
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:DDB1-SDHA (FusionGDB2 ID:21777) |
Fusion Gene Summary for DDB1-SDHA |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: DDB1-SDHA | Fusion gene ID: 21777 | Hgene | Tgene | Gene symbol | DDB1 | SDHA | Gene ID | 1642 | 6389 |
| Gene name | damage specific DNA binding protein 1 | succinate dehydrogenase complex flavoprotein subunit A | |
| Synonyms | DDBA|UV-DDB1|XAP1|XPCE|XPE|XPE-BF | CMD1GG|FP|PGL5|SDH1|SDH2|SDHF | |
| Cytomap | 11q12.2 | 5p15.33 | |
| Type of gene | protein-coding | protein-coding | |
| Description | DNA damage-binding protein 1DDB p127 subunitDNA damage-binding protein aHBV X-associated protein 1UV-DDB 1UV-damaged DNA-binding factorUV-damaged DNA-binding protein 1XAP-1XPE-binding factordamage-specific DNA binding protein 1, 127kDaxeroderma | succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrialflavoprotein subunit of complex IIsuccinate dehydrogenase complex, subunit A, flavoprotein (Fp) | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | Q16531 | Q5VUM1 | |
| Ensembl transtripts involved in fusion gene | ENST00000301764, ENST00000450997, ENST00000451943, ENST00000538470, ENST00000545930, | ENST00000507522, ENST00000264932, ENST00000504309, ENST00000510361, | |
| Fusion gene scores | * DoF score | 12 X 17 X 6=1224 | 5 X 7 X 5=175 |
| # samples | 16 | 7 | |
| ** MAII score | log2(16/1224*10)=-2.93545974780529 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/175*10)=-1.32192809488736 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: DDB1 [Title/Abstract] AND SDHA [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | DDB1(61080971)-SDHA(235259), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | DDB1 | GO:0006511 | ubiquitin-dependent protein catabolic process | 11673459 |
| Hgene | DDB1 | GO:0016567 | protein ubiquitination | 26431207|28886238 |
| Hgene | DDB1 | GO:0035518 | histone H2A monoubiquitination | 22334663 |
| Hgene | DDB1 | GO:0051702 | interaction with symbiont | 23137809 |
| Hgene | DDB1 | GO:0070914 | UV-damage excision repair | 22334663 |
| Tgene | SDHA | GO:0006105 | succinate metabolic process | 7550341 |
| Tgene | SDHA | GO:0022904 | respiratory electron transport chain | 7550341 |
| Tgene | SDHA | GO:0055114 | oxidation-reduction process | 7550341 |
 Fusion gene breakpoints across DDB1 (5'-gene) Fusion gene breakpoints across DDB1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
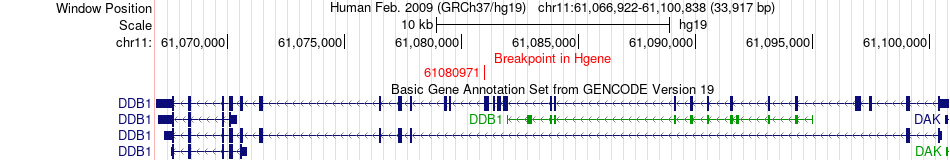 |
 Fusion gene breakpoints across SDHA (3'-gene) Fusion gene breakpoints across SDHA (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
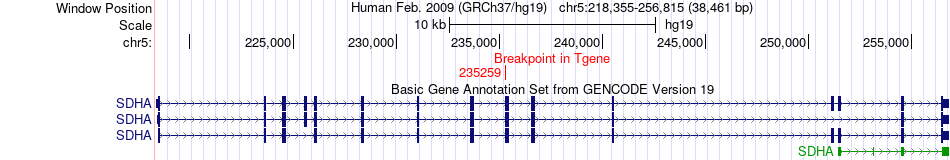 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-3B-A9HQ-01A | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| ChimerDB4 | SARC | TCGA-3R-A8YX-01A | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
Top |
