
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:DCUN1D2-PTPN1 (FusionGDB2 ID:21716) |
Fusion Gene Summary for DCUN1D2-PTPN1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: DCUN1D2-PTPN1 | Fusion gene ID: 21716 | Hgene | Tgene | Gene symbol | DCUN1D2 | PTPN1 | Gene ID | 55208 | 5770 |
| Gene name | defective in cullin neddylation 1 domain containing 2 | protein tyrosine phosphatase non-receptor type 1 | |
| Synonyms | C13orf17 | PTP1B | |
| Cytomap | 13q34 | 20q13.13 | |
| Type of gene | protein-coding | protein-coding | |
| Description | DCN1-like protein 2DCN1, defective in cullin neddylation 1, domain containing 2DCUN1 domain-containing protein 2defective in cullin neddylation protein 1-like protein 2 | tyrosine-protein phosphatase non-receptor type 1protein tyrosine phosphatase, placentalprotein-tyrosine phosphatase 1B | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | Q6PH85 | Q99952 | |
| Ensembl transtripts involved in fusion gene | ENST00000332592, ENST00000375399, ENST00000478244, ENST00000460318, | ENST00000371621, ENST00000541713, | |
| Fusion gene scores | * DoF score | 13 X 6 X 9=702 | 14 X 14 X 5=980 |
| # samples | 18 | 19 | |
| ** MAII score | log2(18/702*10)=-1.96347412397489 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(19/980*10)=-2.36678233067162 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: DCUN1D2 [Title/Abstract] AND PTPN1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | DCUN1D2(114144981)-PTPN1(49195704), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | PTPN1 | GO:0030100 | regulation of endocytosis | 21135139 |
| Tgene | PTPN1 | GO:0030968 | endoplasmic reticulum unfolded protein response | 22169477 |
| Tgene | PTPN1 | GO:0035335 | peptidyl-tyrosine dephosphorylation | 21135139 |
| Tgene | PTPN1 | GO:0061098 | positive regulation of protein tyrosine kinase activity | 21216966 |
| Tgene | PTPN1 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response | 21216966 |
 Fusion gene breakpoints across DCUN1D2 (5'-gene) Fusion gene breakpoints across DCUN1D2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
