
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ADCY3-HADHA (FusionGDB2 ID:2228) |
Fusion Gene Summary for ADCY3-HADHA |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ADCY3-HADHA | Fusion gene ID: 2228 | Hgene | Tgene | Gene symbol | ADCY3 | HADHA | Gene ID | 114 | 3030 |
| Gene name | adenylate cyclase 8 | hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha | |
| Synonyms | AC8|ADCY3|HBAC1 | ECHA|GBP|HADH|LCEH|LCHAD|MTPA|TP-ALPHA | |
| Cytomap | 8q24.22 | 2p23.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | adenylate cyclase type 8ATP pyrophosphate-lyase 8HEL-S-172mPadenylate cyclase 8 (brain)adenylate cyclase type VIIIadenylyl cyclase 8adenylyl cyclase-8, brainca(2+)/calmodulin-activated adenylyl cyclaseepididymis secretory sperm binding protein Li | trifunctional enzyme subunit alpha, mitochondrial3-ketoacyl-Coenzyme A (CoA) thiolase, alpha subunit3-oxoacyl-CoA thiolase78 kDa gastrin-binding proteingastrin-binding proteinhydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase ( | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | O60266 | P40939 | |
| Ensembl transtripts involved in fusion gene | ENST00000260600, ENST00000405392, ENST00000450524, | ENST00000457468, ENST00000461025, ENST00000380649, | |
| Fusion gene scores | * DoF score | 7 X 6 X 5=210 | 7 X 6 X 7=294 |
| # samples | 7 | 9 | |
| ** MAII score | log2(7/210*10)=-1.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/294*10)=-1.70781924850669 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ADCY3 [Title/Abstract] AND HADHA [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ADCY3(25141182)-HADHA(26417507), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | HADHA | GO:0035965 | cardiolipin acyl-chain remodeling | 23152787 |
 Fusion gene breakpoints across ADCY3 (5'-gene) Fusion gene breakpoints across ADCY3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
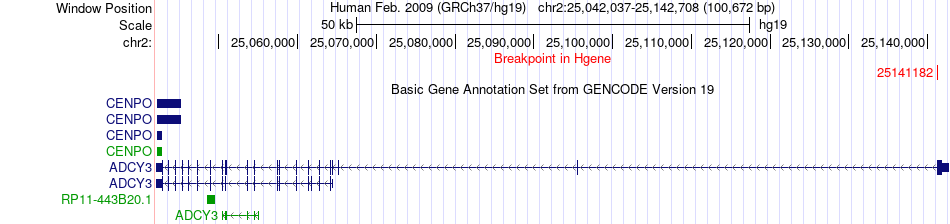 |
 Fusion gene breakpoints across HADHA (3'-gene) Fusion gene breakpoints across HADHA (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
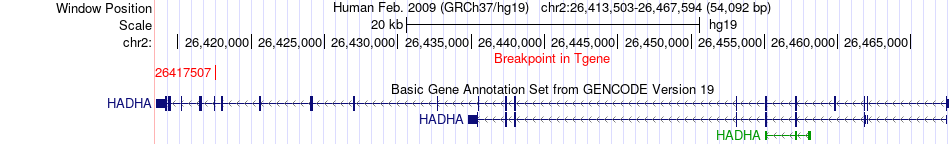 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUSC | TCGA-43-3394-01A | ADCY3 | chr2 | 25141182 | - | HADHA | chr2 | 26417507 | - |
Top |
Fusion Gene ORF analysis for ADCY3-HADHA |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000260600 | ENST00000457468 | ADCY3 | chr2 | 25141182 | - | HADHA | chr2 | 26417507 | - |
| 5CDS-intron | ENST00000260600 | ENST00000461025 | ADCY3 | chr2 | 25141182 | - | HADHA | chr2 | 26417507 | - |
| In-frame | ENST00000260600 | ENST00000380649 | ADCY3 | chr2 | 25141182 | - | HADHA | chr2 | 26417507 | - |
| intron-3CDS | ENST00000405392 | ENST00000380649 | ADCY3 | chr2 | 25141182 | - | HADHA | chr2 | 26417507 | - |
| intron-3CDS | ENST00000450524 | ENST00000380649 | ADCY3 | chr2 | 25141182 | - | HADHA | chr2 | 26417507 | - |
| intron-intron | ENST00000405392 | ENST00000457468 | ADCY3 | chr2 | 25141182 | - | HADHA | chr2 | 26417507 | - |
