
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:DDX52-ABCF1 (FusionGDB2 ID:22036) |
Fusion Gene Summary for DDX52-ABCF1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: DDX52-ABCF1 | Fusion gene ID: 22036 | Hgene | Tgene | Gene symbol | DDX52 | ABCF1 | Gene ID | 11056 | 23 |
| Gene name | DExD-box helicase 52 | ATP binding cassette subfamily F member 1 | |
| Synonyms | HUSSY19|ROK1 | ABC27|ABC50 | |
| Cytomap | 17q12 | 6p21.33 | |
| Type of gene | protein-coding | protein-coding | |
| Description | probable ATP-dependent RNA helicase DDX52DEAD (Asp-Glu-Ala-Asp) box polypeptide 52DEAD box protein 52DEAD-box helicase 52 | ATP-binding cassette sub-family F member 1ATP-binding cassette 50 (TNF-alpha stimulated)ATP-binding cassette, sub-family F (GCN20), member 1TNF-alpha-stimulated ABC proteinTNFalpha-inducible ATP-binding protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9Y2R4 | Q8NE71 | |
| Ensembl transtripts involved in fusion gene | ENST00000349699, ENST00000394367, | ENST00000383587, ENST00000383588, ENST00000400609, ENST00000412443, ENST00000416932, ENST00000419893, ENST00000420257, ENST00000421042, ENST00000421608, ENST00000423247, ENST00000426219, ENST00000448939, ENST00000452530, ENST00000454094, ENST00000456791, ENST00000457078, ENST00000457111, ENST00000457970, ENST00000470464, ENST00000475528, ENST00000485585, ENST00000486105, ENST00000494413, ENST00000495194, ENST00000546690, ENST00000552867, ENST00000326195, ENST00000376545, ENST00000396515, | |
| Fusion gene scores | * DoF score | 12 X 16 X 5=960 | 9 X 5 X 7=315 |
| # samples | 14 | 9 | |
| ** MAII score | log2(14/960*10)=-2.77760757866355 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/315*10)=-1.8073549220576 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: DDX52 [Title/Abstract] AND ABCF1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | DDX52(36002139)-ABCF1(30545186), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across DDX52 (5'-gene) Fusion gene breakpoints across DDX52 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
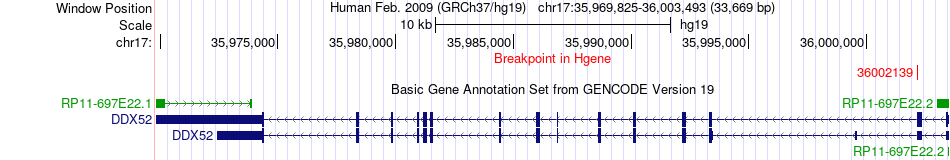 |
 Fusion gene breakpoints across ABCF1 (3'-gene) Fusion gene breakpoints across ABCF1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-04-1356-01A | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
Top |
Fusion Gene ORF analysis for DDX52-ABCF1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000349699 | ENST00000383587 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000383588 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000400609 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000412443 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000416932 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000419893 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000420257 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000421042 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000421608 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000423247 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000426219 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000448939 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000452530 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000454094 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000456791 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000457078 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000457111 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000457970 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000470464 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000475528 