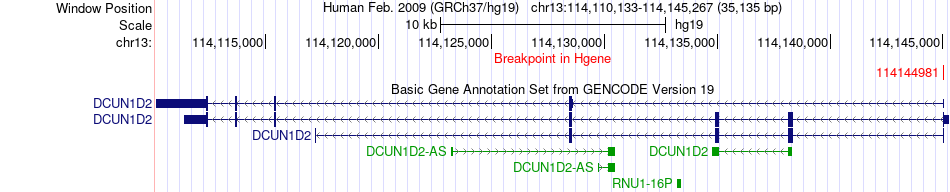 |
 Fusion gene breakpoints across PTPN1 (3'-gene) Fusion gene breakpoints across PTPN1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
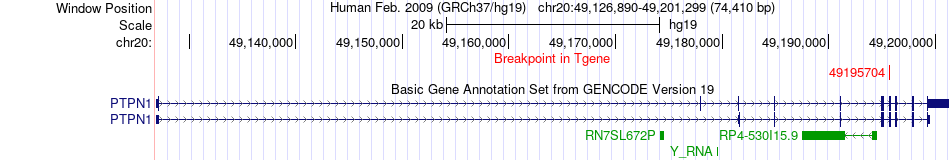 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-29-1688 | DCUN1D2 | chr13 | 114144981 | - | PTPN1 | chr20 | 49195704 | + |
Top |
Fusion Gene ORF analysis for DCUN1D2-PTPN1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5UTR-3CDS | ENST00000332592 | ENST00000371621 | DCUN1D2 | chr13 | 114144981 | - | PTPN1 | chr20 | 49195704 | + |
| 5UTR-3CDS | ENST00000332592 | ENST00000541713 | DCUN1D2 | chr13 | 114144981 | - | PTPN1 | chr20 | 49195704 | + |
| In-frame | ENST00000375399 | ENST00000371621 | DCUN1D2 | chr13 | 114144981 | - | PTPN1 | chr20 | 49195704 | + |
| In-frame | ENST00000375399 | ENST00000541713 | DCUN1D2 | chr13 | 114144981 | - | PTPN1 | chr20 | 49195704 | + |
| In-frame | ENST00000478244 | ENST00000371621 | DCUN1D2 | chr13 | 114144981 | - | PTPN1 | chr20 | 49195704 | + |
| In-frame | ENST00000478244 | ENST00000541713 | DCUN1D2 | chr13 | 114144981 | - | PTPN1 | chr20 | 49195704 | + |
| intron-3CDS | ENST00000460318 | ENST00000371621 | DCUN1D2 | chr13 | 114144981 | - | PTPN1 | chr20 | 49195704 | + |
| intron-3CDS | ENST00000460318 | ENST00000541713 | DCUN1D2 | chr13 | 114144981 | - | PTPN1 | chr20 | 49195704 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000478244 | DCUN1D2 | chr13 | 114144981 | - | ENST00000371621 | PTPN1 | chr20 | 49195704 | + | 2939 | 286 | 283 | 891 | 202 |
| ENST00000478244 | DCUN1D2 | chr13 | 114144981 | - | ENST00000541713 | PTPN1 | chr20 | 49195704 | + | 1162 | 286 | 283 | 891 | 202 |
| ENST00000375399 | DCUN1D2 | chr13 | 114144981 | - | ENST00000371621 | PTPN1 | chr20 | 49195704 | + | 2695 | 42 | 39 | 647 | 202 |
| ENST00000375399 | DCUN1D2 | chr13 | 114144981 | - | ENST00000541713 | PTPN1 | chr20 | 49195704 | + | 918 | 42 | 39 | 647 | 202 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000478244 | ENST00000371621 | DCUN1D2 | chr13 | 114144981 | - | PTPN1 | chr20 | 49195704 | + | 0.002518663 | 0.9974814 |