| intron-intron | ENST00000405392 | ENST00000461025 | ADCY3 | chr2 | 25141182 | - | HADHA | chr2 | 26417507 | - |
| intron-intron | ENST00000450524 | ENST00000457468 | ADCY3 | chr2 | 25141182 | - | HADHA | chr2 | 26417507 | - |
| intron-intron | ENST00000450524 | ENST00000461025 | ADCY3 | chr2 | 25141182 | - | HADHA | chr2 | 26417507 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000260600 | ADCY3 | chr2 | 25141182 | - | ENST00000380649 | HADHA | chr2 | 26417507 | - | 2814 | 1527 | 852 | 2198 | 448 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000260600 | ENST00000380649 | ADCY3 | chr2 | 25141182 | - | HADHA | chr2 | 26417507 | - | 0.002141016 | 0.99785894 |
Top |
Fusion Genomic Features for ADCY3-HADHA |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
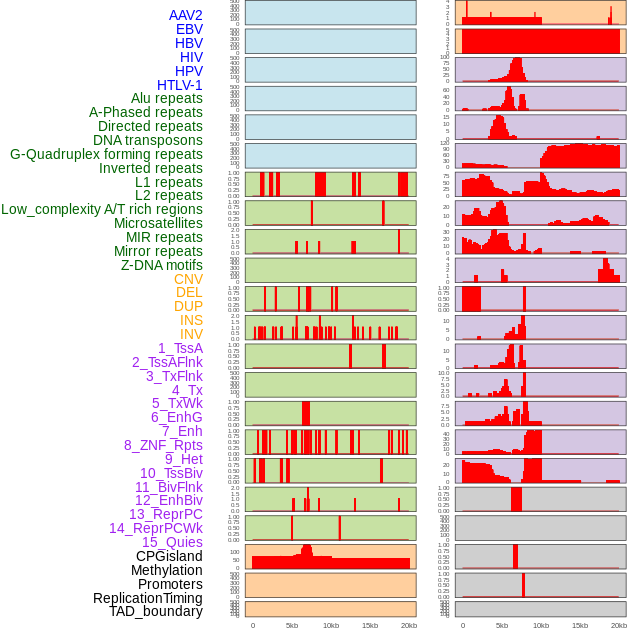 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ADCY3-HADHA |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:25141182/chr2:26417507) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ADCY3 | HADHA |
| FUNCTION: Catalyzes the formation of the signaling molecule cAMP in response to G-protein signaling. Participates in signaling cascades triggered by odorant receptors via its function in cAMP biosynthesis. Required for the perception of odorants. Required for normal sperm motility and normal male fertility. Plays a role in regulating insulin levels and body fat accumulation in response to a high fat diet. {ECO:0000250|UniProtKB:Q8VHH7}. | FUNCTION: Mitochondrial trifunctional enzyme catalyzes the last three of the four reactions of the mitochondrial beta-oxidation pathway (PubMed:8135828, PubMed:1550553, PubMed:29915090, PubMed:30850536). The mitochondrial beta-oxidation pathway is the major energy-producing process in tissues and is performed through four consecutive reactions breaking down fatty acids into acetyl-CoA (PubMed:29915090). Among the enzymes involved in this pathway, the trifunctional enzyme exhibits specificity for long-chain fatty acids (PubMed:30850536). Mitochondrial trifunctional enzyme is a heterotetrameric complex composed of two proteins, the trifunctional enzyme subunit alpha/HADHA described here carries the 2,3-enoyl-CoA hydratase and the 3-hydroxyacyl-CoA dehydrogenase activities while the trifunctional enzyme subunit beta/HADHB bears the 3-ketoacyl-CoA thiolase activity (PubMed:8135828, PubMed:29915090, PubMed:30850536). Independently of the subunit beta, the trifunctional enzyme subunit alpha/HADHA also has a monolysocardiolipin acyltransferase activity (PubMed:23152787). It acylates monolysocardiolipin into cardiolipin, a major mitochondrial membrane phospholipid which plays a key role in apoptosis and supports mitochondrial respiratory chain complexes in the generation of ATP (PubMed:23152787). Allows the acylation of monolysocardiolipin with different acyl-CoA substrates including oleoyl-CoA for which it displays the highest activity (PubMed:23152787). {ECO:0000269|PubMed:1550553, ECO:0000269|PubMed:23152787, ECO:0000269|PubMed:29915090, ECO:0000269|PubMed:30850536, ECO:0000269|PubMed:8135828, ECO:0000303|PubMed:29915090, ECO:0000303|PubMed:30850536}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 1_79 | 225 | 1145.0 | Topological domain | Cytoplasmic |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 105_125 | 225 | 1145.0 | Transmembrane | Helical |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 139_159 | 225 | 1145.0 | Transmembrane | Helical |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 173_193 | 225 | 1145.0 | Transmembrane | Helical |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 80_100 | 225 | 1145.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 1062_1064 | 225 | 1145.0 | Nucleotide binding | ATP |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 1069_1073 | 225 | 1145.0 | Nucleotide binding | ATP |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 324_329 | 225 | 1145.0 | Nucleotide binding | ATP |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 366_368 | 225 | 1145.0 | Nucleotide binding | ATP |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 402_632 | 225 | 1145.0 | Topological domain | Cytoplasmic |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 854_1144 | 225 | 1145.0 | Topological domain | Cytoplasmic |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 226_246 | 225 | 1145.0 | Transmembrane | Helical |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 381_401 | 225 | 1145.0 | Transmembrane | Helical |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 633_653 | 225 | 1145.0 | Transmembrane | Helical |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 664_684 | 225 | 1145.0 | Transmembrane | Helical |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 708_728 | 225 | 1145.0 | Transmembrane | Helical |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 754_774 | 225 | 1145.0 | Transmembrane | Helical |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 775_795 | 225 | 1145.0 | Transmembrane | Helical |
| Hgene | ADCY3 | chr2:25141182 | chr2:26417507 | ENST00000260600 | - | 1 | 21 | 833_853 | 225 | 1145.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for ADCY3-HADHA |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >2228_2228_1_ADCY3-HADHA_ADCY3_chr2_25141182_ENST00000260600_HADHA_chr2_26417507_ENST00000380649_length(transcript)=2814nt_BP=1527nt CGGGCCTGACGGCGGGAGATCGCGCTCCGGCCGGGCCCCGAGGCGCTGCCGGGGCGGCGCGGCTCTGCGGGGAGCAGCCCTCGGGCGCGC CGAGCTCTCCCGCCCCAGCCCGCGCGGGGACCGTCCCGGAGCACGCGGTGGCCGAGTTCCCGCACAGGTAAACGCAGGGGCAGGGCCAGG CTTCCGGGGGGGAAGACAGCGCCCCGAGCGTGGGAAGGGGGCCGGATCGGAGCCCGAGCCTCCAAGGGTTTGGTGGAAAAAGGAAGTAAA CAGAAGGAAGGCTCAGGAATCCCCCACGATGGTCCTGATTTCTTTCCGGCACATTTTATTTACAGCTTCTAAGGACTGGGCCTGGAGGGA ATTTAGGTGAGGAAGTCGGGGACTGAGAAAGAAGAGATGACAGAGAGAAACCGGTTACCAGGGATGGAAAGGCGGCTAGGCCCTTCCACA AGCCAGGGACAGGAAATTGAGTGTCTGGCGTGGACAGGACTAAGTGTGCCTGCGGTGGGCGGTGGTGTGAGCCGGGGGTGGCAGGGAAGG