Fusion Gene ORF analysis for DDB1-SDHA |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000301764 | ENST00000507522 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| In-frame | ENST00000301764 | ENST00000264932 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| In-frame | ENST00000301764 | ENST00000504309 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| In-frame | ENST00000301764 | ENST00000510361 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| intron-3CDS | ENST00000450997 | ENST00000264932 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| intron-3CDS | ENST00000450997 | ENST00000504309 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| intron-3CDS | ENST00000450997 | ENST00000510361 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| intron-3CDS | ENST00000451943 | ENST00000264932 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| intron-3CDS | ENST00000451943 | ENST00000504309 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| intron-3CDS | ENST00000451943 | ENST00000510361 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| intron-3CDS | ENST00000538470 | ENST00000264932 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| intron-3CDS | ENST00000538470 | ENST00000504309 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| intron-3CDS | ENST00000538470 | ENST00000510361 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| intron-3CDS | ENST00000545930 | ENST00000264932 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| intron-3CDS | ENST00000545930 | ENST00000504309 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| intron-3CDS | ENST00000545930 | ENST00000510361 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| intron-intron | ENST00000450997 | ENST00000507522 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| intron-intron | ENST00000451943 | ENST00000507522 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| intron-intron | ENST00000538470 | ENST00000507522 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
| intron-intron | ENST00000545930 | ENST00000507522 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000301764 | DDB1 | chr11 | 61080971 | - | ENST00000264932 | SDHA | chr5 | 235259 | + | 3678 | 2467 | 314 | 3397 | 1027 |
| ENST00000301764 | DDB1 | chr11 | 61080971 | - | ENST00000504309 | SDHA | chr5 | 235259 | + | 3417 | 2467 | 314 | 3154 | 946 |
| ENST00000301764 | DDB1 | chr11 | 61080971 | - | ENST00000510361 | SDHA | chr5 | 235259 | + | 3657 | 2467 | 314 | 3397 | 1027 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000301764 | ENST00000264932 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + | 0.001496136 | 0.9985039 |
| ENST00000301764 | ENST00000504309 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + | 0.001520213 | 0.9984798 |
| ENST00000301764 | ENST00000510361 | DDB1 | chr11 | 61080971 | - | SDHA | chr5 | 235259 | + | 0.001544213 | 0.99845576 |
Top |
Fusion Genomic Features for DDB1-SDHA |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| DDB1 | chr11 | 61080970 | - | SDHA | chr5 | 235258 | + | 3.69E-05 | 0.99996316 |
| DDB1 | chr11 | 61080970 | - | SDHA | chr5 | 235258 | + | 3.69E-05 | 0.99996316 |
| DDB1 | chr11 | 61080970 | - | SDHA | chr5 | 235258 | + | 3.69E-05 | 0.99996316 |
| DDB1 | chr11 | 61080970 | - | SDHA | chr5 | 235258 | + | 3.69E-05 | 0.99996316 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
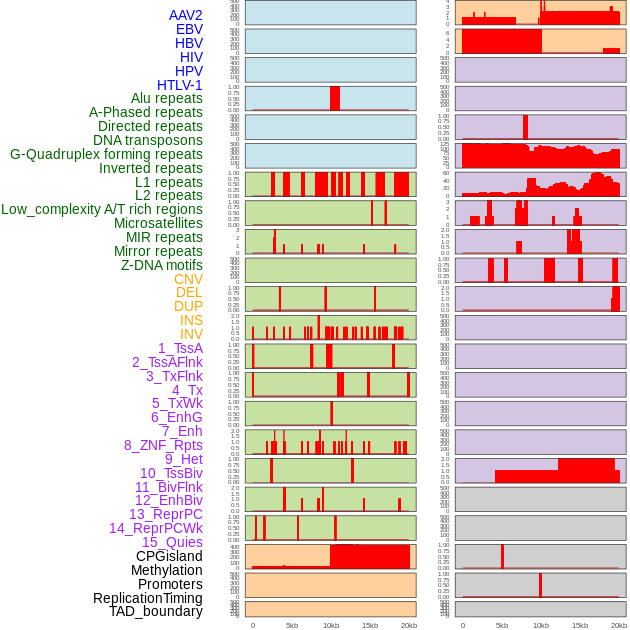 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
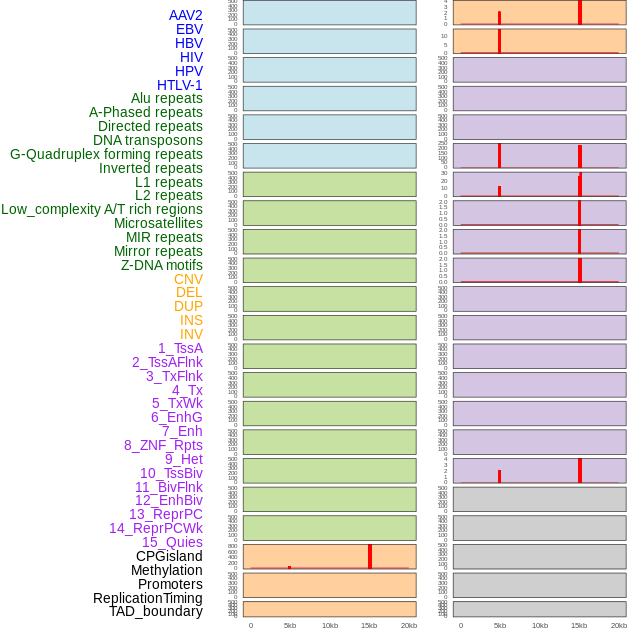 |
Top |
Fusion Protein Features for DDB1-SDHA |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:61080971/chr5:235259) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| DDB1 | SDHA |