| ENST00000478244 | ENST00000541713 | DCUN1D2 | chr13 | 114144981 | - | PTPN1 | chr20 | 49195704 | + | 0.007281539 | 0.99271846 |
| ENST00000375399 | ENST00000371621 | DCUN1D2 | chr13 | 114144981 | - | PTPN1 | chr20 | 49195704 | + | 0.002758105 | 0.99724185 |
| ENST00000375399 | ENST00000541713 | DCUN1D2 | chr13 | 114144981 | - | PTPN1 | chr20 | 49195704 | + | 0.004711625 | 0.9952884 |
Top |
Fusion Genomic Features for DCUN1D2-PTPN1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| DCUN1D2 | chr13 | 114144981 | - | PTPN1 | chr20 | 49195704 | + | 5.79E-06 | 0.99999416 |
| DCUN1D2 | chr13 | 114144981 | - | PTPN1 | chr20 | 49195704 | + | 5.79E-06 | 0.99999416 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
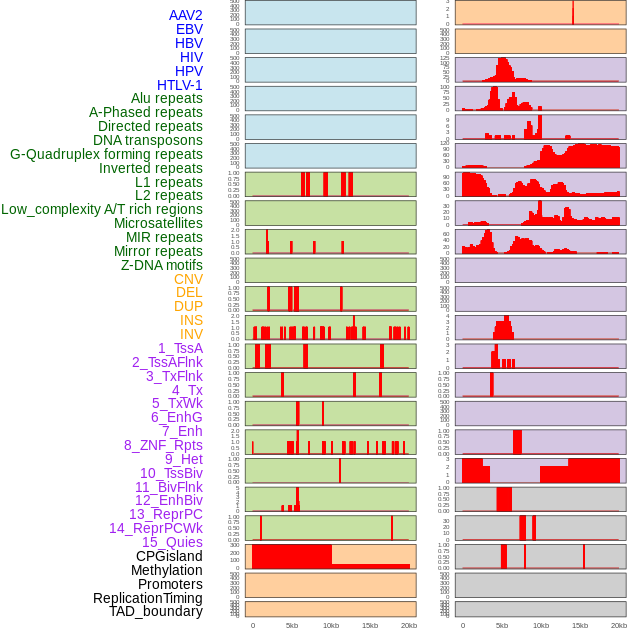 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
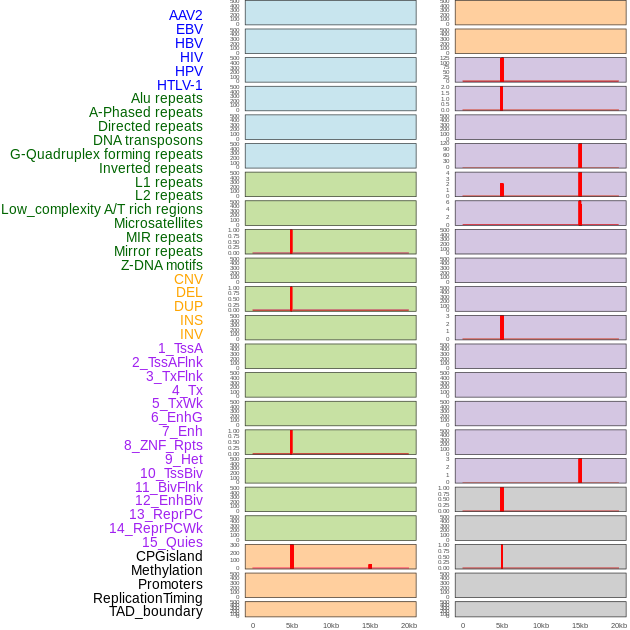 |
Top |
Fusion Protein Features for DCUN1D2-PTPN1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr13:114144981/chr20:49195704) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| DCUN1D2 | PTPN1 |
| FUNCTION: Contributes to the neddylation of all cullins by transfering NEDD8 from N-terminally acetylated NEDD8-conjugating E2s enzyme to different cullin C-terminal domain-RBX complexes and plays an essential role in the regulation of SCF (SKP1-CUL1-F-box protein)-type complexes activity. {ECO:0000269|PubMed:19617556, ECO:0000269|PubMed:23201271, ECO:0000269|PubMed:26906416}. | FUNCTION: Differentially dephosphorylate autophosphorylated tyrosine kinases which are known to be overexpressed in tumor tissues. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DCUN1D2 | chr13:114144981 | chr20:49195704 | ENST00000332592 | - | 1 | 5 | 60_248 | 0 | 127.0 | Domain | DCUN1 |
| Hgene | DCUN1D2 | chr13:114144981 | chr20:49195704 | ENST00000332592 | - | 1 | 5 | 8_45 | 0 | 127.0 | Domain | Note=UBA-like |
| Hgene | DCUN1D2 | chr13:114144981 | chr20:49195704 | ENST00000375399 | - | 1 | 5 | 60_248 | 1 | 187.0 | Domain | DCUN1 |
| Hgene | DCUN1D2 | chr13:114144981 | chr20:49195704 | ENST00000375399 | - | 1 | 5 | 8_45 | 1 | 187.0 | Domain | Note=UBA-like |