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000485585 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000486105 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000494413 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000495194 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000546690 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5CDS-intron | ENST00000349699 | ENST00000552867 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-3CDS | ENST00000394367 | ENST00000326195 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-3CDS | ENST00000394367 | ENST00000376545 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-3CDS | ENST00000394367 | ENST00000396515 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000383587 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000383588 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000400609 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000412443 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000416932 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000419893 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000420257 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000421042 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000421608 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000423247 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000426219 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000448939 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000452530 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000454094 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000456791 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000457078 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000457111 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000457970 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000470464 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000475528 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000485585 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000486105 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000494413 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000495194 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000546690 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| 5UTR-intron | ENST00000394367 | ENST00000552867 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| In-frame | ENST00000349699 | ENST00000326195 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| In-frame | ENST00000349699 | ENST00000376545 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
| In-frame | ENST00000349699 | ENST00000396515 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000349699 | DDX52 | chr17 | 36002139 | - | ENST00000326195 | ABCF1 | chr6 | 30545186 | + | 3626 | 330 | 44 | 2794 | 916 |
| ENST00000349699 | DDX52 | chr17 | 36002139 | - | ENST00000376545 | ABCF1 | chr6 | 30545186 | + | 3293 | 330 | 44 | 2680 | 878 |
| ENST00000349699 | DDX52 | chr17 | 36002139 | - | ENST00000396515 | ABCF1 | chr6 | 30545186 | + | 1803 | 330 | 44 | 973 | 309 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000349699 | ENST00000326195 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + | 0.002296992 | 0.997703 |
| ENST00000349699 | ENST00000376545 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + | 0.00342132 | 0.99657863 |
| ENST00000349699 | ENST00000396515 | DDX52 | chr17 | 36002139 | - | ABCF1 | chr6 | 30545186 | + | 0.000614816 | 0.9993851 |
Top |