| FUNCTION: Protein, which is both involved in DNA repair and protein ubiquitination, as part of the UV-DDB complex and DCX (DDB1-CUL4-X-box) complexes, respectively (PubMed:15448697, PubMed:14739464, PubMed:16260596, PubMed:16482215, PubMed:17079684, PubMed:16407242, PubMed:16407252, PubMed:16940174). Core component of the UV-DDB complex (UV-damaged DNA-binding protein complex), a complex that recognizes UV-induced DNA damage and recruit proteins of the nucleotide excision repair pathway (the NER pathway) to initiate DNA repair (PubMed:15448697, PubMed:16260596, PubMed:16407242, PubMed:16940174). The UV-DDB complex preferentially binds to cyclobutane pyrimidine dimers (CPD), 6-4 photoproducts (6-4 PP), apurinic sites and short mismatches (PubMed:15448697, PubMed:16260596, PubMed:16407242, PubMed:16940174). Also functions as a component of numerous distinct DCX (DDB1-CUL4-X-box) E3 ubiquitin-protein ligase complexes which mediate the ubiquitination and subsequent proteasomal degradation of target proteins (PubMed:14739464, PubMed:16407252, PubMed:16482215, PubMed:17079684, PubMed:25043012, PubMed:25108355, PubMed:18332868, PubMed:18381890, PubMed:19966799, PubMed:22118460, PubMed:28886238). The functional specificity of the DCX E3 ubiquitin-protein ligase complex is determined by the variable substrate recognition component recruited by DDB1 (PubMed:14739464, PubMed:16407252, PubMed:16482215, PubMed:17079684, PubMed:25043012, PubMed:25108355, PubMed:18332868, PubMed:18381890, PubMed:19966799, PubMed:22118460). DCX(DDB2) (also known as DDB1-CUL4-ROC1, CUL4-DDB-ROC1 and CUL4-DDB-RBX1) may ubiquitinate histone H2A, histone H3 and histone H4 at sites of UV-induced DNA damage (PubMed:16678110, PubMed:17041588, PubMed:16473935, PubMed:18593899). The ubiquitination of histones may facilitate their removal from the nucleosome and promote subsequent DNA repair (PubMed:16678110, PubMed:17041588, PubMed:16473935, PubMed:18593899). DCX(DDB2) also ubiquitinates XPC, which may enhance DNA-binding by XPC and promote NER (PubMed:15882621). DCX(DTL) plays a role in PCNA-dependent polyubiquitination of CDT1 and MDM2-dependent ubiquitination of TP53 in response to radiation-induced DNA damage and during DNA replication (PubMed:17041588). DCX(ERCC8) (the CSA complex) plays a role in transcription-coupled repair (TCR) (PubMed:12732143). The DDB1-CUL4A-DTL E3 ligase complex regulates the circadian clock function by mediating the ubiquitination and degradation of CRY1 (PubMed:26431207). DDB1-mediated CRY1 degradation promotes FOXO1 protein stability and FOXO1-mediated gluconeogenesis in the liver (By similarity). {ECO:0000250|UniProtKB:Q3U1J4, ECO:0000269|PubMed:12732143, ECO:0000269|PubMed:14739464, ECO:0000269|PubMed:15448697, ECO:0000269|PubMed:15882621, ECO:0000269|PubMed:16260596, ECO:0000269|PubMed:16407242, ECO:0000269|PubMed:16407252, ECO:0000269|PubMed:16473935, ECO:0000269|PubMed:16482215, ECO:0000269|PubMed:16678110, ECO:0000269|PubMed:16940174, ECO:0000269|PubMed:17041588, ECO:0000269|PubMed:17079684, ECO:0000269|PubMed:18332868, ECO:0000269|PubMed:18381890, ECO:0000269|PubMed:18593899, ECO:0000269|PubMed:19966799, ECO:0000269|PubMed:22118460, ECO:0000269|PubMed:25043012, ECO:0000269|PubMed:25108355, ECO:0000269|PubMed:26431207, ECO:0000269|PubMed:28886238}. | FUNCTION: Plays