| Hgene | DCUN1D2 | chr13:114144981 | chr20:49195704 | ENST00000478244 | - | 1 | 7 | 60_248 | 1 | 260.0 | Domain | DCUN1 |
| Hgene | DCUN1D2 | chr13:114144981 | chr20:49195704 | ENST00000478244 | - | 1 | 7 | 8_45 | 1 | 260.0 | Domain | Note=UBA-like |
| Tgene | PTPN1 | chr13:114144981 | chr20:49195704 | ENST00000371621 | 5 | 10 | 3_277 | 234 | 436.0 | Domain | Tyrosine-protein phosphatase | |
| Tgene | PTPN1 | chr13:114144981 | chr20:49195704 | ENST00000371621 | 5 | 10 | 215_221 | 234 | 436.0 | Region | Substrate binding |
Top |
Fusion Gene Sequence for DCUN1D2-PTPN1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >21716_21716_1_DCUN1D2-PTPN1_DCUN1D2_chr13_114144981_ENST00000375399_PTPN1_chr20_49195704_ENST00000371621_length(transcript)=2695nt_BP=42nt GAAGGCCGGGGCCGGCCAGAAGCGGGCGGCGCGGGGGAGATGATGGACAAGAGGAAAGACCCTTCTTCCGTTGATATCAAGAAAGTGCTG TTAGAAATGAGGAAGTTTCGGATGGGGCTGATCCAGACAGCCGACCAGCTGCGCTTCTCCTACCTGGCTGTGATCGAAGGTGCCAAATTC ATCATGGGGGACTCTTCCGTGCAGGATCAGTGGAAGGAGCTTTCCCACGAGGACCTGGAGCCCCCACCCGAGCATATCCCCCCACCTCCC CGGCCACCCAAACGAATCCTGGAGCCACACAATGGGAAATGCAGGGAGTTCTTCCCAAATCACCAGTGGGTGAAGGAAGAGACCCAGGAG GATAAAGACTGCCCCATCAAGGAAGAAAAAGGAAGCCCCTTAAATGCCGCACCCTACGGCATCGAAAGCATGAGTCAAGACACTGAAGTT AGAAGTCGGGTCGTGGGGGGAAGTCTTCGAGGTGCCCAGGCTGCCTCCCCAGCCAAAGGGGAGCCGTCACTGCCCGAGAAGGACGAGGAC CATGCACTGAGTTACTGGAAGCCCTTCCTGGTCAACATGTGCGTGGCTACGGTCCTCACGGCCGGCGCTTACCTCTGCTACAGGTTCCTG TTCAACAGCAACACATAGCCTGACCCTCCTCCACTCCACCTCCACCCACTGTCCGCCTCTGCCCGCAGAGCCCACGCCCGACTAGCAGGC ATGCCGCGGTAGGTAAGGGCCGCCGGACCGCGTAGAGAGCCGGGCCCCGGACGGACGTTGGTTCTGCACTAAAACCCATCTTCCCCGGAT GTGTGTCTCACCCCTCATCCTTTTACTTTTTGCCCCTTCCACTTTGAGTACCAAATCCACAAGCCATTTTTTGAGGAGAGTGAAAGAGAG TACCATGCTGGCGGCGCAGAGGGAAGGGGCCTACACCCGTCTTGGGGCTCGCCCCACCCAGGGCTCCCTCCTGGAGCATCCCAGGCGGGC GGCACGCCAACAGCCCCCCCCTTGAATCTGCAGGGAGCAACTCTCCACTCCATATTTATTTAAACAATTTTTTCCCCAAAGGCATCCATA GTGCACTAGCATTTTCTTGAACCAATAATGTATTAAAATTTTTTGATGTCAGCCTTGCATCAAGGGCTTTATCAAAAAGTACAATAATAA ATCCTCAGGTAGTACTGGGAATGGAAGGCTTTGCCATGGGCCTGCTGCGTCAGACCAGTACTGGGAAGGAGGACGGTTGTAAGCAGTTGT