
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:DCLRE1C-NMT2 (FusionGDB2 ID:21629) |
Fusion Gene Summary for DCLRE1C-NMT2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: DCLRE1C-NMT2 | Fusion gene ID: 21629 | Hgene | Tgene | Gene symbol | DCLRE1C | NMT2 | Gene ID | 64421 | 9397 |
| Gene name | DNA cross-link repair 1C | N-myristoyltransferase 2 | |
| Synonyms | A-SCID|DCLREC1C|RS-SCID|SCIDA|SNM1C | - | |
| Cytomap | 10p13 | 10p13 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein artemisDNA cross-link repair 1C (PSO2 homolog, S. cerevisiae)SNM1 homolog CSNM1-like proteinsevere combined immunodeficiency, type a (Athabascan) | glycylpeptide N-tetradecanoyltransferase 2NMT 2glycylpeptide N-tetradecanoyltransferase 2 variant 3glycylpeptide N-tetradecanoyltransferase 2 variant 4myristoyl-CoA:protein N-myristoyltransferase 2peptide N-myristoyltransferase 2type II N-myristoylt | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q96SD1 | O60551 | |
| Ensembl transtripts involved in fusion gene | ENST00000378278, ENST00000378289, ENST00000357717, ENST00000378246, ENST00000378254, ENST00000378255, ENST00000378258, ENST00000396817, ENST00000453695, ENST00000378242, ENST00000378249, ENST00000492201, | ENST00000466201, ENST00000540259, ENST00000378150, ENST00000378165, ENST00000535341, | |
| Fusion gene scores | * DoF score | 3 X 3 X 3=27 | 4 X 3 X 4=48 |
| # samples | 3 | 5 | |
| ** MAII score | log2(3/27*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(5/48*10)=0.0588936890535686 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: DCLRE1C [Title/Abstract] AND NMT2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | DCLRE1C(14991035)-NMT2(15170457), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | NMT2 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation | 25255805 |
 Fusion gene breakpoints across DCLRE1C (5'-gene) Fusion gene breakpoints across DCLRE1C (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
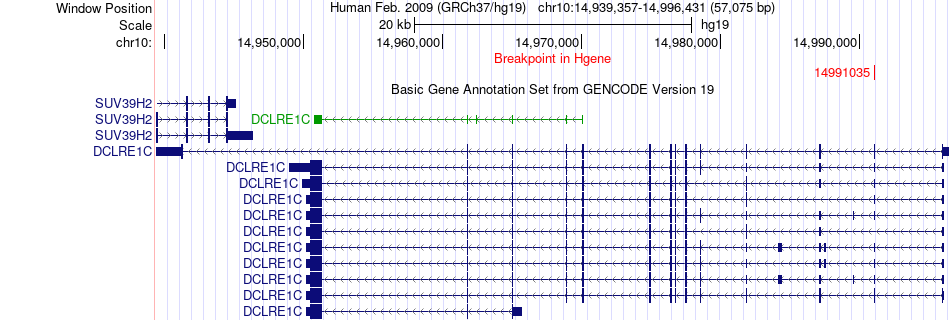 |
 Fusion gene breakpoints across NMT2 (3'-gene) Fusion gene breakpoints across NMT2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
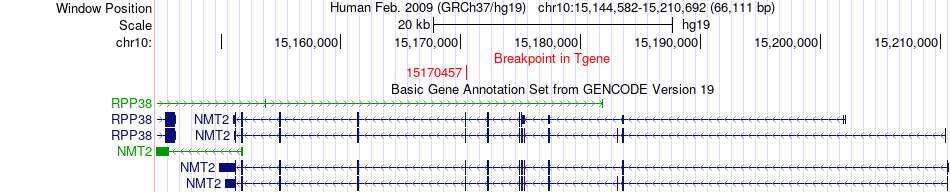 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BLCA | TCGA-GV-A3JW-01A | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
Top |
Fusion Gene ORF analysis for DCLRE1C-NMT2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000378278 | ENST00000466201 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5CDS-intron | ENST00000378278 | ENST00000540259 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5CDS-intron | ENST00000378289 | ENST00000466201 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5CDS-intron | ENST00000378289 | ENST00000540259 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000357717 | ENST00000378150 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000357717 | ENST00000378165 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000357717 | ENST00000535341 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000378246 | ENST00000378150 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000378246 | ENST00000378165 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000378246 | ENST00000535341 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000378254 | ENST00000378150 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000378254 | ENST00000378165 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000378254 | ENST00000535341 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000378255 | ENST00000378150 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000378255 | ENST00000378165 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000378255 | ENST00000535341 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000378258 | ENST00000378150 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000378258 | ENST00000378165 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000378258 | ENST00000535341 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000396817 | ENST00000378150 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000396817 | ENST00000378165 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000396817 | ENST00000535341 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000453695 | ENST00000378150 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000453695 | ENST00000378165 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-3CDS | ENST00000453695 | ENST00000535341 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-intron | ENST00000357717 | ENST00000466201 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-intron | ENST00000357717 | ENST00000540259 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-intron | ENST00000378246 | ENST00000466201 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-intron | ENST00000378246 | ENST00000540259 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-intron | ENST00000378254 | ENST00000466201 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-intron | ENST00000378254 | ENST00000540259 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-intron | ENST00000378255 | ENST00000466201 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-intron | ENST00000378255 | ENST00000540259 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-intron | ENST00000378258 | ENST00000466201 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-intron | ENST00000378258 | ENST00000540259 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-intron | ENST00000396817 | ENST00000466201 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-intron | ENST00000396817 | ENST00000540259 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-intron | ENST00000453695 | ENST00000466201 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| 5UTR-intron | ENST00000453695 | ENST00000540259 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| In-frame | ENST00000378278 | ENST00000378150 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| In-frame | ENST00000378278 | ENST00000378165 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| In-frame | ENST00000378278 | ENST00000535341 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| In-frame | ENST00000378289 | ENST00000378150 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| In-frame | ENST00000378289 | ENST00000378165 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| In-frame | ENST00000378289 | ENST00000535341 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| intron-3CDS | ENST00000378242 | ENST00000378150 