Fusion Genomic Features for DDX52-ABCF1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| DDX52 | chr17 | 36002138 | - | ABCF1 | chr6 | 30545185 | + | 0.00023471 | 0.99976534 |
| DDX52 | chr17 | 36002138 | - | ABCF1 | chr6 | 30545185 | + | 0.00023471 | 0.99976534 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
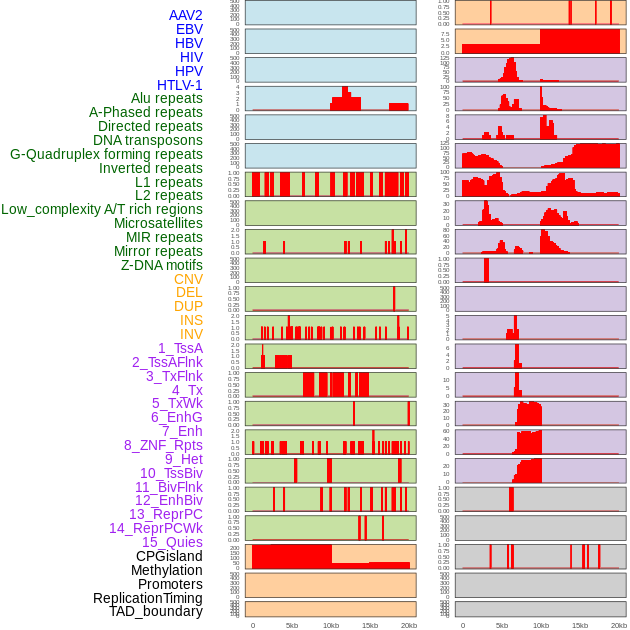 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
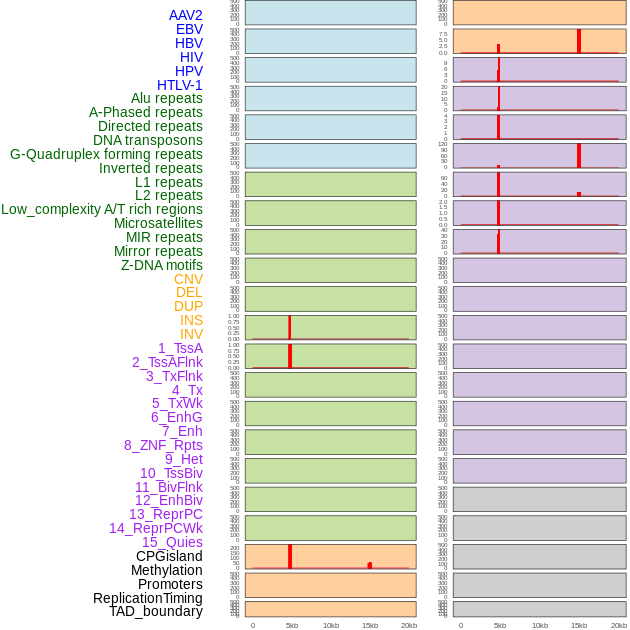 |
Top |
Fusion Protein Features for DDX52-ABCF1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:36002139/chr6:30545186) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| DDX52 | ABCF1 |
| FUNCTION: Isoform 2 is required for efficient Cap- and IRES-mediated mRNA translation initiation. Isoform 2 is not involved in the ribosome biogenesis. {ECO:0000269|PubMed:19570978}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000326195 | 0 | 25 | 141_243 | 24 | 846.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000376545 | 0 | 24 | 141_243 | 24 | 808.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000383587 | 0 | 24 | 141_243 | 0 | 808.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000383588 | 0 | 25 | 141_243 | 0 | 846.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000412443 | 0 | 24 | 141_243 | 0 | 808.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000419893 | 0 | 25 | 141_243 | 0 | 846.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000420257 | 0 | 24 | 141_243 | 0 | 808.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000421042 | 0 | 24 | 141_243 | 0 | 808.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000423247 | 0 | 25 | 141_243 | 0 | 846.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000426219 | 0 | 25 | 141_243 | 0 | 846.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000448939 | 0 | 24 | 141_243 | 0 | 808.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000452530 | 0 | 24 | 141_243 | 0 | 808.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000457078 | 0 | 25 | 141_243 | 0 | 846.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000457111 | 0 | 25 | 141_243 | 0 | 846.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000326195 | 0 | 25 | 304_548 | 24 | 846.0 | Domain | ABC transporter 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000326195 | 0 | 25 | 625_840 | 24 | 846.0 | Domain | ABC transporter 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000376545 | 0 | 24 | 304_548 | 24 | 808.0 | Domain | ABC transporter 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000376545 | 0 | 24 | 625_840 | 24 | 808.0 | Domain | ABC transporter 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000383587 | 0 | 24 | 304_548 | 0 | 808.0 | Domain | ABC transporter 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000383587 | 0 | 24 | 625_840 | 0 | 808.0 | Domain | ABC transporter 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000383588 | 0 | 25 | 304_548 | 0 | 846.0 | Domain | ABC transporter 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000383588 | 0 | 25 | 625_840 | 0 | 846.0 | Domain | ABC transporter 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000412443 | 0 | 24 | 304_548 | 0 | 808.0 | Domain | ABC transporter 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000412443 | 0 | 24 | 625_840 | 0 | 808.0 | Domain | ABC transporter 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000419893 | 0 | 25 | 304_548 | 0 | 846.0 | Domain | ABC transporter 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000419893 | 0 | 25 | 625_840 | 0 | 846.0 | Domain | ABC transporter 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000420257 | 0 | 24 | 304_548 | 0 | 808.0 | Domain | ABC transporter 