TATTTAGTGATATTGTGGGTAACGTGAGAAGATAGAACAATGCTATAATATATAATGAACACGTGGGTATTTAATAAGAAACATGATGTG AGATTACTTTGTCCCGCTTATTCTCCTCCCTGTTATCTGCTAGATCTAGTTCTCAATCACTGCTCCCCCGTGTGTATTAGAATGCATGTA AGGTCTTCTTGTGTCCTGATGAAAAATATGTGCTTGAAATGAGAAACTTTGATCTCTGCTTACTAATGTGCCCCATGTCCAAGTCCAACC TGCCTGTGCATGACCTGATCATTACATGGCTGTGGTTCCTAAGCCTGTTGCTGAAGTCATTGTCGCTCAGCAATAGGGTGCAGTTTTCCA GGAATAGGCATTTGCCTAATTCCTGGCATGACACTCTAGTGACTTCCTGGTGAGGCCCAGCCTGTCCTGGTACAGCAGGGTCTTGCTGTA ACTCAGACATTCCAAGGGTATGGGAAGCCATATTCACACCTCACGCTCTGGACATGATTTAGGGAAGCAGGGACACCCCCCGCCCCCCAC CTTTGGGATCAGCCTCCGCCATTCCAAGTCAACACTCTTCTTGAGCAGACCGTGATTTGGAAGAGAGGCACCTGCTGGAAACCACACTTC TTGAAACAGCCTGGGTGACGGTCCTTTAGGCAGCCTGCCGCCGTCTCTGTCCCGGTTCACCTTGCCGAGAGAGGCGCGTCTGCCCCACCC TCAAACCCTGTGGGGCCTGATGGTGCTCACGACTCTTCCTGCAAAGGGAACTGAAGACCTCCACATTAAGTGGCTTTTTAACATGAAAAA CACGGCAGCTGTAGCTCCCGAGCTACTCTCTTGCCAGCATTTTCACATTTTGCCTTTCTCGTGGTAGAAGCCAGTACAGAGAAATTCTGT GGTGGGAACATTCGAGGTGTCACCCTGCAGAGCTATGGTGAGGTGTGGATAAGGCTTAGGTGCCAGGCTGTAAGCATTCTGAGCTGGGCT TGTTGTTTTTAAGTCCTGTATATGTATGTAGTAGTTTGGGTGTGTATATATAGTAGCATTTCAAAATGGACGTACTGGTTTAACCTCCTA TCCTTGGAGAGCAGCTGGCTCTCCACCTTGTTACACATTATGTTAGAGAGGTAGCGAGCTGCTCTGCTATATGCCTTAAGCCAATATTTA CTCATCAGGTCATTATTTTTTACAATGGCCATGGAATAAACCATTTTTACAAAAATAAAAACAAAAAAAGCAAGGTGTTTTGGTATAATA CCTTTTCAGGTGTGTGTGGATACGTGGCTGCATGACCGGGTGGGTGGGGGGGAGTGTCTCAGGGTCTTCTGTGACCTCACAGAACTGTCA >21716_21716_1_DCUN1D2-PTPN1_DCUN1D2_chr13_114144981_ENST00000375399_PTPN1_chr20_49195704_ENST00000371621_length(amino acids)=202AA_BP=1 MMDKRKDPSSVDIKKVLLEMRKFRMGLIQTADQLRFSYLAVIEGAKFIMGDSSVQDQWKELSHEDLEPPPEHIPPPPRPPKRILEPHNGK CREFFPNHQWVKEETQEDKDCPIKEEKGSPLNAAPYGIESMSQDTEVRSRVVGGSLRGAQAASPAKGEPSLPEKDEDHALSYWKPFLVNM -------------------------------------------------------------- >21716_21716_2_DCUN1D2-PTPN1_DCUN1D2_chr13_114144981_ENST00000375399_PTPN1_chr20_49195704_ENST00000541713_length(transcript)=918nt_BP=42nt GAAGGCCGGGGCCGGCCAGAAGCGGGCGGCGCGGGGGAGATGATGGACAAGAGGAAAGACCCTTCTTCCGTTGATATCAAGAAAGTGCTG TTAGAAATGAGGAAGTTTCGGATGGGGCTGATCCAGACAGCCGACCAGCTGCGCTTCTCCTACCTGGCTGTGATCGAAGGTGCCAAATTC ATCATGGGGGACTCTTCCGTGCAGGATCAGTGGAAGGAGCTTTCCCACGAGGACCTGGAGCCCCCACCCGAGCATATCCCCCCACCTCCC CGGCCACCCAAACGAATCCTGGAGCCACACAATGGGAAATGCAGGGAGTTCTTCCCAAATCACCAGTGGGTGAAGGAAGAGACCCAGGAG GATAAAGACTGCCCCATCAAGGAAGAAAAAGGAAGCCCCTTAAATGCCGCACCCTACGGCATCGAAAGCATGAGTCAAGACACTGAAGTT AGAAGTCGGGTCGTGGGGGGAAGTCTTCGAGGTGCCCAGGCTGCCTCCCCAGCCAAAGGGGAGCCGTCACTGCCCGAGAAGGACGAGGAC CATGCACTGAGTTACTGGAAGCCCTTCCTGGTCAACATGTGCGTGGCTACGGTCCTCACGGCCGGCGCTTACCTCTGCTACAGGTTCCTG TTCAACAGCAACACATAGCCTGACCCTCCTCCACTCCACCTCCACCCACTGTCCGCCTCTGCCCGCAGAGCCCACGCCCGACTAGCAGGC ATGCCGCGGTAGGTAAGGGCCGCCGGACCGCGTAGAGAGCCGGGCCCCGGACGGACGTTGGTTCTGCACTAAAACCCATCTTCCCCGGAT GTGTGTCTCACCCCTCATCCTTTTACTTTTTGCCCCTTCCACTTTGAGTACCAAATCCACAAGCCATTTTTTGAGGAGAGTGAAAGAGAG >21716_21716_2_DCUN1D2-PTPN1_DCUN1D2_chr13_114144981_ENST00000375399_PTPN1_chr20_49195704_ENST00000541713_length(amino acids)=202AA_BP=1 MMDKRKDPSSVDIKKVLLEMRKFRMGLIQTADQLRFSYLAVIEGAKFIMGDSSVQDQWKELSHEDLEPPPEHIPPPPRPPKRILEPHNGK CREFFPNHQWVKEETQEDKDCPIKEEKGSPLNAAPYGIESMSQDTEVRSRVVGGSLRGAQAASPAKGEPSLPEKDEDHALSYWKPFLVNM -------------------------------------------------------------- >21716_21716_3_DCUN1D2-PTPN1_DCUN1D2_chr13_114144981_ENST00000478244_PTPN1_chr20_49195704_ENST00000371621_length(transcript)=2939nt_BP=286nt AGCACTTCCTGGCCGGCGTGGGGCCTGGCAGCCGCAGGCTGGCGTTGGACCCCGCAGGGGCGAGCCAAGGCCCTCGCAGAGGGCTGGACG CGCAGGGGCTGTGGGGCGCGAGCTCCCGTGGGCCCCCGGGCGGCGCTGAGCATGCCGGGAGTGGGAGTCCCCGCGGCCGGCTCCCGGCAC TGCCCCGGAGCCGAGGGCCGCGGCCGTCCGAGGACGAGGGCGGAGCCGCCGCCGCGCCTGCGCAGAAGGCCGGGGCCGGCCAGAAGCGGG CGGCGCGGGGGAGATGATGGACAAGAGGAAAGACCCTTCTTCCGTTGATATCAAGAAAGTGCTGTTAGAAATGAGGAAGTTTCGGATGGG GCTGATCCAGACAGCCGACCAGCTGCGCTTCTCCTACCTGGCTGTGATCGAAGGTGCCAAATTCATCATGGGGGACTCTTCCGTGCAGGA TCAGTGGAAGGAGCTTTCCCACGAGGACCTGGAGCCCCCACCCGAGCATATCCCCCCACCTCCCCGGCCACCCAAACGAATCCTGGAGCC ACACAATGGGAAATGCAGGGAGTTCTTCCCAAATCACCAGTGGGTGAAGGAAGAGACCCAGGAGGATAAAGACTGCCCCATCAAGGAAGA AAAAGGAAGCCCCTTAAATGCCGCACCCTACGGCATCGAAAGCATGAGTCAAGACACTGAAGTTAGAAGTCGGGTCGTGGGGGGAAGTCT TCGAGGTGCCCAGGCTGCCTCCCCAGCCAAAGGGGAGCCGTCACTGCCCGAGAAGGACGAGGACCATGCACTGAGTTACTGGAAGCCCTT CCTGGTCAACATGTGCGTGGCTACGGTCCTCACGGCCGGCGCTTACCTCTGCTACAGGTTCCTGTTCAACAGCAACACATAGCCTGACCC