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| intron-3CDS | ENST00000378242 | ENST00000378165 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| intron-3CDS | ENST00000378242 | ENST00000535341 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| intron-3CDS | ENST00000378249 | ENST00000378150 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| intron-3CDS | ENST00000378249 | ENST00000378165 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| intron-3CDS | ENST00000378249 | ENST00000535341 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| intron-3CDS | ENST00000492201 | ENST00000378150 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| intron-3CDS | ENST00000492201 | ENST00000378165 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| intron-3CDS | ENST00000492201 | ENST00000535341 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| intron-intron | ENST00000378242 | ENST00000466201 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| intron-intron | ENST00000378242 | ENST00000540259 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| intron-intron | ENST00000378249 | ENST00000466201 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| intron-intron | ENST00000378249 | ENST00000540259 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| intron-intron | ENST00000492201 | ENST00000466201 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
| intron-intron | ENST00000492201 | ENST00000540259 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000378289 | DCLRE1C | chr10 | 14991035 | - | ENST00000378165 | NMT2 | chr10 | 15170457 | - | 2519 | 583 | 302 | 1189 | 295 |
| ENST00000378289 | DCLRE1C | chr10 | 14991035 | - | ENST00000378150 | NMT2 | chr10 | 15170457 | - | 2091 | 583 | 302 | 1189 | 295 |
| ENST00000378289 | DCLRE1C | chr10 | 14991035 | - | ENST00000535341 | NMT2 | chr10 | 15170457 | - | 1286 | 583 | 302 | 1189 | 295 |
| ENST00000378278 | DCLRE1C | chr10 | 14991035 | - | ENST00000378165 | NMT2 | chr10 | 15170457 | - | 2135 | 199 | 38 | 805 | 255 |
| ENST00000378278 | DCLRE1C | chr10 | 14991035 | - | ENST00000378150 | NMT2 | chr10 | 15170457 | - | 1707 | 199 | 38 | 805 | 255 |
| ENST00000378278 | DCLRE1C | chr10 | 14991035 | - | ENST00000535341 | NMT2 | chr10 | 15170457 | - | 902 | 199 | 38 | 805 | 255 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000378289 | ENST00000378165 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - | 0.002161421 | 0.99783856 |
| ENST00000378289 | ENST00000378150 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - | 0.003159076 | 0.99684095 |
| ENST00000378289 | ENST00000535341 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - | 0.00742845 | 0.99257153 |
| ENST00000378278 | ENST00000378165 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - | 0.001583955 | 0.99841607 |
| ENST00000378278 | ENST00000378150 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - | 0.001867527 | 0.9981325 |
| ENST00000378278 | ENST00000535341 | DCLRE1C | chr10 | 14991035 | - | NMT2 | chr10 | 15170457 | - | 0.005055732 | 0.9949443 |
Top |
Fusion Genomic Features for DCLRE1C-NMT2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
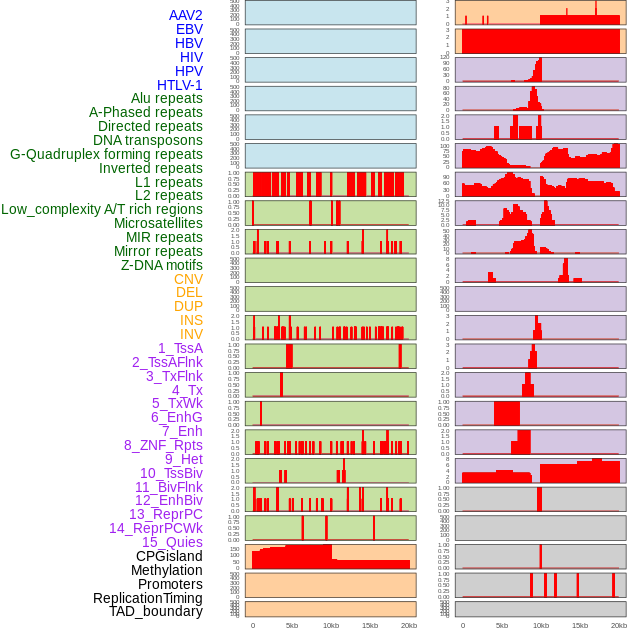 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for DCLRE1C-NMT2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:14991035/chr10:15170457) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| DCLRE1C | NMT2 |