an essential role in the assembly of succinate dehydrogenase (SDH), an enzyme complex (also referred to as respiratory complex II) that is a component of both the tricarboxylic acid (TCA) cycle and the mitochondrial electron transport chain, and which couples the oxidation of succinate to fumarate with the reduction of ubiquinone (coenzyme Q) to ubiquinol (PubMed:24954416). Binds to the flavoprotein subunit SDHA in its FAD-bound form, blocking the generation of excess reactive oxigen species (ROS) and facilitating its assembly with the iron-sulfur protein subunit SDHB into the SDH catalytic dimer (By similarity). {ECO:0000250|UniProtKB:P38345, ECO:0000269|PubMed:24954416}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DDB1 | chr11:61080971 | chr5:235259 | ENST00000301764 | - | 16 | 27 | 13_356 | 689 | 1141.0 | Region | Note=WD repeat beta-propeller A |
| Tgene | SDHA | chr11:61080971 | chr5:235259 | ENST00000264932 | 7 | 15 | 456_457 | 354 | 665.0 | Nucleotide binding | FAD |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DDB1 | chr11:61080971 | chr5:235259 | ENST00000301764 | - | 16 | 27 | 709_1043 | 689 | 1141.0 | Region | Note=WD repeat beta-propeller C |
| Tgene | SDHA | chr11:61080971 | chr5:235259 | ENST00000264932 | 7 | 15 | 68_73 | 354 | 665.0 | Nucleotide binding | FAD | |
| Tgene | SDHA | chr11:61080971 | chr5:235259 | ENST00000264932 | 7 | 15 | 91_106 | 354 | 665.0 | Nucleotide binding | FAD |
Top |
Fusion Gene Sequence for DDB1-SDHA |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >21777_21777_1_DDB1-SDHA_DDB1_chr11_61080971_ENST00000301764_SDHA_chr5_235259_ENST00000264932_length(transcript)=3678nt_BP=2467nt GGGAGTCTGCGGGCGAGCTCGGAGGACACTGGCGCACCTCCCCCGCCGGCTCACGGCCGCATCCGCCACATCCGAAGGCGCCGCGACTTC CCGGAGACCCCGAAGCGTGCGCTCTCATCCGGGTCCTGCGGGCGCTGGCGGAAAGAGGCGAGGCGGTGCCTCCGGGGGCGGGGCCTCCTT CGGTTGGCGGCCTCGGGCTTCGGGAGTCCTCCAAGAGGCCAGGTGAGGCCGTCCCGTGATGCCCCGCGCCCCGGCCGCTCTGGCCTGCAA CGTGTCTCTGGGGCGGAGGCAGCGGCAGTGGAGTTCGCTGCGCGCTGTTGGGGGCCACCTGTCTTTTCGCTTGTGTCCCTCTTTCTAGTG TCGCGCTCGAGTCCCGACGGGCCGCTCCAAGCCTCGACATGTCGTACAACTACGTGGTAACGGCCCAGAAGCCCACCGCCGTGAACGGCT GCGTGACCGGACACTTTACTTCGGCCGAAGACTTAAACCTGTTGATTGCCAAAAACACGAGATTAGAGATCTATGTGGTCACCGCCGAGG GGCTTCGGCCCGTCAAAGAGGTGGGCATGTATGGGAAGATTGCGGTCATGGAGCTTTTCAGGCCCAAGGGGGAGAGCAAGGACCTGCTGT TTATCTTGACAGCGAAGTACAATGCCTGCATCCTGGAGTATAAACAGAGTGGCGAGAGCATTGACATCATTACGCGAGCCCATGGCAATG TCCAGGACCGCATTGGCCGCCCCTCAGAGACCGGCATTATTGGCATCATTGACCCTGAGTGCCGGATGATTGGCCTGCGTCTCTATGATG GCCTTTTCAAGGTTATTCCACTAGATCGCGATAATAAAGAACTCAAGGCCTTCAACATCCGCCTGGAGGAGCTGCATGTCATTGATGTCA AGTTCCTATATGGTTGCCAAGCACCTACTATTTGCTTTGTCTACCAGGACCCTCAGGGGCGGCACGTAAAAACCTATGAGGTGTCTCTCC GAGAAAAGGAATTCAATAAGGGCCCTTGGAAACAGGAAAATGTCGAAGCTGAAGCTTCCATGGTGATCGCAGTCCCAGAGCCCTTTGGGG GGGCCATCATCATTGGACAGGAGTCAATCACCTATCACAATGGTGACAAATACCTGGCTATTGCCCCTCCTATCATCAAGCAAAGCACGA TTGTGTGCCACAATCGAGTGGACCCTAATGGCTCAAGATACCTGCTGGGAGACATGGAAGGCCGGCTCTTCATGCTGCTTTTGGAGAAGG AGGAACAGATGGATGGCACCGTCACTCTCAAGGATCTCCGTGTAGAACTCCTTGGAGAGACCTCTATTGCTGAGTGCTTGACATACCTTG ATAATGGTGTTGTGTTTGTCGGGTCTCGCCTGGGTGACTCCCAGCTTGTGAAGCTCAACGTTGACAGTAATGAACAAGGCTCCTATGTAG TGGCCATGGAAACCTTTACCAACTTAGGACCCATTGTCGATATGTGCGTGGTGGACCTGGAGAGGCAGGGGCAGGGGCAGCTGGTCACTT GCTCTGGGGCTTTCAAGGAAGGTTCTTTGCGGATCATCCGGAATGGAATTGGAATCCACGAGCATGCCAGCATTGACTTACCAGGCATCA