGGCCCGCGCTGAAGAGAGCTGCCGAAGAGGAGGATGAAGCGGATCCCAAA >21097_21097_6_CYP4F3-PTMS_CYP4F3_chr19_15752423_ENST00000586182_PTMS_chr12_6878769_ENST00000389462_length(amino acids)=188AA_BP=0 MPHFLSSAVWDPLHPPLRQLSSARALHHPLLLLPHLLLPLHLPPLHPRQFLLLPPQPRSPPPVGTRRGSSLRLAACLAHHHHQWDWSLHH FFLSLFPACLLLHLLLLLLQVQAQVTQEPIPFRGLRETPEAAAVVIEGIGPGQDAGQEPGGPNQQQEQPRGCCHWPKAQRGQAQLWHPCP -------------------------------------------------------------- >21097_21097_7_CYP4F3-PTMS_CYP4F3_chr19_15752423_ENST00000591058_PTMS_chr12_6878769_ENST00000309083_length(transcript)=1010nt_BP=245nt AGAAGGGGAGAGGAGGTTGTGTGGGACAAGGTGCTCCTGACAGAAGGATGCCACAGCTGAGCCTGTCCTCGCTGGGCCTTTGGCCAATGG CAGCATCCCCGTGGCTGCTCCTGCTGCTGGTTGGGGCCTCCTGGCTCCTGGCCCGCATCCTGGCCTGGACCTATACCTTCTATGACAACT GCTGCCGCCTCCGGTGTTTCCCGCAACCCCCGAAACGGAATTGGTTCTTGGGTCACCTGGGCCTGGACCTGAAGGAGAAGAAGGAGAAGG TGGAGGAGAAGGCAAGCCGGAAAGAGCGAAAGAAAGAAGTGGTGGAGGAGGAGGAGAACGGGGCTGAGGAGGAAGAAGAAGAAACTGCCG AGGATGGAGAGGAGGAAGATGAAGGGGAAGAAGAAGATGAGGAAGAAGAAGAAGAGGATGATGAAGGGCCCGCGCTGAAGAGAGCTGCCG AAGAGGAGGATGAAGCGGATCCCAAACGGCAGAAGACAGAAAATGGGGCATCGGCGTGAGCCCCTGCCAACAGGCTGGGGTTGGGAGGCC TCTCTGGGCCTGGAGGTGGGGGTGGGGGCAGCCAAGTCCAGCCACTCTTCACCTGGCTCCCTGCTCTGGGCCCTGCACCAGAGCTGCCAC CCTCTTCTTTCTCCCCAGCCTTCTCATTTCCGCCTCTCCAGACACTGCGCCCTCCACCCTCACTCTGCCATTGTTCCACCTCCTGACCTG CTCCATCTGAGCTCTCCAGCTGGCCCCCAATTGCTCCTCTCTCTCTTTGCTCTCTTTCTCCCTCCCCTACCAGCCTCATTCTTCCTCCGG TAGCCTCTCCCACCTAACCTCTGCATCCCCCAGCCTCATGTCCTGCCCCATCCCTATCCTGCCTGATCCCTGGATCTCCCTCAGATCCCC TCTTCTCAGACAGCGCCAGGCCGGGGTGGGGCCGGGGTTGGGGCCGAGCCCCACAGCTGCCCCCCTCCCCTCCCTTTTTGTATAATTTAA >21097_21097_7_CYP4F3-PTMS_CYP4F3_chr19_15752423_ENST00000591058_PTMS_chr12_6878769_ENST00000309083_length(amino acids)=193AA_BP=1 MDLAAPTPTSRPREASQPQPVGRGSRRCPIFCLLPFGIRFILLFGSSLQRGPFIILFFFFLIFFFPFIFLLSILGSFFFFLLSPVLLLLH HFFLSLFPACLLLHLLLLLLQVQAQVTQEPIPFRGLRETPEAAAVVIEGIGPGQDAGQEPGGPNQQQEQPRGCCHWPKAQRGQAQLWHPS -------------------------------------------------------------- >21097_21097_8_CYP4F3-PTMS_CYP4F3_chr19_15752423_ENST00000591058_PTMS_chr12_6878769_ENST00000389462_length(transcript)=642nt_BP=245nt AGAAGGGGAGAGGAGGTTGTGTGGGACAAGGTGCTCCTGACAGAAGGATGCCACAGCTGAGCCTGTCCTCGCTGGGCCTTTGGCCAATGG CAGCATCCCCGTGGCTGCTCCTGCTGCTGGTTGGGGCCTCCTGGCTCCTGGCCCGCATCCTGGCCTGGACCTATACCTTCTATGACAACT GCTGCCGCCTCCGGTGTTTCCCGCAACCCCCGAAACGGAATTGGTTCTTGGGTCACCTGGGCCTGGACCTGAAGGAGAAGAAGGAGAAGG TGGAGGAGAAGGCAAGCCGGAAAGAGCGAAAGAAAGAAGTGGTGGAGGCTCCAGTCCCATTGGTGGTGATGGTGGGCAAGGCATGCAGCC AGCCTGAGGCTACTCCCTCTCCTGGTCCCCACAGGAGGAGGAGAACGGGGCTGAGGAGGAAGAAGAAGAAACTGCCGAGGATGGAGAGGA GGAAGATGAAGGGGAAGAAGAAGATGAGGAAGAAGAAGAAGAGGATGATGAAGGGCCCGCGCTGAAGAGAGCTGCCGAAGAGGAGGATGA AGCGGATCCCAAACGGCAGAAGACAGAAAATGGGGCATCGGCGTGAGCCCCTGCCAACAGGCTGGGGTTGGGAGGCCTCTCTGGGCCTGG >21097_21097_8_CYP4F3-PTMS_CYP4F3_chr19_15752423_ENST00000591058_PTMS_chr12_6878769_ENST00000389462_length(amino acids)=192AA_BP=0 MPHFLSSAVWDPLHPPLRQLSSARALHHPLLLLPHLLLPLHLPPLHPRQFLLLPPQPRSPPPVGTRRGSSLRLAACLAHHHHQWDWSLHH FFLSLFPACLLLHLLLLLLQVQAQVTQEPIPFRGLRETPEAAAVVIEGIGPGQDAGQEPGGPNQQQEQPRGCCHWPKAQRGQAQLWHPSV -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CYP4F3-PTMS |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CYP4F3-PTMS |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CYP4F3-PTMS |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