| FUNCTION: Required for V(D)J recombination, the process by which exons encoding the antigen-binding domains of immunoglobulins and T-cell receptor proteins are assembled from individual V, (D), and J gene segments. V(D)J recombination is initiated by the lymphoid specific RAG endonuclease complex, which generates site specific DNA double strand breaks (DSBs). These DSBs present two types of DNA end structures: hairpin sealed coding ends and phosphorylated blunt signal ends. These ends are independently repaired by the non homologous end joining (NHEJ) pathway to form coding and signal joints respectively. This protein exhibits single-strand specific 5'-3' exonuclease activity in isolation and acquires endonucleolytic activity on 5' and 3' hairpins and overhangs when in a complex with PRKDC. The latter activity is required specifically for the resolution of closed hairpins prior to the formation of the coding joint. May also be required for the repair of complex DSBs induced by ionizing radiation, which require substantial end-processing prior to religation by NHEJ. {ECO:0000269|PubMed:11336668, ECO:0000269|PubMed:11955432, ECO:0000269|PubMed:12055248, ECO:0000269|PubMed:14744996, ECO:0000269|PubMed:15071507, ECO:0000269|PubMed:15456891, ECO:0000269|PubMed:15468306, ECO:0000269|PubMed:15574326, ECO:0000269|PubMed:15574327, ECO:0000269|PubMed:15811628, ECO:0000269|PubMed:15936993}. | FUNCTION: Adds a myristoyl group to the N-terminal glycine residue of certain cellular and viral proteins. {ECO:0000269|PubMed:25255805, ECO:0000269|PubMed:9506952}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | NMT2 | chr10:14991035 | chr10:15170457 | ENST00000378165 | 6 | 12 | 46_56 | 296 | 499.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | NMT2 | chr10:14991035 | chr10:15170457 | ENST00000378165 | 6 | 12 | 120_122 | 296 | 499.0 | Region | Myristoyl-CoA binding | |
| Tgene | NMT2 | chr10:14991035 | chr10:15170457 | ENST00000378165 | 6 | 12 | 250_252 | 296 | 499.0 | Region | Myristoyl-CoA binding | |
| Tgene | NMT2 | chr10:14991035 | chr10:15170457 | ENST00000378165 | 6 | 12 | 258_262 | 296 | 499.0 | Region | Myristoyl-CoA binding |
Top |
Fusion Gene Sequence for DCLRE1C-NMT2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >21629_21629_1_DCLRE1C-NMT2_DCLRE1C_chr10_14991035_ENST00000378278_NMT2_chr10_15170457_ENST00000378150_length(transcript)=1707nt_BP=199nt GCGGTTTTGGGGTCCCGGACTCTGGGATCGGCGGCGCTATGAGTTCTTTCGAGGGGCAGATGGCCGAGTATCCAACTATCTCCATAGACC GCTTCGATAGGGAGAACCTGAGGGCCCGCGCCTACTTCCTGTCCCACTGCCACAAAGATCACATGAAAGGATTAAGAGCCCCTACCTTGA AAAGAAGGTTGGAGTGCAGATACTGGCATCGATCACTAAACCCCAGAAAATTGGTAGAAGTGAAATTTTCTCACTTGAGTAGAAATATGA CTTTACAGAGAACAATGAAGCTATACAGACTTCCAGATGTTACAAAGACTTCAGGTTTGAGACCAATGGAACCAAAAGATATCAAATCAG TTCGAGAATTAATCAACACTTACCTGAAGCAGTTTCATCTGGCTCCAGTGATGGATGAAGAGGAAGTAGCCCACTGGTTCCTCCCCCGGG AGCACATTATTGACACGTTTGTAGTGGAGAGCCCCAACGGTAAACTGACTGATTTCCTGAGCTTCTATACGCTCCCCTCCACGGTGATGC ACCACCCTGCTCACAAGAGCCTCAAAGCCGCCTACTCATTCTACAACATCCACACAGAGACGCCCCTGCTGGACCTCATGAGCGACGCGC TCATCCTGGCTAAATCGAAAGGATTTGATGTATTCAATGCACTGGATTTGATGGAAAATAAGACATTCTTGGAAAAACTCAAGTTTGGTA TAGGAGATGGCAATTTGCAGTATTACCTGTACAATTGGAGGTGTCCAGGTACAGATTCTGAAAAGGTTGGACTAGTACTACAATAGATGG