AAGGATTATGGCCACTGCGGTCTGACCCTAATCGTGAGACTGATGACACTTTGGTGCTCTCTTTTGTGGGCCAGACAAGAGTTCTCATGT TAAATGGAGAGGAGGTAGAAGAAACCGAACTGATGGGTTTCGTGGATGATCAGCAGACTTTCTTCTGTGGCAACGTGGCTCATCAGCAGC TTATCCAGATCACTTCAGCATCGGTGAGGTTGGTCTCTCAAGAACCCAAAGCTCTGGTCAGTGAATGGAAGGAGCCTCAGGCCAAGAACA TCAGTGTGGCCTCCTGCAATAGCAGCCAGGTGGTGGTGGCTGTAGGCAGGGCCCTCTACTATCTGCAGATCCATCCTCAGGAGCTCCGGC AGATCAGCCACACAGAGATGGAACATGAAGTGGCTTGCTTGGACATCACCCCATTAGGAGACAGCAATGGACTGTCCCCTCTTTGTGCCA TTGGCCTCTGGACGGACATCTCGGCTCGTATCTTGAAGTTGCCCTCTTTTGAACTACTGCACAAGGAGATGCTGGGTGGAGAGATCATTC CTCGCTCCATCCTGATGACCACCTTTGAGAGTAGCCATTACCTCCTTTGTGCCTTGGGAGATGGAGCGCTTTTCTACTTTGGGCTCAACA TTGAGACAGGTCTGTTGAGCGACCGTAAGAAGGTGACTTTGGGCACCCAGCCCACCGTATTGAGGACTTTTCGTTCTCTTTCTACCACCA ACGTCTTTGCTTGTTCTGACCGCCCCACTGTCATCTATAGCAGCAACCACAAATTGGTCTTCTCAAATGTCAACCTCAAGGAAGTGAACT ACATGTGTCCCCTCAATTCAGATGGCTATCCTGACAGAGGCTGTGGCCCTGAGAAAGATCACGTCTACCTGCAGCTGCACCACCTACCTC CAGAGCAGCTGGCCACGCGCCTGCCTGGCATTTCAGAGACAGCCATGATCTTCGCTGGCGTGGACGTCACGAAGGAGCCGATCCCTGTCC TCCCCACCGTGCATTATAACATGGGCGGCATTCCCACCAACTACAAGGGGCAGGTCCTGAGGCACGTGAATGGCCAGGATCAGATTGTGC CCGGCCTGTACGCCTGTGGGGAGGCCGCCTGTGCCTCGGTACATGGTGCCAACCGCCTCGGGGCAAACTCGCTCTTGGACCTGGTTGTCT TTGGTCGGGCATGTGCCCTGAGCATCGAAGAGTCATGCAGGCCTGGAGATAAAGTCCCTCCAATTAAACCAAACGCTGGGGAAGAATCTG TCATGAATCTTGACAAATTGAGATTTGCTGATGGAAGCATAAGAACATCGGAACTGCGACTCAGCATGCAGAAGTCAATGCAAAATCATG CTGCCGTGTTCCGTGTGGGAAGCGTGTTGCAAGAAGGTTGTGGGAAAATCAGCAAGCTCTATGGAGACCTAAAGCACCTGAAGACGTTCG ACCGGGGAATGGTCTGGAACACGGACCTGGTGGAGACCCTGGAGCTGCAGAACCTGATGCTGTGTGCGCTGCAGACCATCTACGGAGCAG AGGCACGGAAGGAGTCACGGGGCGCGCATGCCAGGGAAGACTACAAGGTGCGGATTGATGAGTACGATTACTCCAAGCCCATCCAGGGGC AACAGAAGAAGCCCTTTGAGGAGCACTGGAGGAAGCACACCCTGTCCTATGTGGACGTTGGCACTGGGAAGGTCACTCTGGAATATAGAC CCGTGATCGACAAAACTTTGAACGAGGCTGACTGTGCCACCGTCCCGCCAGCCATTCGCTCCTACTGATGAGACAAGATGTGGTGATGAC AGAATCAGCTTTTGTAATTATGTATAATAGCTCATGCATGTGTCCATGTCATAACTGTCTTCATACGCTTCTGCACTCTGGGGAAGAAGG AGTACATTGAAGGGAGATTGGCACCTAGTGGCTGGGAGCTTGCCAGGAACCCAGTGGCCAGGGAGCGTGGCACTTACCTTTGTCCCTTGC >21777_21777_1_DDB1-SDHA_DDB1_chr11_61080971_ENST00000301764_SDHA_chr5_235259_ENST00000264932_length(amino acids)=1027AA_BP=717 MLGATCLFACVPLSSVALESRRAAPSLDMSYNYVVTAQKPTAVNGCVTGHFTSAEDLNLLIAKNTRLEIYVVTAEGLRPVKEVGMYGKIA VMELFRPKGESKDLLFILTAKYNACILEYKQSGESIDIITRAHGNVQDRIGRPSETGIIGIIDPECRMIGLRLYDGLFKVIPLDRDNKEL KAFNIRLEELHVIDVKFLYGCQAPTICFVYQDPQGRHVKTYEVSLREKEFNKGPWKQENVEAEASMVIAVPEPFGGAIIIGQESITYHNG DKYLAIAPPIIKQSTIVCHNRVDPNGSRYLLGDMEGRLFMLLLEKEEQMDGTVTLKDLRVELLGETSIAECLTYLDNGVVFVGSRLGDSQ LVKLNVDSNEQGSYVVAMETFTNLGPIVDMCVVDLERQGQGQLVTCSGAFKEGSLRIIRNGIGIHEHASIDLPGIKGLWPLRSDPNRETD DTLVLSFVGQTRVLMLNGEEVEETELMGFVDDQQTFFCGNVAHQQLIQITSASVRLVSQEPKALVSEWKEPQAKNISVASCNSSQVVVAV GRALYYLQIHPQELRQISHTEMEHEVACLDITPLGDSNGLSPLCAIGLWTDISARILKLPSFELLHKEMLGGEIIPRSILMTTFESSHYL LCALGDGALFYFGLNIETGLLSDRKKVTLGTQPTVLRTFRSLSTTNVFACSDRPTVIYSSNHKLVFSNVNLKEVNYMCPLNSDGYPDRGC GPEKDHVYLQLHHLPPEQLATRLPGISETAMIFAGVDVTKEPIPVLPTVHYNMGGIPTNYKGQVLRHVNGQDQIVPGLYACGEAACASVH GANRLGANSLLDLVVFGRACALSIEESCRPGDKVPPIKPNAGEESVMNLDKLRFADGSIRTSELRLSMQKSMQNHAAVFRVGSVLQEGCG KISKLYGDLKHLKTFDRGMVWNTDLVETLELQNLMLCALQTIYGAEARKESRGAHAREDYKVRIDEYDYSKPIQGQQKKPFEEHWRKHTL -------------------------------------------------------------- >21777_21777_2_DDB1-SDHA_DDB1_chr11_61080971_ENST00000301764_SDHA_chr5_235259_ENST00000504309_length(transcript)=3417nt_BP=2467nt