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000420257 | 0 | 24 | 625_840 | 0 | 808.0 | Domain | ABC transporter 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000421042 | 0 | 24 | 304_548 | 0 | 808.0 | Domain | ABC transporter 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000421042 | 0 | 24 | 625_840 | 0 | 808.0 | Domain | ABC transporter 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000423247 | 0 | 25 | 304_548 | 0 | 846.0 | Domain | ABC transporter 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000423247 | 0 | 25 | 625_840 | 0 | 846.0 | Domain | ABC transporter 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000426219 | 0 | 25 | 304_548 | 0 | 846.0 | Domain | ABC transporter 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000426219 | 0 | 25 | 625_840 | 0 | 846.0 | Domain | ABC transporter 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000448939 | 0 | 24 | 304_548 | 0 | 808.0 | Domain | ABC transporter 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000448939 | 0 | 24 | 625_840 | 0 | 808.0 | Domain | ABC transporter 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000452530 | 0 | 24 | 304_548 | 0 | 808.0 | Domain | ABC transporter 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000452530 | 0 | 24 | 625_840 | 0 | 808.0 | Domain | ABC transporter 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000457078 | 0 | 25 | 304_548 | 0 | 846.0 | Domain | ABC transporter 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000457078 | 0 | 25 | 625_840 | 0 | 846.0 | Domain | ABC transporter 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000457111 | 0 | 25 | 304_548 | 0 | 846.0 | Domain | ABC transporter 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000457111 | 0 | 25 | 625_840 | 0 | 846.0 | Domain | ABC transporter 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000326195 | 0 | 25 | 336_343 | 24 | 846.0 | Nucleotide binding | ATP 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000326195 | 0 | 25 | 658_665 | 24 | 846.0 | Nucleotide binding | ATP 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000376545 | 0 | 24 | 336_343 | 24 | 808.0 | Nucleotide binding | ATP 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000376545 | 0 | 24 | 658_665 | 24 | 808.0 | Nucleotide binding | ATP 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000383587 | 0 | 24 | 336_343 | 0 | 808.0 | Nucleotide binding | ATP 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000383587 | 0 | 24 | 658_665 | 0 | 808.0 | Nucleotide binding | ATP 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000383588 | 0 | 25 | 336_343 | 0 | 846.0 | Nucleotide binding | ATP 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000383588 | 0 | 25 | 658_665 | 0 | 846.0 | Nucleotide binding | ATP 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000412443 | 0 | 24 | 336_343 | 0 | 808.0 | Nucleotide binding | ATP 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000412443 | 0 | 24 | 658_665 | 0 | 808.0 | Nucleotide binding | ATP 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000419893 | 0 | 25 | 336_343 | 0 | 846.0 | Nucleotide binding | ATP 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000419893 | 0 | 25 | 658_665 | 0 | 846.0 | Nucleotide binding | ATP 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000420257 | 0 | 24 | 336_343 | 0 | 808.0 | Nucleotide binding | ATP 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000420257 | 0 | 24 | 658_665 | 0 | 808.0 | Nucleotide binding | ATP 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000421042 | 0 | 24 | 336_343 | 0 | 808.0 | Nucleotide binding | ATP 