ATATTTTTATTTCTAGAACTCTGACATCATCATTTGTTAATATTTAATGATTTCTGGAACTGCCATTCCAAAGAAGAATAAAAGCACAAC TCAAGTGAAATTGAAGTAGTCGATAATCAGAAAAGATGACAAAAGTCCACATGTGACATTTGTACGTTTCTAGCTAGAATGTTAAACTTC ATCCTTTTTTTACTGTTGACCTATTTGTGGGAGGGATGAAAGGCTACAAAGAGCACATTCTTGTAATTTTCAAATTTTTGCCTGTCTCTT GAAGAGGTATTTTTTCACCCTTAGAAAAGGGACTGTTTTTCTATGGACTAAGTAGAAGTTACAGAGTCATGTAAGAAAATCTTTGCTATT GTGTGGAGAGAAATTGCTGCATTTCTGTTTCTTAAGTGATGGTACATTTGTCCATGTAACAGAACAAAAGATCCATTTGGAAATTTTTTT TACTGGTATCACCTTACATGGTATCACTGGGAACATTTATGGAATCAACACTTCTGTAGCTCAGTGACAGGTGGTGGTTGTTTAAGTGCA GAGCAATATTAAAATGTACTGGTGACTAGTACATGTATTTCTCCATAAGCAGAGTTGCCGAATCAATAAACATGCAAGTTTGTACTTTGA TCTTTACCAGGTCAGTGGATTGCTTCAAAAGCAGAGTTTTGTGCAAGCACTCACTGTGCTCTCTGAAAGAGCCTGAGGCCGGGCGTGGTG GCTCACGCCTGTAATCCCAGCACTTTGGGAGGCCAAGGTGGGCAGATCATGAGGTCAGGAGATCGAGACCATTCTGGCCAACATAGTGAA >21629_21629_1_DCLRE1C-NMT2_DCLRE1C_chr10_14991035_ENST00000378278_NMT2_chr10_15170457_ENST00000378150_length(amino acids)=255AA_BP=46 MSSFEGQMAEYPTISIDRFDRENLRARAYFLSHCHKDHMKGLRAPTLKRRLECRYWHRSLNPRKLVEVKFSHLSRNMTLQRTMKLYRLPD VTKTSGLRPMEPKDIKSVRELINTYLKQFHLAPVMDEEEVAHWFLPREHIIDTFVVESPNGKLTDFLSFYTLPSTVMHHPAHKSLKAAYS -------------------------------------------------------------- >21629_21629_2_DCLRE1C-NMT2_DCLRE1C_chr10_14991035_ENST00000378278_NMT2_chr10_15170457_ENST00000378165_length(transcript)=2135nt_BP=199nt GCGGTTTTGGGGTCCCGGACTCTGGGATCGGCGGCGCTATGAGTTCTTTCGAGGGGCAGATGGCCGAGTATCCAACTATCTCCATAGACC GCTTCGATAGGGAGAACCTGAGGGCCCGCGCCTACTTCCTGTCCCACTGCCACAAAGATCACATGAAAGGATTAAGAGCCCCTACCTTGA AAAGAAGGTTGGAGTGCAGATACTGGCATCGATCACTAAACCCCAGAAAATTGGTAGAAGTGAAATTTTCTCACTTGAGTAGAAATATGA CTTTACAGAGAACAATGAAGCTATACAGACTTCCAGATGTTACAAAGACTTCAGGTTTGAGACCAATGGAACCAAAAGATATCAAATCAG TTCGAGAATTAATCAACACTTACCTGAAGCAGTTTCATCTGGCTCCAGTGATGGATGAAGAGGAAGTAGCCCACTGGTTCCTCCCCCGGG AGCACATTATTGACACGTTTGTAGTGGAGAGCCCCAACGGTAAACTGACTGATTTCCTGAGCTTCTATACGCTCCCCTCCACGGTGATGC ACCACCCTGCTCACAAGAGCCTCAAAGCCGCCTACTCATTCTACAACATCCACACAGAGACGCCCCTGCTGGACCTCATGAGCGACGCGC TCATCCTGGCTAAATCGAAAGGATTTGATGTATTCAATGCACTGGATTTGATGGAAAATAAGACATTCTTGGAAAAACTCAAGTTTGGTA TAGGAGATGGCAATTTGCAGTATTACCTGTACAATTGGAGGTGTCCAGGTACAGATTCTGAAAAGGTTGGACTAGTACTACAATAGATGG ATATTTTTATTTCTAGAACTCTGACATCATCATTTGTTAATATTTAATGATTTCTGGAACTGCCATTCCAAAGAAGAATAAAAGCACAAC TCAAGTGAAATTGAAGTAGTCGATAATCAGAAAAGATGACAAAAGTCCACATGTGACATTTGTACGTTTCTAGCTAGAATGTTAAACTTC ATCCTTTTTTTACTGTTGACCTATTTGTGGGAGGGATGAAAGGCTACAAAGAGCACATTCTTGTAATTTTCAAATTTTTGCCTGTCTCTT GAAGAGGTATTTTTTCACCCTTAGAAAAGGGACTGTTTTTCTATGGACTAAGTAGAAGTTACAGAGTCATGTAAGAAAATCTTTGCTATT GTGTGGAGAGAAATTGCTGCATTTCTGTTTCTTAAGTGATGGTACATTTGTCCATGTAACAGAACAAAAGATCCATTTGGAAATTTTTTT TACTGGTATCACCTTACATGGTATCACTGGGAACATTTATGGAATCAACACTTCTGTAGCTCAGTGACAGGTGGTGGTTGTTTAAGTGCA GAGCAATATTAAAATGTACTGGTGACTAGTACATGTATTTCTCCATAAGCAGAGTTGCCGAATCAATAAACATGCAAGTTTGTACTTTGA TCTTTACCAGGTCAGTGGATTGCTTCAAAAGCAGAGTTTTGTGCAAGCACTCACTGTGCTCTCTGAAAGAGCCTGAGGCCGGGCGTGGTG GCTCACGCCTGTAATCCCAGCACTTTGGGAGGCCAAGGTGGGCAGATCATGAGGTCAGGAGATCGAGACCATTCTGGCCAACATAGTGAA ACCCCATCTCTACTAAAAATACAAAAATTAGCCGGGCGTGGTGGCGTGTGCCTGTAGTCCCAGCTGCTTGGGAGGCTGAGGCAGGAGAAT CGCTTGAACCTGAGAGGTGAGGTTGCAGTGAGCCGAGATCGAGCCACTGCACTCCAGCCTGGGTGACAGAGCGTGAGACTCCGTCTCAAA AAAAAAAAAAAAGAAAGAGCCTGTACCCGCGACTAACTAGACTTTTCTGCTGTTGTGCAAAGTTCAGTTTCTTCTCAGCAACATCAAGGA