CCCTGGGTTGGGAGGCTCTTGCAGCAGAGATCCTTTTATGAGGGTTAGGGCGGGTCATGGGGAAACCGCAAGGTAGGAGGGAAGAGGTGC TTCTACTCAGCAATGCCCTTTTCAGTTCTAGCTGATCAGTGCTACCTGTGCTCTGGAAACCCGCTCTGCGTTCCTGCTGGAGGTGGCCTC CCCTCCGCCCCAGACAAGAAGAGGCCCTCAGCCCTCCCCCGGTCTCAGAGAGCCCTGAGAGGAGGCCCAGTCCAGAGCTCTTCCTCCGTT CCCAGTCCACTTCTCTAGGGCCAGTAGCAGACACCAGCCAGTATGCCGAGGAACCAGGGCTTCTCCGAGCCCGAATACTCGGCCGAGTAC TCAGCCGAGTACTCCGTCAGCCTGCCCTCCGACCCTGACCGCGGGGTGGGCCGGACCCATGAAATCTCGGTCCGGAACTCGGGCTCCTGC CTGTGCCTGCCTCGCTTCATGCGGCTGACTTTCGTGCCGGAGTCCTTGGAGAACCTCTACCAGACCTACTTCAAAAGGCAGCGCCACGAG ACCCTGCTGGTGCTGGTGGTCTTTGCAGCCCTCTTTGACTGCTACGTGGTGGTCATGTGTGCTGTGGTCTTCTCCAGCGACAAGCTGGCT TCCCTCGCCGTGGCTGGAATTGGACTGGTGTTGGACATCATCCTCTTCGTGCTCTGCAAAAAGGGGCTGCTCCCGGACCGGGTCACCCGC AGAGTGCTGCCCTACGTGCTGTGGCTGCTCATAACCGCCCAGATCTTCTCCTACCTGGGCCTGAACTTCGCGCGTGCCCACGCGGCTAGT GACACGGTGGGCTGGCAGGTCTTCTTTGTCTTCTCCTTCTTCATCACGCTGCCCCTCAGCCTCAGCCCCATCGTGATCATCTCCGTGGTC TCCTGTGTGGTGCACACGTTGGTCCTGGGGGTCACCGTGGCCCAGCAGCAGCAGGAGGAGCTCAAGGGGATGCAGCTGCTGCGGGAGGAT GGACCTGGCTTCTATACTACCAGGTGTCTTGCGCCCATGATGTCTGAAGTCATCCGAATCCTCCAGGAAGGAGTTGACCCGAAGAAGCTG GATTCCCTGACCACAAGCTTTGGCTTTCCTGTGGGTGCCGCCACACTGGTGGATGAAGTTGGTGTGGATGTAGCGAAACATGTGGCGGAA GATCTGGGCAAAGTCTTTGGGGAGCGGTTTGGAGGTGGAAACCCAGAACTGCTGACACAGATGGTGTCCAAGGGCTTCCTAGGTCGTAAA TCTGGGAAGGGCTTTTACATCTATCAGGAGGGTGTGAAGAGGAAGGATTTGAATTCTGACATGGATAGTATTTTAGCGAGTCTGAAGCTG CCTCCTAAGTCTGAAGTCTCATCAGACGAAGACATCCAGTTCCGCCTGGTGACAAGATTTGTGAATGAGGCAGTCATGTGCCTGCAAGAG GGGATCTTGGCCACACCTGCAGAGGGAGACATCGGAGCCGTCTTTGGGCTTGGCTTCCCGCCTTGTCTGGGAGGGCCTTTCCGCTTTGTG GATCTGTATGGCGCCCAGAAGATAGTGGACCGGCTCAAGAAATATGAAGCTGCCTATGGAAAACAGTTCACCCCATGCCAGCTGCTAGCT GACCATGCTAACAGCCCTAACAAGAAGTTCTACCAGTGAGCAGGCCTCATGCCTCGCTCAGTCAGTGCACTAACCCCAGCTGCCGGCAGT GCTGGTTCTCCAACAGAGTGGTGTCTAGATTTATCAGAGTAACGAGAAGACAAACTCCGGCACTGGGTTTGCTCCCTGATTAAAGTGCCT TCAGCCAAGACCATCTCTCCCTCCTGGTGAAGTGTGACTTCGAATTAGTTTGCACTTCCTGTTGGAAGGTAGAGCCCACTGCTCATTGTA TAAGCCCCGAGGCCTAGAGTGGCAGCCAAGAGCCATCTGAAGCCACCTCTCTGCCTGTTCCTCCCAAGAGGCCAGGGTGGCCAGGGGTGG TGAGGGCAGTTCTGCACCCAGCCAAACACATAACAATAAAAACCAAACTCTGTGTCAGCATCTTTGCCCTTCTGGTTTAAACGCCTCCTT CAAAAAGCAATCTGGAAGAAAGCCCTGTGCTTTGGGGGAGTAAGAATGTGTGTGCAGAATTCTAGGCAGCACCTTAGGGAGGGACTGGGA TGAGAGAAAGTGGGACCTGGTGGGCTCAACCACACACACCTGTCTGTGCAGATGCTTTGCCCAGGCTTCTCACCACGGTGTACCGGGATA >2228_2228_1_ADCY3-HADHA_ADCY3_chr2_25141182_ENST00000260600_HADHA_chr2_26417507_ENST00000380649_length(amino acids)=448AA_BP=225 MPRNQGFSEPEYSAEYSAEYSVSLPSDPDRGVGRTHEISVRNSGSCLCLPRFMRLTFVPESLENLYQTYFKRQRHETLLVLVVFAALFDC YVVVMCAVVFSSDKLASLAVAGIGLVLDIILFVLCKKGLLPDRVTRRVLPYVLWLLITAQIFSYLGLNFARAHAASDTVGWQVFFVFSFF ITLPLSLSPIVIISVVSCVVHTLVLGVTVAQQQQEELKGMQLLREDGPGFYTTRCLAPMMSEVIRILQEGVDPKKLDSLTTSFGFPVGAA TLVDEVGVDVAKHVAEDLGKVFGERFGGGNPELLTQMVSKGFLGRKSGKGFYIYQEGVKRKDLNSDMDSILASLKLPPKSEVSSDEDIQF -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ADCY3-HADHA |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ADCY3-HADHA |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ADCY3-HADHA |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