TCCTCCACTCCACCTCCACCCACTGTCCGCCTCTGCCCGCAGAGCCCACGCCCGACTAGCAGGCATGCCGCGGTAGGTAAGGGCCGCCGG ACCGCGTAGAGAGCCGGGCCCCGGACGGACGTTGGTTCTGCACTAAAACCCATCTTCCCCGGATGTGTGTCTCACCCCTCATCCTTTTAC TTTTTGCCCCTTCCACTTTGAGTACCAAATCCACAAGCCATTTTTTGAGGAGAGTGAAAGAGAGTACCATGCTGGCGGCGCAGAGGGAAG GGGCCTACACCCGTCTTGGGGCTCGCCCCACCCAGGGCTCCCTCCTGGAGCATCCCAGGCGGGCGGCACGCCAACAGCCCCCCCCTTGAA TCTGCAGGGAGCAACTCTCCACTCCATATTTATTTAAACAATTTTTTCCCCAAAGGCATCCATAGTGCACTAGCATTTTCTTGAACCAAT AATGTATTAAAATTTTTTGATGTCAGCCTTGCATCAAGGGCTTTATCAAAAAGTACAATAATAAATCCTCAGGTAGTACTGGGAATGGAA GGCTTTGCCATGGGCCTGCTGCGTCAGACCAGTACTGGGAAGGAGGACGGTTGTAAGCAGTTGTTATTTAGTGATATTGTGGGTAACGTG AGAAGATAGAACAATGCTATAATATATAATGAACACGTGGGTATTTAATAAGAAACATGATGTGAGATTACTTTGTCCCGCTTATTCTCC TCCCTGTTATCTGCTAGATCTAGTTCTCAATCACTGCTCCCCCGTGTGTATTAGAATGCATGTAAGGTCTTCTTGTGTCCTGATGAAAAA TATGTGCTTGAAATGAGAAACTTTGATCTCTGCTTACTAATGTGCCCCATGTCCAAGTCCAACCTGCCTGTGCATGACCTGATCATTACA TGGCTGTGGTTCCTAAGCCTGTTGCTGAAGTCATTGTCGCTCAGCAATAGGGTGCAGTTTTCCAGGAATAGGCATTTGCCTAATTCCTGG CATGACACTCTAGTGACTTCCTGGTGAGGCCCAGCCTGTCCTGGTACAGCAGGGTCTTGCTGTAACTCAGACATTCCAAGGGTATGGGAA GCCATATTCACACCTCACGCTCTGGACATGATTTAGGGAAGCAGGGACACCCCCCGCCCCCCACCTTTGGGATCAGCCTCCGCCATTCCA AGTCAACACTCTTCTTGAGCAGACCGTGATTTGGAAGAGAGGCACCTGCTGGAAACCACACTTCTTGAAACAGCCTGGGTGACGGTCCTT TAGGCAGCCTGCCGCCGTCTCTGTCCCGGTTCACCTTGCCGAGAGAGGCGCGTCTGCCCCACCCTCAAACCCTGTGGGGCCTGATGGTGC TCACGACTCTTCCTGCAAAGGGAACTGAAGACCTCCACATTAAGTGGCTTTTTAACATGAAAAACACGGCAGCTGTAGCTCCCGAGCTAC TCTCTTGCCAGCATTTTCACATTTTGCCTTTCTCGTGGTAGAAGCCAGTACAGAGAAATTCTGTGGTGGGAACATTCGAGGTGTCACCCT GCAGAGCTATGGTGAGGTGTGGATAAGGCTTAGGTGCCAGGCTGTAAGCATTCTGAGCTGGGCTTGTTGTTTTTAAGTCCTGTATATGTA TGTAGTAGTTTGGGTGTGTATATATAGTAGCATTTCAAAATGGACGTACTGGTTTAACCTCCTATCCTTGGAGAGCAGCTGGCTCTCCAC CTTGTTACACATTATGTTAGAGAGGTAGCGAGCTGCTCTGCTATATGCCTTAAGCCAATATTTACTCATCAGGTCATTATTTTTTACAAT GGCCATGGAATAAACCATTTTTACAAAAATAAAAACAAAAAAAGCAAGGTGTTTTGGTATAATACCTTTTCAGGTGTGTGTGGATACGTG GCTGCATGACCGGGTGGGTGGGGGGGAGTGTCTCAGGGTCTTCTGTGACCTCACAGAACTGTCAGACTGTACAGTTTTCCAACTTGCCAT >21716_21716_3_DCUN1D2-PTPN1_DCUN1D2_chr13_114144981_ENST00000478244_PTPN1_chr20_49195704_ENST00000371621_length(amino