TATATGAAGAAAAGGAAATAAAAAACTGTAAAGTCAAAAGAGATGCCTGGTGGAACTTATTCGTGCTATCAGGGCATTGATAGTTTGAAT GTTTTTATTACTTATGCAAAATTGCACATACTCCTCTATGCTGATTTTATCATGAAAACTGTCATGTGTCATGCTAATGGAAAATGTCTT >21629_21629_2_DCLRE1C-NMT2_DCLRE1C_chr10_14991035_ENST00000378278_NMT2_chr10_15170457_ENST00000378165_length(amino acids)=255AA_BP=46 MSSFEGQMAEYPTISIDRFDRENLRARAYFLSHCHKDHMKGLRAPTLKRRLECRYWHRSLNPRKLVEVKFSHLSRNMTLQRTMKLYRLPD VTKTSGLRPMEPKDIKSVRELINTYLKQFHLAPVMDEEEVAHWFLPREHIIDTFVVESPNGKLTDFLSFYTLPSTVMHHPAHKSLKAAYS -------------------------------------------------------------- >21629_21629_3_DCLRE1C-NMT2_DCLRE1C_chr10_14991035_ENST00000378278_NMT2_chr10_15170457_ENST00000535341_length(transcript)=902nt_BP=199nt GCGGTTTTGGGGTCCCGGACTCTGGGATCGGCGGCGCTATGAGTTCTTTCGAGGGGCAGATGGCCGAGTATCCAACTATCTCCATAGACC GCTTCGATAGGGAGAACCTGAGGGCCCGCGCCTACTTCCTGTCCCACTGCCACAAAGATCACATGAAAGGATTAAGAGCCCCTACCTTGA AAAGAAGGTTGGAGTGCAGATACTGGCATCGATCACTAAACCCCAGAAAATTGGTAGAAGTGAAATTTTCTCACTTGAGTAGAAATATGA CTTTACAGAGAACAATGAAGCTATACAGACTTCCAGATGTTACAAAGACTTCAGGTTTGAGACCAATGGAACCAAAAGATATCAAATCAG TTCGAGAATTAATCAACACTTACCTGAAGCAGTTTCATCTGGCTCCAGTGATGGATGAAGAGGAAGTAGCCCACTGGTTCCTCCCCCGGG AGCACATTATTGACACGTTTGTAGTGGAGAGCCCCAACGGTAAACTGACTGATTTCCTGAGCTTCTATACGCTCCCCTCCACGGTGATGC ACCACCCTGCTCACAAGAGCCTCAAAGCCGCCTACTCATTCTACAACATCCACACAGAGACGCCCCTGCTGGACCTCATGAGCGACGCGC TCATCCTGGCTAAATCGAAAGGATTTGATGTATTCAATGCACTGGATTTGATGGAAAATAAGACATTCTTGGAAAAACTCAAGTTTGGTA TAGGAGATGGCAATTTGCAGTATTACCTGTACAATTGGAGGTGTCCAGGTACAGATTCTGAAAAGGTTGGACTAGTACTACAATAGATGG ATATTTTTATTTCTAGAACTCTGACATCATCATTTGTTAATATTTAATGATTTCTGGAACTGCCATTCCAAAGAAGAATAAAAGCACAAC >21629_21629_3_DCLRE1C-NMT2_DCLRE1C_chr10_14991035_ENST00000378278_NMT2_chr10_15170457_ENST00000535341_length(amino acids)=255AA_BP=46 MSSFEGQMAEYPTISIDRFDRENLRARAYFLSHCHKDHMKGLRAPTLKRRLECRYWHRSLNPRKLVEVKFSHLSRNMTLQRTMKLYRLPD VTKTSGLRPMEPKDIKSVRELINTYLKQFHLAPVMDEEEVAHWFLPREHIIDTFVVESPNGKLTDFLSFYTLPSTVMHHPAHKSLKAAYS -------------------------------------------------------------- >21629_21629_4_DCLRE1C-NMT2_DCLRE1C_chr10_14991035_ENST00000378289_NMT2_chr10_15170457_ENST00000378150_length(transcript)=2091nt_BP=583nt CGGAACGAAGAATGATTTCTAAGCGCAGTTCCGCAGCCCACTCACCTCGTCGGCTGGGGCCACCTGCTCTGGGAGTTTCGATTTCCCTTC CCGCGACTGCACCTCCACAGACATGGGCAACGCCTTACCAGAGCAACACCTGTGTTTGTTGGGCGGGAATGAGCCTTGCACTGGGCAGGG CTCAGGGCCCATCGCGTGCAGCGAAGCGCGGGTGATTTAAACCCAAGCAGCGGGCGCCTAGAACCCGACCGGATGCTCCTTGACTTTGCC CCCGGTCTCCGGACTCCTCTGATTGGACGTGGCTGCGTTCGGCCGCCCAATGGCGAGGCAGCGCGCGGCTTCCCGGAAGTGGCGGCGCGG TCAGGGCTGGCCTTGGCTTCAGCTGCGGTTTTGGGGTCCCGGACTCTGGGATCGGCGGCGCTATGAGTTCTTTCGAGGGGCAGATGGCCG AGTATCCAACTATCTCCATAGACCGCTTCGATAGGGAGAACCTGAGGGCCCGCGCCTACTTCCTGTCCCACTGCCACAAAGATCACATGA AAGGATTAAGAGCCCCTACCTTGAAAAGAAGGTTGGAGTGCAGATACTGGCATCGATCACTAAACCCCAGAAAATTGGTAGAAGTGAAAT TTTCTCACTTGAGTAGAAATATGACTTTACAGAGAACAATGAAGCTATACAGACTTCCAGATGTTACAAAGACTTCAGGTTTGAGACCAA TGGAACCAAAAGATATCAAATCAGTTCGAGAATTAATCAACACTTACCTGAAGCAGTTTCATCTGGCTCCAGTGATGGATGAAGAGGAAG TAGCCCACTGGTTCCTCCCCCGGGAGCACATTATTGACACGTTTGTAGTGGAGAGCCCCAACGGTAAACTGACTGATTTCCTGAGCTTCT ATACGCTCCCCTCCACGGTGATGCACCACCCTGCTCACAAGAGCCTCAAAGCCGCCTACTCATTCTACAACATCCACACAGAGACGCCCC TGCTGGACCTCATGAGCGACGCGCTCATCCTGGCTAAATCGAAAGGATTTGATGTATTCAATGCACTGGATTTGATGGAAAATAAGACAT TCTTGGAAAAACTCAAGTTTGGTATAGGAGATGGCAATTTGCAGTATTACCTGTACAATTGGAGGTGTCCAGGTACAGATTCTGAAAAGG