GGGAGTCTGCGGGCGAGCTCGGAGGACACTGGCGCACCTCCCCCGCCGGCTCACGGCCGCATCCGCCACATCCGAAGGCGCCGCGACTTC CCGGAGACCCCGAAGCGTGCGCTCTCATCCGGGTCCTGCGGGCGCTGGCGGAAAGAGGCGAGGCGGTGCCTCCGGGGGCGGGGCCTCCTT CGGTTGGCGGCCTCGGGCTTCGGGAGTCCTCCAAGAGGCCAGGTGAGGCCGTCCCGTGATGCCCCGCGCCCCGGCCGCTCTGGCCTGCAA CGTGTCTCTGGGGCGGAGGCAGCGGCAGTGGAGTTCGCTGCGCGCTGTTGGGGGCCACCTGTCTTTTCGCTTGTGTCCCTCTTTCTAGTG TCGCGCTCGAGTCCCGACGGGCCGCTCCAAGCCTCGACATGTCGTACAACTACGTGGTAACGGCCCAGAAGCCCACCGCCGTGAACGGCT GCGTGACCGGACACTTTACTTCGGCCGAAGACTTAAACCTGTTGATTGCCAAAAACACGAGATTAGAGATCTATGTGGTCACCGCCGAGG GGCTTCGGCCCGTCAAAGAGGTGGGCATGTATGGGAAGATTGCGGTCATGGAGCTTTTCAGGCCCAAGGGGGAGAGCAAGGACCTGCTGT TTATCTTGACAGCGAAGTACAATGCCTGCATCCTGGAGTATAAACAGAGTGGCGAGAGCATTGACATCATTACGCGAGCCCATGGCAATG TCCAGGACCGCATTGGCCGCCCCTCAGAGACCGGCATTATTGGCATCATTGACCCTGAGTGCCGGATGATTGGCCTGCGTCTCTATGATG GCCTTTTCAAGGTTATTCCACTAGATCGCGATAATAAAGAACTCAAGGCCTTCAACATCCGCCTGGAGGAGCTGCATGTCATTGATGTCA AGTTCCTATATGGTTGCCAAGCACCTACTATTTGCTTTGTCTACCAGGACCCTCAGGGGCGGCACGTAAAAACCTATGAGGTGTCTCTCC GAGAAAAGGAATTCAATAAGGGCCCTTGGAAACAGGAAAATGTCGAAGCTGAAGCTTCCATGGTGATCGCAGTCCCAGAGCCCTTTGGGG GGGCCATCATCATTGGACAGGAGTCAATCACCTATCACAATGGTGACAAATACCTGGCTATTGCCCCTCCTATCATCAAGCAAAGCACGA TTGTGTGCCACAATCGAGTGGACCCTAATGGCTCAAGATACCTGCTGGGAGACATGGAAGGCCGGCTCTTCATGCTGCTTTTGGAGAAGG AGGAACAGATGGATGGCACCGTCACTCTCAAGGATCTCCGTGTAGAACTCCTTGGAGAGACCTCTATTGCTGAGTGCTTGACATACCTTG ATAATGGTGTTGTGTTTGTCGGGTCTCGCCTGGGTGACTCCCAGCTTGTGAAGCTCAACGTTGACAGTAATGAACAAGGCTCCTATGTAG TGGCCATGGAAACCTTTACCAACTTAGGACCCATTGTCGATATGTGCGTGGTGGACCTGGAGAGGCAGGGGCAGGGGCAGCTGGTCACTT GCTCTGGGGCTTTCAAGGAAGGTTCTTTGCGGATCATCCGGAATGGAATTGGAATCCACGAGCATGCCAGCATTGACTTACCAGGCATCA AAGGATTATGGCCACTGCGGTCTGACCCTAATCGTGAGACTGATGACACTTTGGTGCTCTCTTTTGTGGGCCAGACAAGAGTTCTCATGT TAAATGGAGAGGAGGTAGAAGAAACCGAACTGATGGGTTTCGTGGATGATCAGCAGACTTTCTTCTGTGGCAACGTGGCTCATCAGCAGC TTATCCAGATCACTTCAGCATCGGTGAGGTTGGTCTCTCAAGAACCCAAAGCTCTGGTCAGTGAATGGAAGGAGCCTCAGGCCAAGAACA TCAGTGTGGCCTCCTGCAATAGCAGCCAGGTGGTGGTGGCTGTAGGCAGGGCCCTCTACTATCTGCAGATCCATCCTCAGGAGCTCCGGC AGATCAGCCACACAGAGATGGAACATGAAGTGGCTTGCTTGGACATCACCCCATTAGGAGACAGCAATGGACTGTCCCCTCTTTGTGCCA TTGGCCTCTGGACGGACATCTCGGCTCGTATCTTGAAGTTGCCCTCTTTTGAACTACTGCACAAGGAGATGCTGGGTGGAGAGATCATTC CTCGCTCCATCCTGATGACCACCTTTGAGAGTAGCCATTACCTCCTTTGTGCCTTGGGAGATGGAGCGCTTTTCTACTTTGGGCTCAACA TTGAGACAGGTCTGTTGAGCGACCGTAAGAAGGTGACTTTGGGCACCCAGCCCACCGTATTGAGGACTTTTCGTTCTCTTTCTACCACCA ACGTCTTTGCTTGTTCTGACCGCCCCACTGTCATCTATAGCAGCAACCACAAATTGGTCTTCTCAAATGTCAACCTCAAGGAAGTGAACT ACATGTGTCCCCTCAATTCAGATGGCTATCCTGACAGAGGCTGTGGCCCTGAGAAAGATCACGTCTACCTGCAGCTGCACCACCTACCTC CAGAGCAGCTGGCCACGCGCCTGCCTGGCATTTCAGAGACAGCCATGATCTTCGCTGGCGTGGACGTCACGAAGGAGCCGATCCCTGTCC TCCCCACCGTGCATTATAACATGGGCGGCATTCCCACCAACTACAAGGGGCAGGTCCTGAGGCACGTGAATGGCCAGGATCAGATTGTGC CCGGCCTGTACGCCTGTGGGGAGGCCGCCTGTGCCTCGGTACATGGTGCCAACCGCCTCGGGGCAAACTCGCTCTTGGACCTGGTTGTCT TTGGTCGGGCATGTGCCCTGAGCATCGAAGAGTCATGCAGGCCTGGAGATAAAGTCCCTCCAATTAAACCAAACGCTGGGGAAGAATCTG TCATGAATCTTGACAAATTGAGATTTGCTGATGGAAGCATAAGAACATCGGAACTGCGACTCAGCATGCAGAAGGTGCGGATTGATGAGT ACGATTACTCCAAGCCCATCCAGGGGCAACAGAAGAAGCCCTTTGAGGAGCACTGGAGGAAGCACACCCTGTCCTATGTGGACGTTGGCA CTGGGAAGGTCACTCTGGAATATAGACCCGTGATCGACAAAACTTTGAACGAGGCTGACTGTGCCACCGTCCCGCCAGCCATTCGCTCCT ACTGATGAGACAAGATGTGGTGATGACAGAATCAGCTTTTGTAATTATGTATAATAGCTCATGCATGTGTCCATGTCATAACTGTCTTCA