acids)=202AA_BP=1 MMDKRKDPSSVDIKKVLLEMRKFRMGLIQTADQLRFSYLAVIEGAKFIMGDSSVQDQWKELSHEDLEPPPEHIPPPPRPPKRILEPHNGK CREFFPNHQWVKEETQEDKDCPIKEEKGSPLNAAPYGIESMSQDTEVRSRVVGGSLRGAQAASPAKGEPSLPEKDEDHALSYWKPFLVNM -------------------------------------------------------------- >21716_21716_4_DCUN1D2-PTPN1_DCUN1D2_chr13_114144981_ENST00000478244_PTPN1_chr20_49195704_ENST00000541713_length(transcript)=1162nt_BP=286nt AGCACTTCCTGGCCGGCGTGGGGCCTGGCAGCCGCAGGCTGGCGTTGGACCCCGCAGGGGCGAGCCAAGGCCCTCGCAGAGGGCTGGACG CGCAGGGGCTGTGGGGCGCGAGCTCCCGTGGGCCCCCGGGCGGCGCTGAGCATGCCGGGAGTGGGAGTCCCCGCGGCCGGCTCCCGGCAC TGCCCCGGAGCCGAGGGCCGCGGCCGTCCGAGGACGAGGGCGGAGCCGCCGCCGCGCCTGCGCAGAAGGCCGGGGCCGGCCAGAAGCGGG CGGCGCGGGGGAGATGATGGACAAGAGGAAAGACCCTTCTTCCGTTGATATCAAGAAAGTGCTGTTAGAAATGAGGAAGTTTCGGATGGG GCTGATCCAGACAGCCGACCAGCTGCGCTTCTCCTACCTGGCTGTGATCGAAGGTGCCAAATTCATCATGGGGGACTCTTCCGTGCAGGA TCAGTGGAAGGAGCTTTCCCACGAGGACCTGGAGCCCCCACCCGAGCATATCCCCCCACCTCCCCGGCCACCCAAACGAATCCTGGAGCC ACACAATGGGAAATGCAGGGAGTTCTTCCCAAATCACCAGTGGGTGAAGGAAGAGACCCAGGAGGATAAAGACTGCCCCATCAAGGAAGA AAAAGGAAGCCCCTTAAATGCCGCACCCTACGGCATCGAAAGCATGAGTCAAGACACTGAAGTTAGAAGTCGGGTCGTGGGGGGAAGTCT TCGAGGTGCCCAGGCTGCCTCCCCAGCCAAAGGGGAGCCGTCACTGCCCGAGAAGGACGAGGACCATGCACTGAGTTACTGGAAGCCCTT CCTGGTCAACATGTGCGTGGCTACGGTCCTCACGGCCGGCGCTTACCTCTGCTACAGGTTCCTGTTCAACAGCAACACATAGCCTGACCC TCCTCCACTCCACCTCCACCCACTGTCCGCCTCTGCCCGCAGAGCCCACGCCCGACTAGCAGGCATGCCGCGGTAGGTAAGGGCCGCCGG ACCGCGTAGAGAGCCGGGCCCCGGACGGACGTTGGTTCTGCACTAAAACCCATCTTCCCCGGATGTGTGTCTCACCCCTCATCCTTTTAC >21716_21716_4_DCUN1D2-PTPN1_DCUN1D2_chr13_114144981_ENST00000478244_PTPN1_chr20_49195704_ENST00000541713_length(amino acids)=202AA_BP=1 MMDKRKDPSSVDIKKVLLEMRKFRMGLIQTADQLRFSYLAVIEGAKFIMGDSSVQDQWKELSHEDLEPPPEHIPPPPRPPKRILEPHNGK CREFFPNHQWVKEETQEDKDCPIKEEKGSPLNAAPYGIESMSQDTEVRSRVVGGSLRGAQAASPAKGEPSLPEKDEDHALSYWKPFLVNM -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for DCUN1D2-PTPN1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for DCUN1D2-PTPN1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for DCUN1D2-PTPN1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