TTGGACTAGTACTACAATAGATGGATATTTTTATTTCTAGAACTCTGACATCATCATTTGTTAATATTTAATGATTTCTGGAACTGCCAT TCCAAAGAAGAATAAAAGCACAACTCAAGTGAAATTGAAGTAGTCGATAATCAGAAAAGATGACAAAAGTCCACATGTGACATTTGTACG TTTCTAGCTAGAATGTTAAACTTCATCCTTTTTTTACTGTTGACCTATTTGTGGGAGGGATGAAAGGCTACAAAGAGCACATTCTTGTAA TTTTCAAATTTTTGCCTGTCTCTTGAAGAGGTATTTTTTCACCCTTAGAAAAGGGACTGTTTTTCTATGGACTAAGTAGAAGTTACAGAG TCATGTAAGAAAATCTTTGCTATTGTGTGGAGAGAAATTGCTGCATTTCTGTTTCTTAAGTGATGGTACATTTGTCCATGTAACAGAACA AAAGATCCATTTGGAAATTTTTTTTACTGGTATCACCTTACATGGTATCACTGGGAACATTTATGGAATCAACACTTCTGTAGCTCAGTG ACAGGTGGTGGTTGTTTAAGTGCAGAGCAATATTAAAATGTACTGGTGACTAGTACATGTATTTCTCCATAAGCAGAGTTGCCGAATCAA TAAACATGCAAGTTTGTACTTTGATCTTTACCAGGTCAGTGGATTGCTTCAAAAGCAGAGTTTTGTGCAAGCACTCACTGTGCTCTCTGA AAGAGCCTGAGGCCGGGCGTGGTGGCTCACGCCTGTAATCCCAGCACTTTGGGAGGCCAAGGTGGGCAGATCATGAGGTCAGGAGATCGA GACCATTCTGGCCAACATAGTGAAACCCCATCTCTACTAAAAATACAAAAATTAGCCGGGCGTGGTGGCGTGTGCCTGTAGTCCCAGCTG >21629_21629_4_DCLRE1C-NMT2_DCLRE1C_chr10_14991035_ENST00000378289_NMT2_chr10_15170457_ENST00000378150_length(amino acids)=295AA_BP=86 MRSAAQWRGSARLPGSGGAVRAGLGFSCGFGVPDSGIGGAMSSFEGQMAEYPTISIDRFDRENLRARAYFLSHCHKDHMKGLRAPTLKRR LECRYWHRSLNPRKLVEVKFSHLSRNMTLQRTMKLYRLPDVTKTSGLRPMEPKDIKSVRELINTYLKQFHLAPVMDEEEVAHWFLPREHI IDTFVVESPNGKLTDFLSFYTLPSTVMHHPAHKSLKAAYSFYNIHTETPLLDLMSDALILAKSKGFDVFNALDLMENKTFLEKLKFGIGD -------------------------------------------------------------- >21629_21629_5_DCLRE1C-NMT2_DCLRE1C_chr10_14991035_ENST00000378289_NMT2_chr10_15170457_ENST00000378165_length(transcript)=2519nt_BP=583nt CGGAACGAAGAATGATTTCTAAGCGCAGTTCCGCAGCCCACTCACCTCGTCGGCTGGGGCCACCTGCTCTGGGAGTTTCGATTTCCCTTC CCGCGACTGCACCTCCACAGACATGGGCAACGCCTTACCAGAGCAACACCTGTGTTTGTTGGGCGGGAATGAGCCTTGCACTGGGCAGGG CTCAGGGCCCATCGCGTGCAGCGAAGCGCGGGTGATTTAAACCCAAGCAGCGGGCGCCTAGAACCCGACCGGATGCTCCTTGACTTTGCC CCCGGTCTCCGGACTCCTCTGATTGGACGTGGCTGCGTTCGGCCGCCCAATGGCGAGGCAGCGCGCGGCTTCCCGGAAGTGGCGGCGCGG TCAGGGCTGGCCTTGGCTTCAGCTGCGGTTTTGGGGTCCCGGACTCTGGGATCGGCGGCGCTATGAGTTCTTTCGAGGGGCAGATGGCCG AGTATCCAACTATCTCCATAGACCGCTTCGATAGGGAGAACCTGAGGGCCCGCGCCTACTTCCTGTCCCACTGCCACAAAGATCACATGA AAGGATTAAGAGCCCCTACCTTGAAAAGAAGGTTGGAGTGCAGATACTGGCATCGATCACTAAACCCCAGAAAATTGGTAGAAGTGAAAT TTTCTCACTTGAGTAGAAATATGACTTTACAGAGAACAATGAAGCTATACAGACTTCCAGATGTTACAAAGACTTCAGGTTTGAGACCAA TGGAACCAAAAGATATCAAATCAGTTCGAGAATTAATCAACACTTACCTGAAGCAGTTTCATCTGGCTCCAGTGATGGATGAAGAGGAAG TAGCCCACTGGTTCCTCCCCCGGGAGCACATTATTGACACGTTTGTAGTGGAGAGCCCCAACGGTAAACTGACTGATTTCCTGAGCTTCT ATACGCTCCCCTCCACGGTGATGCACCACCCTGCTCACAAGAGCCTCAAAGCCGCCTACTCATTCTACAACATCCACACAGAGACGCCCC TGCTGGACCTCATGAGCGACGCGCTCATCCTGGCTAAATCGAAAGGATTTGATGTATTCAATGCACTGGATTTGATGGAAAATAAGACAT TCTTGGAAAAACTCAAGTTTGGTATAGGAGATGGCAATTTGCAGTATTACCTGTACAATTGGAGGTGTCCAGGTACAGATTCTGAAAAGG TTGGACTAGTACTACAATAGATGGATATTTTTATTTCTAGAACTCTGACATCATCATTTGTTAATATTTAATGATTTCTGGAACTGCCAT TCCAAAGAAGAATAAAAGCACAACTCAAGTGAAATTGAAGTAGTCGATAATCAGAAAAGATGACAAAAGTCCACATGTGACATTTGTACG TTTCTAGCTAGAATGTTAAACTTCATCCTTTTTTTACTGTTGACCTATTTGTGGGAGGGATGAAAGGCTACAAAGAGCACATTCTTGTAA TTTTCAAATTTTTGCCTGTCTCTTGAAGAGGTATTTTTTCACCCTTAGAAAAGGGACTGTTTTTCTATGGACTAAGTAGAAGTTACAGAG TCATGTAAGAAAATCTTTGCTATTGTGTGGAGAGAAATTGCTGCATTTCTGTTTCTTAAGTGATGGTACATTTGTCCATGTAACAGAACA AAAGATCCATTTGGAAATTTTTTTTACTGGTATCACCTTACATGGTATCACTGGGAACATTTATGGAATCAACACTTCTGTAGCTCAGTG