TACGCTTCTGCACTCTGGGGAAGAAGGAGTACATTGAAGGGAGATTGGCACCTAGTGGCTGGGAGCTTGCCAGGAACCCAGTGGCCAGGG >21777_21777_2_DDB1-SDHA_DDB1_chr11_61080971_ENST00000301764_SDHA_chr5_235259_ENST00000504309_length(amino acids)=946AA_BP=717 MLGATCLFACVPLSSVALESRRAAPSLDMSYNYVVTAQKPTAVNGCVTGHFTSAEDLNLLIAKNTRLEIYVVTAEGLRPVKEVGMYGKIA VMELFRPKGESKDLLFILTAKYNACILEYKQSGESIDIITRAHGNVQDRIGRPSETGIIGIIDPECRMIGLRLYDGLFKVIPLDRDNKEL KAFNIRLEELHVIDVKFLYGCQAPTICFVYQDPQGRHVKTYEVSLREKEFNKGPWKQENVEAEASMVIAVPEPFGGAIIIGQESITYHNG DKYLAIAPPIIKQSTIVCHNRVDPNGSRYLLGDMEGRLFMLLLEKEEQMDGTVTLKDLRVELLGETSIAECLTYLDNGVVFVGSRLGDSQ LVKLNVDSNEQGSYVVAMETFTNLGPIVDMCVVDLERQGQGQLVTCSGAFKEGSLRIIRNGIGIHEHASIDLPGIKGLWPLRSDPNRETD DTLVLSFVGQTRVLMLNGEEVEETELMGFVDDQQTFFCGNVAHQQLIQITSASVRLVSQEPKALVSEWKEPQAKNISVASCNSSQVVVAV GRALYYLQIHPQELRQISHTEMEHEVACLDITPLGDSNGLSPLCAIGLWTDISARILKLPSFELLHKEMLGGEIIPRSILMTTFESSHYL LCALGDGALFYFGLNIETGLLSDRKKVTLGTQPTVLRTFRSLSTTNVFACSDRPTVIYSSNHKLVFSNVNLKEVNYMCPLNSDGYPDRGC GPEKDHVYLQLHHLPPEQLATRLPGISETAMIFAGVDVTKEPIPVLPTVHYNMGGIPTNYKGQVLRHVNGQDQIVPGLYACGEAACASVH GANRLGANSLLDLVVFGRACALSIEESCRPGDKVPPIKPNAGEESVMNLDKLRFADGSIRTSELRLSMQKVRIDEYDYSKPIQGQQKKPF -------------------------------------------------------------- >21777_21777_3_DDB1-SDHA_DDB1_chr11_61080971_ENST00000301764_SDHA_chr5_235259_ENST00000510361_length(transcript)=3657nt_BP=2467nt GGGAGTCTGCGGGCGAGCTCGGAGGACACTGGCGCACCTCCCCCGCCGGCTCACGGCCGCATCCGCCACATCCGAAGGCGCCGCGACTTC CCGGAGACCCCGAAGCGTGCGCTCTCATCCGGGTCCTGCGGGCGCTGGCGGAAAGAGGCGAGGCGGTGCCTCCGGGGGCGGGGCCTCCTT CGGTTGGCGGCCTCGGGCTTCGGGAGTCCTCCAAGAGGCCAGGTGAGGCCGTCCCGTGATGCCCCGCGCCCCGGCCGCTCTGGCCTGCAA CGTGTCTCTGGGGCGGAGGCAGCGGCAGTGGAGTTCGCTGCGCGCTGTTGGGGGCCACCTGTCTTTTCGCTTGTGTCCCTCTTTCTAGTG TCGCGCTCGAGTCCCGACGGGCCGCTCCAAGCCTCGACATGTCGTACAACTACGTGGTAACGGCCCAGAAGCCCACCGCCGTGAACGGCT GCGTGACCGGACACTTTACTTCGGCCGAAGACTTAAACCTGTTGATTGCCAAAAACACGAGATTAGAGATCTATGTGGTCACCGCCGAGG GGCTTCGGCCCGTCAAAGAGGTGGGCATGTATGGGAAGATTGCGGTCATGGAGCTTTTCAGGCCCAAGGGGGAGAGCAAGGACCTGCTGT TTATCTTGACAGCGAAGTACAATGCCTGCATCCTGGAGTATAAACAGAGTGGCGAGAGCATTGACATCATTACGCGAGCCCATGGCAATG TCCAGGACCGCATTGGCCGCCCCTCAGAGACCGGCATTATTGGCATCATTGACCCTGAGTGCCGGATGATTGGCCTGCGTCTCTATGATG GCCTTTTCAAGGTTATTCCACTAGATCGCGATAATAAAGAACTCAAGGCCTTCAACATCCGCCTGGAGGAGCTGCATGTCATTGATGTCA AGTTCCTATATGGTTGCCAAGCACCTACTATTTGCTTTGTCTACCAGGACCCTCAGGGGCGGCACGTAAAAACCTATGAGGTGTCTCTCC GAGAAAAGGAATTCAATAAGGGCCCTTGGAAACAGGAAAATGTCGAAGCTGAAGCTTCCATGGTGATCGCAGTCCCAGAGCCCTTTGGGG GGGCCATCATCATTGGACAGGAGTCAATCACCTATCACAATGGTGACAAATACCTGGCTATTGCCCCTCCTATCATCAAGCAAAGCACGA TTGTGTGCCACAATCGAGTGGACCCTAATGGCTCAAGATACCTGCTGGGAGACATGGAAGGCCGGCTCTTCATGCTGCTTTTGGAGAAGG AGGAACAGATGGATGGCACCGTCACTCTCAAGGATCTCCGTGTAGAACTCCTTGGAGAGACCTCTATTGCTGAGTGCTTGACATACCTTG ATAATGGTGTTGTGTTTGTCGGGTCTCGCCTGGGTGACTCCCAGCTTGTGAAGCTCAACGTTGACAGTAATGAACAAGGCTCCTATGTAG TGGCCATGGAAACCTTTACCAACTTAGGACCCATTGTCGATATGTGCGTGGTGGACCTGGAGAGGCAGGGGCAGGGGCAGCTGGTCACTT GCTCTGGGGCTTTCAAGGAAGGTTCTTTGCGGATCATCCGGAATGGAATTGGAATCCACGAGCATGCCAGCATTGACTTACCAGGCATCA AAGGATTATGGCCACTGCGGTCTGACCCTAATCGTGAGACTGATGACACTTTGGTGCTCTCTTTTGTGGGCCAGACAAGAGTTCTCATGT TAAATGGAGAGGAGGTAGAAGAAACCGAACTGATGGGTTTCGTGGATGATCAGCAGACTTTCTTCTGTGGCAACGTGGCTCATCAGCAGC TTATCCAGATCACTTCAGCATCGGTGAGGTTGGTCTCTCAAGAACCCAAAGCTCTGGTCAGTGAATGGAAGGAGCCTCAGGCCAAGAACA TCAGTGTGGCCTCCTGCAATAGCAGCCAGGTGGTGGTGGCTGTAGGCAGGGCCCTCTACTATCTGCAGATCCATCCTCAGGAGCTCCGGC