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000421042 | 0 | 24 | 658_665 | 0 | 808.0 | Nucleotide binding | ATP 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000423247 | 0 | 25 | 336_343 | 0 | 846.0 | Nucleotide binding | ATP 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000423247 | 0 | 25 | 658_665 | 0 | 846.0 | Nucleotide binding | ATP 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000426219 | 0 | 25 | 336_343 | 0 | 846.0 | Nucleotide binding | ATP 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000426219 | 0 | 25 | 658_665 | 0 | 846.0 | Nucleotide binding | ATP 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000448939 | 0 | 24 | 336_343 | 0 | 808.0 | Nucleotide binding | ATP 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000448939 | 0 | 24 | 658_665 | 0 | 808.0 | Nucleotide binding | ATP 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000452530 | 0 | 24 | 336_343 | 0 | 808.0 | Nucleotide binding | ATP 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000452530 | 0 | 24 | 658_665 | 0 | 808.0 | Nucleotide binding | ATP 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000457078 | 0 | 25 | 336_343 | 0 | 846.0 | Nucleotide binding | ATP 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000457078 | 0 | 25 | 658_665 | 0 | 846.0 | Nucleotide binding | ATP 2 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000457111 | 0 | 25 | 336_343 | 0 | 846.0 | Nucleotide binding | ATP 1 | |
| Tgene | ABCF1 | chr17:36002139 | chr6:30545186 | ENST00000457111 | 0 | 25 | 658_665 | 0 | 846.0 | Nucleotide binding | ATP 2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DDX52 | chr17:36002139 | chr6:30545186 | ENST00000349699 | - | 2 | 15 | 543_592 | 95 | 600.0 | Compositional bias | Note=Lys-rich |
| Hgene | DDX52 | chr17:36002139 | chr6:30545186 | ENST00000349699 | - | 2 | 15 | 196_374 | 95 | 600.0 | Domain | Helicase ATP-binding |
| Hgene | DDX52 | chr17:36002139 | chr6:30545186 | ENST00000349699 | - | 2 | 15 | 385_546 | 95 | 600.0 | Domain | Helicase C-terminal |
| Hgene | DDX52 | chr17:36002139 | chr6:30545186 | ENST00000349699 | - | 2 | 15 | 165_193 | 95 | 600.0 | Motif | Note=Q motif |
| Hgene | DDX52 | chr17:36002139 | chr6:30545186 | ENST00000349699 | - | 2 | 15 | 318_321 | 95 | 600.0 | Motif | Note=DEAD box |
| Hgene | DDX52 | chr17:36002139 | chr6:30545186 | ENST00000349699 | - | 2 | 15 | 209_216 | 95 | 600.0 | Nucleotide binding | Note=ATP |
Top |
Fusion Gene Sequence for DDX52-ABCF1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >22036_22036_1_DDX52-ABCF1_DDX52_chr17_36002139_ENST00000349699_ABCF1_chr6_30545186_ENST00000326195_length(transcript)=3626nt_BP=330nt AGTCTGCGCAGGCGCCGTAGAACTGTGGCGCTTTCTGGGTAAAGATGGACGTCCACGATCTCTTTCGCCGGCTCGGCGCGGGGGCCAAAT TCGACACGAGACGCTTCTCGGCAGACGCAGCTCGATTCCAGATAGGAAAAAGGAAATATGACTTTGATTCTTCGGAGGTGCTTCAGGGAC TGGACTTTTTTGGAAACAAGAAGTCTGTCCCAGGTGTGTGTGGAGCATCACAAACACATCAGAAGCCCCAAAATGGAGAGAAAAAAGAAG AGAGCCTAACTGAAAGGAAGAGGGAGCAGAGCAAGAAAAAAAGGAAGACGATGACTTCAGACAAAGTGGTGAAGAAAGGGAAGAAGGACA AGAAGATCAAAAAAACGTTCTTTGAAGAGCTGGCAGTAGAAGATAAACAGGCTGGGGAAGAAGAGAAAGTGCTCAAGGAGAAGGAGCAGC AGCAGCAGCAACAGCAACAGCAGCAAAAAAAAAAGCGAGATACCCGAAAAGGCAGGCGGAAGAAGGATGTGGATGATGATGGAGAAGAGA AAGAGCTCATGGAGCGTCTTAAGAAGCTCTCAGTGCCAACCAGTGATGAGGAGGATGAAGTACCCGCCCCAAAACCCCGCGGAGGGAAGA AAACCAAGGGTGGTAATGTTTTTGCAGCCCTGATTCAGGATCAGAGTGAGGAAGAGGAGGAGGAAGAAAAACATCCTCCTAAGCCTGCCA AGCCGGAGAAGAATCGGATCAATAAGGCCGTATCTGAGGAACAGCAGCCTGCACTCAAGGGCAAAAAGGGAAAGGAAGAGAAGTCAAAAG GGAAGGCTAAGCCTCAAAATAAATTCGCTGCTCTGGACAATGAAGAGGAGGATAAAGAAGAAGAAATTATAAAGGAAAAGGAGCCTCCCA AACAAGGGAAGGAGAAGGCCAAGAAGGCAGAGCAGGGTTCAGAGGAAGAAGGAGAAGGGGAAGAAGAGGAGGAGGAAGGAGGAGAGTCTA