ACAGGTGGTGGTTGTTTAAGTGCAGAGCAATATTAAAATGTACTGGTGACTAGTACATGTATTTCTCCATAAGCAGAGTTGCCGAATCAA TAAACATGCAAGTTTGTACTTTGATCTTTACCAGGTCAGTGGATTGCTTCAAAAGCAGAGTTTTGTGCAAGCACTCACTGTGCTCTCTGA AAGAGCCTGAGGCCGGGCGTGGTGGCTCACGCCTGTAATCCCAGCACTTTGGGAGGCCAAGGTGGGCAGATCATGAGGTCAGGAGATCGA GACCATTCTGGCCAACATAGTGAAACCCCATCTCTACTAAAAATACAAAAATTAGCCGGGCGTGGTGGCGTGTGCCTGTAGTCCCAGCTG CTTGGGAGGCTGAGGCAGGAGAATCGCTTGAACCTGAGAGGTGAGGTTGCAGTGAGCCGAGATCGAGCCACTGCACTCCAGCCTGGGTGA CAGAGCGTGAGACTCCGTCTCAAAAAAAAAAAAAAAGAAAGAGCCTGTACCCGCGACTAACTAGACTTTTCTGCTGTTGTGCAAAGTTCA GTTTCTTCTCAGCAACATCAAGGATATATGAAGAAAAGGAAATAAAAAACTGTAAAGTCAAAAGAGATGCCTGGTGGAACTTATTCGTGC TATCAGGGCATTGATAGTTTGAATGTTTTTATTACTTATGCAAAATTGCACATACTCCTCTATGCTGATTTTATCATGAAAACTGTCATG >21629_21629_5_DCLRE1C-NMT2_DCLRE1C_chr10_14991035_ENST00000378289_NMT2_chr10_15170457_ENST00000378165_length(amino acids)=295AA_BP=86 MRSAAQWRGSARLPGSGGAVRAGLGFSCGFGVPDSGIGGAMSSFEGQMAEYPTISIDRFDRENLRARAYFLSHCHKDHMKGLRAPTLKRR LECRYWHRSLNPRKLVEVKFSHLSRNMTLQRTMKLYRLPDVTKTSGLRPMEPKDIKSVRELINTYLKQFHLAPVMDEEEVAHWFLPREHI IDTFVVESPNGKLTDFLSFYTLPSTVMHHPAHKSLKAAYSFYNIHTETPLLDLMSDALILAKSKGFDVFNALDLMENKTFLEKLKFGIGD -------------------------------------------------------------- >21629_21629_6_DCLRE1C-NMT2_DCLRE1C_chr10_14991035_ENST00000378289_NMT2_chr10_15170457_ENST00000535341_length(transcript)=1286nt_BP=583nt CGGAACGAAGAATGATTTCTAAGCGCAGTTCCGCAGCCCACTCACCTCGTCGGCTGGGGCCACCTGCTCTGGGAGTTTCGATTTCCCTTC CCGCGACTGCACCTCCACAGACATGGGCAACGCCTTACCAGAGCAACACCTGTGTTTGTTGGGCGGGAATGAGCCTTGCACTGGGCAGGG CTCAGGGCCCATCGCGTGCAGCGAAGCGCGGGTGATTTAAACCCAAGCAGCGGGCGCCTAGAACCCGACCGGATGCTCCTTGACTTTGCC CCCGGTCTCCGGACTCCTCTGATTGGACGTGGCTGCGTTCGGCCGCCCAATGGCGAGGCAGCGCGCGGCTTCCCGGAAGTGGCGGCGCGG TCAGGGCTGGCCTTGGCTTCAGCTGCGGTTTTGGGGTCCCGGACTCTGGGATCGGCGGCGCTATGAGTTCTTTCGAGGGGCAGATGGCCG AGTATCCAACTATCTCCATAGACCGCTTCGATAGGGAGAACCTGAGGGCCCGCGCCTACTTCCTGTCCCACTGCCACAAAGATCACATGA AAGGATTAAGAGCCCCTACCTTGAAAAGAAGGTTGGAGTGCAGATACTGGCATCGATCACTAAACCCCAGAAAATTGGTAGAAGTGAAAT TTTCTCACTTGAGTAGAAATATGACTTTACAGAGAACAATGAAGCTATACAGACTTCCAGATGTTACAAAGACTTCAGGTTTGAGACCAA TGGAACCAAAAGATATCAAATCAGTTCGAGAATTAATCAACACTTACCTGAAGCAGTTTCATCTGGCTCCAGTGATGGATGAAGAGGAAG TAGCCCACTGGTTCCTCCCCCGGGAGCACATTATTGACACGTTTGTAGTGGAGAGCCCCAACGGTAAACTGACTGATTTCCTGAGCTTCT ATACGCTCCCCTCCACGGTGATGCACCACCCTGCTCACAAGAGCCTCAAAGCCGCCTACTCATTCTACAACATCCACACAGAGACGCCCC TGCTGGACCTCATGAGCGACGCGCTCATCCTGGCTAAATCGAAAGGATTTGATGTATTCAATGCACTGGATTTGATGGAAAATAAGACAT TCTTGGAAAAACTCAAGTTTGGTATAGGAGATGGCAATTTGCAGTATTACCTGTACAATTGGAGGTGTCCAGGTACAGATTCTGAAAAGG TTGGACTAGTACTACAATAGATGGATATTTTTATTTCTAGAACTCTGACATCATCATTTGTTAATATTTAATGATTTCTGGAACTGCCAT >21629_21629_6_DCLRE1C-NMT2_DCLRE1C_chr10_14991035_ENST00000378289_NMT2_chr10_15170457_ENST00000535341_length(amino acids)=295AA_BP=86 MRSAAQWRGSARLPGSGGAVRAGLGFSCGFGVPDSGIGGAMSSFEGQMAEYPTISIDRFDRENLRARAYFLSHCHKDHMKGLRAPTLKRR LECRYWHRSLNPRKLVEVKFSHLSRNMTLQRTMKLYRLPDVTKTSGLRPMEPKDIKSVRELINTYLKQFHLAPVMDEEEVAHWFLPREHI IDTFVVESPNGKLTDFLSFYTLPSTVMHHPAHKSLKAAYSFYNIHTETPLLDLMSDALILAKSKGFDVFNALDLMENKTFLEKLKFGIGD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for DCLRE1C-NMT2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for DCLRE1C-NMT2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for DCLRE1C-NMT2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