AGATCAGCCACACAGAGATGGAACATGAAGTGGCTTGCTTGGACATCACCCCATTAGGAGACAGCAATGGACTGTCCCCTCTTTGTGCCA TTGGCCTCTGGACGGACATCTCGGCTCGTATCTTGAAGTTGCCCTCTTTTGAACTACTGCACAAGGAGATGCTGGGTGGAGAGATCATTC CTCGCTCCATCCTGATGACCACCTTTGAGAGTAGCCATTACCTCCTTTGTGCCTTGGGAGATGGAGCGCTTTTCTACTTTGGGCTCAACA TTGAGACAGGTCTGTTGAGCGACCGTAAGAAGGTGACTTTGGGCACCCAGCCCACCGTATTGAGGACTTTTCGTTCTCTTTCTACCACCA ACGTCTTTGCTTGTTCTGACCGCCCCACTGTCATCTATAGCAGCAACCACAAATTGGTCTTCTCAAATGTCAACCTCAAGGAAGTGAACT ACATGTGTCCCCTCAATTCAGATGGCTATCCTGACAGAGGCTGTGGCCCTGAGAAAGATCACGTCTACCTGCAGCTGCACCACCTACCTC CAGAGCAGCTGGCCACGCGCCTGCCTGGCATTTCAGAGACAGCCATGATCTTCGCTGGCGTGGACGTCACGAAGGAGCCGATCCCTGTCC TCCCCACCGTGCATTATAACATGGGCGGCATTCCCACCAACTACAAGGGGCAGGTCCTGAGGCACGTGAATGGCCAGGATCAGATTGTGC CCGGCCTGTACGCCTGTGGGGAGGCCGCCTGTGCCTCGGTACATGGTGCCAACCGCCTCGGGGCAAACTCGCTCTTGGACCTGGTTGTCT TTGGTCGGGCATGTGCCCTGAGCATCGAAGAGTCATGCAGGCCTGGAGATAAAGTCCCTCCAATTAAACCAAACGCTGGGGAAGAATCTG TCATGAATCTTGACAAATTGAGATTTGCTGATGGAAGCATAAGAACATCGGAACTGCGACTCAGCATGCAGAAGTCAATGCAAAATCATG CTGCCGTGTTCCGTGTGGGAAGCGTGTTGCAAGAAGGTTGTGGGAAAATCAGCAAGCTCTATGGAGACCTAAAGCACCTGAAGACGTTCG ACCGGGGAATGGTCTGGAACACGGACCTGGTGGAGACCCTGGAGCTGCAGAACCTGATGCTGTGTGCGCTGCAGACCATCTACGGAGCAG AGGCACGGAAGGAGTCACGGGGCGCGCATGCCAGGGAAGACTACAAGGTGCGGATTGATGAGTACGATTACTCCAAGCCCATCCAGGGGC AACAGAAGAAGCCCTTTGAGGAGCACTGGAGGAAGCACACCCTGTCCTATGTGGACGTTGGCACTGGGAAGGTCACTCTGGAATATAGAC CCGTGATCGACAAAACTTTGAACGAGGCTGACTGTGCCACCGTCCCGCCAGCCATTCGCTCCTACTGATGAGACAAGATGTGGTGATGAC AGAATCAGCTTTTGTAATTATGTATAATAGCTCATGCATGTGTCCATGTCATAACTGTCTTCATACGCTTCTGCACTCTGGGGAAGAAGG AGTACATTGAAGGGAGATTGGCACCTAGTGGCTGGGAGCTTGCCAGGAACCCAGTGGCCAGGGAGCGTGGCACTTACCTTTGTCCCTTGC >21777_21777_3_DDB1-SDHA_DDB1_chr11_61080971_ENST00000301764_SDHA_chr5_235259_ENST00000510361_length(amino acids)=1027AA_BP=717 MLGATCLFACVPLSSVALESRRAAPSLDMSYNYVVTAQKPTAVNGCVTGHFTSAEDLNLLIAKNTRLEIYVVTAEGLRPVKEVGMYGKIA VMELFRPKGESKDLLFILTAKYNACILEYKQSGESIDIITRAHGNVQDRIGRPSETGIIGIIDPECRMIGLRLYDGLFKVIPLDRDNKEL KAFNIRLEELHVIDVKFLYGCQAPTICFVYQDPQGRHVKTYEVSLREKEFNKGPWKQENVEAEASMVIAVPEPFGGAIIIGQESITYHNG DKYLAIAPPIIKQSTIVCHNRVDPNGSRYLLGDMEGRLFMLLLEKEEQMDGTVTLKDLRVELLGETSIAECLTYLDNGVVFVGSRLGDSQ LVKLNVDSNEQGSYVVAMETFTNLGPIVDMCVVDLERQGQGQLVTCSGAFKEGSLRIIRNGIGIHEHASIDLPGIKGLWPLRSDPNRETD DTLVLSFVGQTRVLMLNGEEVEETELMGFVDDQQTFFCGNVAHQQLIQITSASVRLVSQEPKALVSEWKEPQAKNISVASCNSSQVVVAV GRALYYLQIHPQELRQISHTEMEHEVACLDITPLGDSNGLSPLCAIGLWTDISARILKLPSFELLHKEMLGGEIIPRSILMTTFESSHYL LCALGDGALFYFGLNIETGLLSDRKKVTLGTQPTVLRTFRSLSTTNVFACSDRPTVIYSSNHKLVFSNVNLKEVNYMCPLNSDGYPDRGC GPEKDHVYLQLHHLPPEQLATRLPGISETAMIFAGVDVTKEPIPVLPTVHYNMGGIPTNYKGQVLRHVNGQDQIVPGLYACGEAACASVH GANRLGANSLLDLVVFGRACALSIEESCRPGDKVPPIKPNAGEESVMNLDKLRFADGSIRTSELRLSMQKSMQNHAAVFRVGSVLQEGCG KISKLYGDLKHLKTFDRGMVWNTDLVETLELQNLMLCALQTIYGAEARKESRGAHAREDYKVRIDEYDYSKPIQGQQKKPFEEHWRKHTL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for DDB1-SDHA |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | DDB1 | chr11:61080971 | chr5:235259 | ENST00000301764 | - | 16 | 27 | 771_1140 | 689.6666666666666 | 1141.0 | CDT1 and CUL4A |
| Hgene | DDB1 | chr11:61080971 | chr5:235259 | ENST00000301764 | - | 16 | 27 | 2_768 | 689.6666666666666 | 1141.0 | CDT1 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for DDB1-SDHA |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for DDB1-SDHA |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