AGGCAGATGATCCCTATGCTCATCTTAGCAAAAAGGAGAAGAAAAAGCTGAAAAAACAGATGGAGTATGAGCGCCAAGTGGCTTCATTAA AAGCAGCCAATGCAGCTGAAAATGACTTCTCCGTGTCCCAGGCGGAGATGTCCTCCCGCCAAGCCATGTTAGAAAATGCATCTGACATCA AGCTGGAGAAGTTCAGCATCTCCGCTCATGGCAAGGAGCTGTTCGTCAATGCAGACCTGTACATTGTAGCCGGCCGCCGCTACGGGCTGG TAGGACCCAATGGCAAGGGCAAGACCACACTCCTCAAGCACATTGCCAACCGAGCCCTGAGCATCCCTCCCAACATTGATGTGTTGCTGT GTGAGCAGGAGGTGGTAGCAGATGAGACACCAGCAGTCCAGGCTGTTCTTCGAGCTGACACCAAGCGATTGAAGCTGCTGGAAGAGGAGC GGCGGCTTCAGGGACAGCTGGAACAAGGGGATGACACAGCTGCTGAGAGGCTAGAGAAGGTGTATGAGGAATTGCGGGCCACTGGGGCGG CAGCTGCAGAGGCCAAAGCACGGCGGATCCTGGCTGGCCTGGGCTTTGACCCTGAAATGCAGAATCGACCCACACAGAAGTTCTCAGGGG GCTGGCGCATGCGTGTCTCCCTGGCCAGGGCACTGTTCATGGAGCCCACACTGCTGATGCTGGATGAGCCCACCAACCACCTGGACCTCA ACGCTGTCATCTGGCTTAATAACTACCTCCAGGGCTGGCGGAAGACCTTGCTGATCGTCTCCCATGACCAGGGCTTCTTGGATGATGTCT GCACTGATATCATCCACCTCGATGCCCAGCGGCTCCACTACTATAGGGGCAATTACATGACCTTCAAAAAGATGTACCAGCAGAAGCAGA AAGAACTGCTGAAACAGTATGAGAAGCAAGAGAAAAAGCTGAAGGAGCTGAAGGCAGGCGGGAAGTCCACCAAGCAGGCGGAAAAACAAA CGAAGGAAGCCCTGACTCGGAAGCAGCAGAAATGCCGACGGAAAAACCAAGATGAGGAATCCCAGGAGGCCCCTGAGCTCCTGAAGCGCC CTAAGGAGTACACTGTGCGCTTCACTTTTCCAGACCCCCCACCACTCAGCCCTCCAGTGCTGGGTCTGCATGGTGTGACATTCGGCTACC AGGGACAGAAACCACTCTTTAAGAACTTGGATTTTGGCATCGACATGGATTCAAGGATTTGCATTGTGGGCCCTAATGGTGTGGGGAAGA GTACGCTACTCCTGCTGCTGACTGGCAAGCTGACACCGACCCATGGGGAAATGAGAAAGAACCACCGGCTGAAAATTGGCTTCTTCAACC AGCAGTATGCAGAGCAGCTGCGCATGGAGGAGACGCCCACTGAGTACCTGCAGCGGGGCTTCAACCTGCCCTACCAGGATGCCCGCAAGT GCCTGGGCCGCTTCGGCCTGGAGAGTCACGCCCACACCATCCAGATCTGCAAACTCTCTGGTGGTCAGAAGGCGCGAGTTGTGTTTGCTG AGCTGGCCTGTCGGGAACCTGATGTCCTCATCTTGGACGAGCCAACCAATAACCTGGACATAGAGTCTATTGATGCTCTAGGGGAGGCCA TCAATGAATACAAGGGTGCTGTGATCGTTGTCAGCCATGATGCCCGACTCATCACAGAAACCAATTGCCAGCTGTGGGTGGTGGAGGAGC AGAGTGTTAGCCAAATCGATGGTGACTTTGAAGACTACAAGCGGGAGGTGTTGGAGGCCCTGGGTGAAGTCATGGTCAGCCGGCCCCGAG AGTGAGCTTTCCTTCCCAGAAGTCTCCCGAGAGACATATTTGTGTGGCCTAGAAGTCCTCTGTGGTCTCCCCTCCTCTGAAGACTGCCTC TGGCCTGCAGCTGACCTGGCAACCATTCAGGCACATGAAGGTGGAGTGTGACCTTGATGTGACCGGGATCCCACTCTGATTGCATCCATT TCTCTGAAAGACTTGTTTGTTCTGCTTCTCTTCATATAACTGAGCTGGCCTTATCCTTGGCATCCCCCTAAACAAACAAGAGGTGACCAC CTTATTGTGAGGTTCCATCCAGCCAAGTTTATGTGGCCTATTGTCTCAGGACTCTCATCACTCAGAAGCCTGCCTCTGATTTACCCTACA GCTTCAGGCCCAGCTGCCCCCCAGTCTTTGGGTGGTGCTGTTCTTTTCTGGTGGATTTAATGCTGACTCACTGGTACAAACAGCTGTTGA AGCTCAGAGCTGGAGGTGAGCTTCTGAGGCCTTTGCCATTATCCAGCCCAAGATTTGGTGCCTGCAGCCTCTTGTCTGGTTGAGGACTTG GGGCAGGAAAGGAATGCTGCTGAACTTGAATTTCCCTTTACAAGGGGAAGAAATAAAGGAAAGGAGTTGCTGCCGACCTGTCACTGTTTG GAGATTGATGGGAGTTGGAACTGTTCTCAGTCTTGATTTGCTTTATTCAGTTTTCTAGCAGCTTTTAATAGTCCCCTCTTCCCCACTAAA TGGATCTTGTTTGCAGTCTTGCTGACAGTGTTTGCTGTTTAAGGATCATAGGATTCCTTTCCCCCAACCCTTCACGCAAGGAAAAAGCAA >22036_22036_1_DDX52-ABCF1_DDX52_chr17_36002139_ENST00000349699_ABCF1_chr6_30545186_ENST00000326195_length(amino acids)=916AA_BP=86 MDVHDLFRRLGAGAKFDTRRFSADAARFQIGKRKYDFDSSEVLQGLDFFGNKKSVPGVCGASQTHQKPQNGEKKEESLTERKREQSKKKR KTMTSDKVVKKGKKDKKIKKTFFEELAVEDKQAGEEEKVLKEKEQQQQQQQQQQKKKRDTRKGRRKKDVDDDGEEKELMERLKKLSVPTS DEEDEVPAPKPRGGKKTKGGNVFAALIQDQSEEEEEEEKHPPKPAKPEKNRINKAVSEEQQPALKGKKGKEEKSKGKAKPQNKFAALDNE EEDKEEEIIKEKEPPKQGKEKAKKAEQGSEEEGEGEEEEEEGGESKADDPYAHLSKKEKKKLKKQMEYERQVASLKAANAAENDFSVSQA EMSSRQAMLENASDIKLEKFSISAHGKELFVNADLYIVAGRRYGLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADETPAVQA VLRADTKRLKLLEEERRLQGQLEQGDDTAAERLEKVYEELRATGAAAAEAKARRILAGLGFDPEMQNRPTQKFSGGWRMRVSLARALFME PTLLMLDEPTNHLDLNAVIWLNNYLQGWRKTLLIVSHDQGFLDDVCTDIIHLDAQRLHYYRGNYMTFKKMYQQKQKELLKQYEKQEKKLK ELKAGGKSTKQAEKQTKEALTRKQQKCRRKNQDEESQEAPELLKRPKEYTVRFTFPDPPPLSPPVLGLHGVTFGYQGQKPLFKNLDFGID MDSRICIVGPNGVGKSTLLLLLTGKLTPTHGEMRKNHRLKIGFFNQQYAEQLRMEETPTEYLQRGFNLPYQDARKCLGRFGLESHAHTIQ ICKLSGGQKARVVFAELACREPDVLILDEPTNNLDIESIDALGEAINEYKGAVIVVSHDARLITETNCQLWVVEEQSVSQIDGDFEDYKR -------------------------------------------------------------- >22036_22036_2_DDX52-ABCF1_DDX52_chr17_36002139_ENST00000349699_ABCF1_chr6_30545186_ENST00000376545_length(transcript)=3293nt_BP=330nt AGTCTGCGCAGGCGCCGTAGAACTGTGGCGCTTTCTGGGTAAAGATGGACGTCCACGATCTCTTTCGCCGGCTCGGCGCGGGGGCCAAAT TCGACACGAGACGCTTCTCGGCAGACGCAGCTCGATTCCAGATAGGAAAAAGGAAATATGACTTTGATTCTTCGGAGGTGCTTCAGGGAC TGGACTTTTTTGGAAACAAGAAGTCTGTCCCAGGTGTGTGTGGAGCATCACAAACACATCAGAAGCCCCAAAATGGAGAGAAAAAAGAAG AGAGCCTAACTGAAAGGAAGAGGGAGCAGAGCAAGAAAAAAAGGAAGACGATGACTTCAGACAAAGTGGTGAAGAAAGGGAAGAAGGACA AGAAGATCAAAAAAACGTTCTTTGAAGAGCTGGCAGTAGAAGATAAACAGGCTGGGGAAGAAGAGAAAGTGCTCAAGGAGAAGGAGCAGC AGCAGCAGCAACAGCAACAGCAGCAAAAAAAAAAGCGAGATACCCGAAAAGGCAGGCGGAAGAAGGATGTGGATGATGATGGAGAAGAGA AAGAGCTCATGGAGCGTCTTAAGAAGCTCTCAGTGCCAACCAGTGATGAGGAGGATGAAGTACCCGCCCCAAAACCCCGCGGAGGGAAGA AAACCAAGGGTGGTAATGTTTTTGCAGCCCTGATTCAGGATCAGAGTGAGGAAGAGGAGGAGGAAGAAAAACATCCTCCTAAGCCTGCCA AGCCGGAGAAGAATCGGATCAATAAGGCCGTATCTGAGGAACAGCAGCCTGCACTCAAGGGCAAAAAGGGAAAGGAAGAGAAGTCAAAAG GGAAGGCTAAGCCTCAAAATAAATTCGCTGCTCTGGACAATGAAGAGGAGGATAAAGAAGAAGAAATTATAAAGGAAAAGGAGCCTCCCA AACAAGGGAAGGAGAAGGCCAAGAAGGCAGAGCAGATGGAGTATGAGCGCCAAGTGGCTTCATTAAAAGCAGCCAATGCAGCTGAAAATG ACTTCTCCGTGTCCCAGGCGGAGATGTCCTCCCGCCAAGCCATGTTAGAAAATGCATCTGACATCAAGCTGGAGAAGTTCAGCATCTCCG CTCATGGCAAGGAGCTGTTCGTCAATGCAGACCTGTACATTGTAGCCGGCCGCCGCTACGGGCTGGTAGGACCCAATGGCAAGGGCAAGA CCACACTCCTCAAGCACATTGCCAACCGAGCCCTGAGCATCCCTCCCAACATTGATGTGTTGCTGTGTGAGCAGGAGGTGGTAGCAGATG AGACACCAGCAGTCCAGGCTGTTCTTCGAGCTGACACCAAGCGATTGAAGCTGCTGGAAGAGGAGCGGCGGCTTCAGGGACAGCTGGAAC AAGGGGATGACACAGCTGCTGAGAGGCTAGAGAAGGTGTATGAGGAATTGCGGGCCACTGGGGCGGCAGCTGCAGAGGCCAAAGCACGGC GGATCCTGGCTGGCCTGGGCTTTGACCCTGAAATGCAGAATCGACCCACACAGAAGTTCTCAGGGGGCTGGCGCATGCGTGTCTCCCTGG CCAGGGCACTGTTCATGGAGCCCACACTGCTGATGCTGGATGAGCCCACCAACCACCTGGACCTCAACGCTGTCATCTGGCTTAATAACT ACCTCCAGGGCTGGCGGAAGACCTTGCTGATCGTCTCCCATGACCAGGGCTTCTTGGATGATGTCTGCACTGATATCATCCACCTCGATG CCCAGCGGCTCCACTACTATAGGGGCAATTACATGACCTTCAAAAAGATGTACCAGCAGAAGCAGAAAGAACTGCTGAAACAGTATGAGA AGCAAGAGAAAAAGCTGAAGGAGCTGAAGGCAGGCGGGAAGTCCACCAAGCAGGCGGAAAAACAAACGAAGGAAGCCCTGACTCGGAAGC AGCAGAAATGCCGACGGAAAAACCAAGATGAGGAATCCCAGGAGGCCCCTGAGCTCCTGAAGCGCCCTAAGGAGTACACTGTGCGCTTCA CTTTTCCAGACCCCCCACCACTCAGCCCTCCAGTGCTGGGTCTGCATGGTGTGACATTCGGCTACCAGGGACAGAAACCACTCTTTAAGA ACTTGGATTTTGGCATCGACATGGATTCAAGGATTTGCATTGTGGGCCCTAATGGTGTGGGGAAGAGTACGCTACTCCTGCTGCTGACTG GCAAGCTGACACCGACCCATGGGGAAATGAGAAAGAACCACCGGCTGAAAATTGGCTTCTTCAACCAGCAGTATGCAGAGCAGCTGCGCA TGGAGGAGACGCCCACTGAGTACCTGCAGCGGGGCTTCAACCTGCCCTACCAGGATGCCCGCAAGTGCCTGGGCCGCTTCGGCCTGGAGA GTCACGCCCACACCATCCAGATCTGCAAACTCTCTGGTGGTCAGAAGGCGCGAGTTGTGTTTGCTGAGCTGGCCTGTCGGGAACCTGATG TCCTCATCTTGGACGAGCCAACCAATAACCTGGACATAGAGTCTATTGATGCTCTAGGGGAGGCCATCAATGAATACAAGGGTGCTGTGA TCGTTGTCAGCCATGATGCCCGACTCATCACAGAAACCAATTGCCAGCTGTGGGTGGTGGAGGAGCAGAGTGTTAGCCAAATCGATGGTG ACTTTGAAGACTACAAGCGGGAGGTGTTGGAGGCCCTGGGTGAAGTCATGGTCAGCCGGCCCCGAGAGTGAGCTTTCCTTCCCAGAAGTC TCCCGAGAGACATATTTGTGTGGCCTAGAAGTCCTCTGTGGTCTCCCCTCCTCTGAAGACTGCCTCTGGCCTGCAGCTGACCTGGCAACC ATTCAGGCACATGAAGGTGGAGTGTGACCTTGATGTGACCGGGATCCCACTCTGATTGCATCCATTTCTCTGAAAGACTTGTTTGTTCTG CTTCTCTTCATATAACTGAGCTGGCCTTATCCTTGGCATCCCCCTAAACAAACAAGAGGTGACCACCTTATTGTGAGGTTCCATCCAGCC AAGTTTATGTGGCCTATTGTCTCAGGACTCTCATCACTCAGAAGCCTGCCTCTGATTTACCCTACAGCTTCAGGCCCAGCTGCCCCCCAG TCTTTGGGTGGTGCTGTTCTTTTCTGGTGGATTTAATGCTGACTCACTGGTACAAACAGCTGTTGAAGCTCAGAGCTGGAGGTGAGCTTC TGAGGCCTTTGCCATTATCCAGCCCAAGATTTGGTGCCTGCAGCCTCTTGTCTGGTTGAGGACTTGGGGCAGGAAAGGAATGCTGCTGAA >22036_22036_2_DDX52-ABCF1_DDX52_chr17_36002139_ENST00000349699_ABCF1_chr6_30545186_ENST00000376545_length(amino acids)=878AA_BP=86 MDVHDLFRRLGAGAKFDTRRFSADAARFQIGKRKYDFDSSEVLQGLDFFGNKKSVPGVCGASQTHQKPQNGEKKEESLTERKREQSKKKR KTMTSDKVVKKGKKDKKIKKTFFEELAVEDKQAGEEEKVLKEKEQQQQQQQQQQKKKRDTRKGRRKKDVDDDGEEKELMERLKKLSVPTS DEEDEVPAPKPRGGKKTKGGNVFAALIQDQSEEEEEEEKHPPKPAKPEKNRINKAVSEEQQPALKGKKGKEEKSKGKAKPQNKFAALDNE EEDKEEEIIKEKEPPKQGKEKAKKAEQMEYERQVASLKAANAAENDFSVSQAEMSSRQAMLENASDIKLEKFSISAHGKELFVNADLYIV AGRRYGLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADETPAVQAVLRADTKRLKLLEEERRLQGQLEQGDDTAAERLEKVYE ELRATGAAAAEAKARRILAGLGFDPEMQNRPTQKFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVIWLNNYLQGWRKTLLIVSHD QGFLDDVCTDIIHLDAQRLHYYRGNYMTFKKMYQQKQKELLKQYEKQEKKLKELKAGGKSTKQAEKQTKEALTRKQQKCRRKNQDEESQE APELLKRPKEYTVRFTFPDPPPLSPPVLGLHGVTFGYQGQKPLFKNLDFGIDMDSRICIVGPNGVGKSTLLLLLTGKLTPTHGEMRKNHR LKIGFFNQQYAEQLRMEETPTEYLQRGFNLPYQDARKCLGRFGLESHAHTIQICKLSGGQKARVVFAELACREPDVLILDEPTNNLDIES -------------------------------------------------------------- >22036_22036_3_DDX52-ABCF1_DDX52_chr17_36002139_ENST00000349699_ABCF1_chr6_30545186_ENST00000396515_length(transcript)=1803nt_BP=330nt AGTCTGCGCAGGCGCCGTAGAACTGTGGCGCTTTCTGGGTAAAGATGGACGTCCACGATCTCTTTCGCCGGCTCGGCGCGGGGGCCAAAT TCGACACGAGACGCTTCTCGGCAGACGCAGCTCGATTCCAGATAGGAAAAAGGAAATATGACTTTGATTCTTCGGAGGTGCTTCAGGGAC TGGACTTTTTTGGAAACAAGAAGTCTGTCCCAGGTGTGTGTGGAGCATCACAAACACATCAGAAGCCCCAAAATGGAGAGAAAAAAGAAG AGAGCCTAACTGAAAGGAAGAGGGAGCAGAGCAAGAAAAAAAGGAAGACGATGACTTCAGACAAAGTGGTGAAGAAAGGGAAGAAGGACA AGAAGATCAAAAAAACGTTCTTTGAAGAGCTGGCAGTAGAAGATAAACAGGCTGGGGAAGAAGAGAAAGTGCTCAAGGAGAAGGAGCAGC AGCAGCAGCAACAGCAACAGCAGCAAAAAAAAAAGCGAGATACCCGAAAAGGCAGGCGGAAGAAGGATGTGGATGATGATGGAGAAGAGA AAGAGCTCATGGAGCGTCTTAAGAAGCTCTCACGGGGCTTCAACCTGCCCTACCAGGATGCCCGCAAGTGCCTGGGCCGCTTCGGCCTGG AGAGTCACGCCCACACCATCCAGATCTGCAAACTCTCTGGTGGTCAGAAGGCGCGAGTTGTGTTTGCTGAGCTGGCCTGTCGGGAACCTG ATGTCCTCATCTTGGACGAGCCAACCAATAACCTGGACATAGAGTCTATTGATGCTCTAGGGGAGGCCATCAATGAATACAAGGGTGCTG TGATCGTTGTCAGCCATGATGCCCGACTCATCACAGAAACCAATTGCCAGCTGTGGGTGGTGGAGGAGCAGAGTGTTAGCCAAATCGATG GTGACTTTGAAGACTACAAGCGGGAGGTGTTGGAGGCCCTGGGTGAAGTCATGGTCAGCCGGCCCCGAGAGTGAGCTTTCCTTCCCAGAA GTCTCCCGAGAGACATATTTGTGTGGCCTAGAAGTCCTCTGTGGTCTCCCCTCCTCTGAAGACTGCCTCTGGCCTGCAGCTGACCTGGCA ACCATTCAGGCACATGAAGGTGGAGTGTGACCTTGATGTGACCGGGATCCCACTCTGATTGCATCCATTTCTCTGAAAGACTTGTTTGTT CTGCTTCTCTTCATATAACTGAGCTGGCCTTATCCTTGGCATCCCCCTAAACAAACAAGAGGTGACCACCTTATTGTGAGGTTCCATCCA GCCAAGTTTATGTGGCCTATTGTCTCAGGACTCTCATCACTCAGAAGCCTGCCTCTGATTTACCCTACAGCTTCAGGCCCAGCTGCCCCC CAGTCTTTGGGTGGTGCTGTTCTTTTCTGGTGGATTTAATGCTGACTCACTGGTACAAACAGCTGTTGAAGCTCAGAGCTGGAGGTGAGC TTCTGAGGCCTTTGCCATTATCCAGCCCAAGATTTGGTGCCTGCAGCCTCTTGTCTGGTTGAGGACTTGGGGCAGGAAAGGAATGCTGCT GAACTTGAATTTCCCTTTACAAGGGGAAGAAATAAAGGAAAGGAGTTGCTGCCGACCTGTCACTGTTTGGAGATTGATGGGAGTTGGAAC TGTTCTCAGTCTTGATTTGCTTTATTCAGTTTTCTAGCAGCTTTTAATAGTCCCCTCTTCCCCACTAAATGGATCTTGTTTGCAGTCTTG CTGACAGTGTTTGCTGTTTAAGGATCATAGGATTCCTTTCCCCCAACCCTTCACGCAAGGAAAAAGCAAAGTGATTCATACCTTCTATCT >22036_22036_3_DDX52-ABCF1_DDX52_chr17_36002139_ENST00000349699_ABCF1_chr6_30545186_ENST00000396515_length(amino acids)=309AA_BP=86 MDVHDLFRRLGAGAKFDTRRFSADAARFQIGKRKYDFDSSEVLQGLDFFGNKKSVPGVCGASQTHQKPQNGEKKEESLTERKREQSKKKR KTMTSDKVVKKGKKDKKIKKTFFEELAVEDKQAGEEEKVLKEKEQQQQQQQQQQKKKRDTRKGRRKKDVDDDGEEKELMERLKKLSRGFN LPYQDARKCLGRFGLESHAHTIQICKLSGGQKARVVFAELACREPDVLILDEPTNNLDIESIDALGEAINEYKGAVIVVSHDARLITETN -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for DDX52-ABCF1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for DDX52-ABCF1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for DDX52-ABCF1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
