
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:DGKD-NDUFA10 (FusionGDB2 ID:22450) |
Fusion Gene Summary for DGKD-NDUFA10 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: DGKD-NDUFA10 | Fusion gene ID: 22450 | Hgene | Tgene | Gene symbol | DGKD | NDUFA10 | Gene ID | 8527 | 4705 |
| Gene name | diacylglycerol kinase delta | NADH:ubiquinone oxidoreductase subunit A10 | |
| Synonyms | DGK-delta|DGKdelta|dgkd-2 | CI-42KD|CI-42k|MC1DN22 | |
| Cytomap | 2q37.1 | 2q37.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | diacylglycerol kinase deltaDAG kinase deltadiacylglycerol kinase, delta 130kDadiglyceride kinase delta | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrialNADH-ubiquinone oxidoreductase 42 kDa subunitcomplex I 42kDa subunit | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q16760 | O95299 | |
| Ensembl transtripts involved in fusion gene | ENST00000264057, ENST00000409813, ENST00000489613, | ENST00000307300, ENST00000404554, ENST00000407129, ENST00000471378, ENST00000252711, | |
| Fusion gene scores | * DoF score | 21 X 6 X 10=1260 | 9 X 7 X 4=252 |
| # samples | 23 | 9 | |
| ** MAII score | log2(23/1260*10)=-2.4537179674429 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/252*10)=-1.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: DGKD [Title/Abstract] AND NDUFA10 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | DGKD(234299129)-NDUFA10(240961757), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | DGKD | GO:0010033 | response to organic substance | 12200442 |
 Fusion gene breakpoints across DGKD (5'-gene) Fusion gene breakpoints across DGKD (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
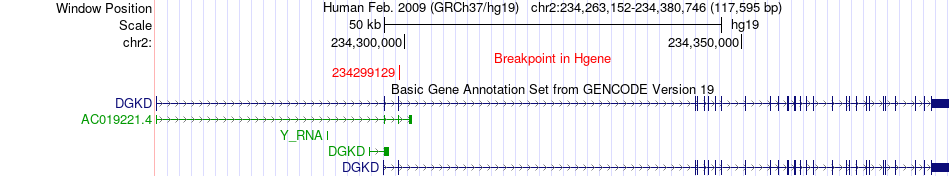 |
 Fusion gene breakpoints across NDUFA10 (3'-gene) Fusion gene breakpoints across NDUFA10 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
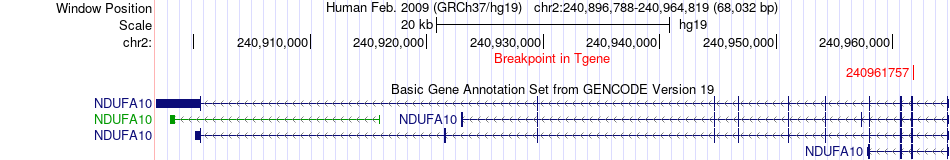 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-DX-A3LY-01B | DGKD | chr2 | 234299129 | - | NDUFA10 | chr2 | 240961757 | - |
| ChimerDB4 | SARC | TCGA-DX-A3LY-01B | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - |
Top |
Fusion Gene ORF analysis for DGKD-NDUFA10 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000264057 | ENST00000307300 | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - |
| 5CDS-intron | ENST00000264057 | ENST00000404554 | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - |
| 5CDS-intron | ENST00000264057 | ENST00000407129 | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - |
| 5CDS-intron | ENST00000264057 | ENST00000471378 | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - |
| 5CDS-intron | ENST00000409813 | ENST00000307300 | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - |
| 5CDS-intron | ENST00000409813 | ENST00000404554 | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - |
| 5CDS-intron | ENST00000409813 | ENST00000407129 | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - |
| 5CDS-intron | ENST00000409813 | ENST00000471378 | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - |
| In-frame | ENST00000264057 | ENST00000252711 | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - |
| In-frame | ENST00000409813 | ENST00000252711 | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - |
| intron-3CDS | ENST00000489613 | ENST00000252711 | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - |
| intron-intron | ENST00000489613 | ENST00000307300 | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - |
| intron-intron | ENST00000489613 | ENST00000404554 | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - |
| intron-intron | ENST00000489613 | ENST00000407129 | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - |
| intron-intron | ENST00000489613 | ENST00000471378 | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000264057 | DGKD | chr2 | 234299129 | + | ENST00000252711 | NDUFA10 | chr2 | 240961757 | - | 5099 | 360 | 12 | 1352 | 446 |
| ENST00000409813 | DGKD | chr2 | 234299129 | + | ENST00000252711 | NDUFA10 | chr2 | 240961757 | - | 5034 | 295 | 79 | 1287 | 402 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000264057 | ENST00000252711 | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - | 0.000290365 | 0.99970967 |
| ENST00000409813 | ENST00000252711 | DGKD | chr2 | 234299129 | + | NDUFA10 | chr2 | 240961757 | - | 0.000139563 | 0.9998604 |
Top |
Fusion Genomic Features for DGKD-NDUFA10 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
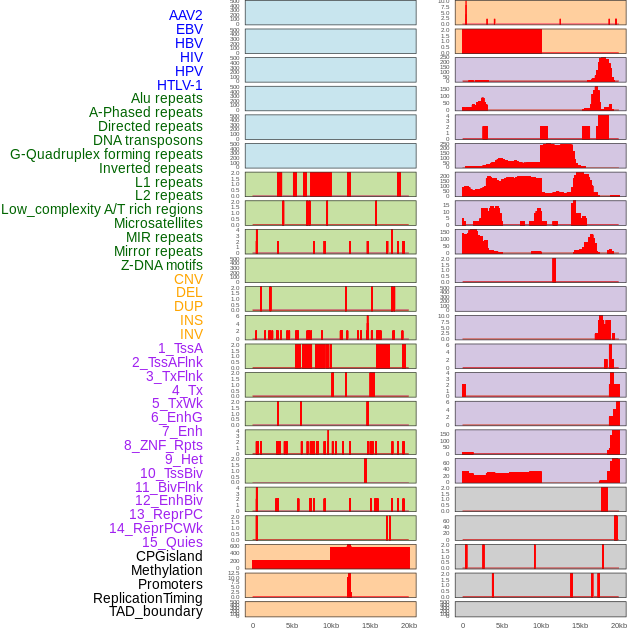 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for DGKD-NDUFA10 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:234299129/chr2:240961757) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| DGKD | NDUFA10 |
| FUNCTION: Diacylglycerol kinase that converts diacylglycerol/DAG into phosphatidic acid/phosphatidate/PA and regulates the respective levels of these two bioactive lipids (PubMed:12200442, PubMed:23949095). Thereby, acts as a central switch between the signaling pathways activated by these second messengers with different cellular targets and opposite effects in numerous biological processes (Probable). By controlling the levels of diacylglycerol, regulates for instance the PKC and EGF receptor signaling pathways and plays a crucial role during development (By similarity). May also regulate clathrin-dependent endocytosis (PubMed:17880279). {ECO:0000250|UniProtKB:E9PUQ8, ECO:0000269|PubMed:12200442, ECO:0000269|PubMed:17880279, ECO:0000269|PubMed:23949095, ECO:0000305}. | FUNCTION: Accessory subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I), that is believed not to be involved in catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone. {ECO:0000269|PubMed:27626371}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DGKD | chr2:234299129 | chr2:240961757 | ENST00000264057 | + | 3 | 30 | 8_36 | 116 | 1215.0 | Compositional bias | Note=Pro-rich |
| Hgene | DGKD | chr2:234299129 | chr2:240961757 | ENST00000409813 | + | 2 | 29 | 8_36 | 72 | 1171.0 | Compositional bias | Note=Pro-rich |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DGKD | chr2:234299129 | chr2:240961757 | ENST00000264057 | + | 3 | 30 | 1145_1208 | 116 | 1215.0 | Domain | SAM |
| Hgene | DGKD | chr2:234299129 | chr2:240961757 | ENST00000264057 | + | 3 | 30 | 317_451 | 116 | 1215.0 | Domain | DAGKc |
| Hgene | DGKD | chr2:234299129 | chr2:240961757 | ENST00000264057 | + | 3 | 30 | 53_146 | 116 | 1215.0 | Domain | PH |
| Hgene | DGKD | chr2:234299129 | chr2:240961757 | ENST00000409813 | + | 2 | 29 | 1145_1208 | 72 | 1171.0 | Domain | SAM |
| Hgene | DGKD | chr2:234299129 | chr2:240961757 | ENST00000409813 | + | 2 | 29 | 317_451 | 72 | 1171.0 | Domain | DAGKc |
| Hgene | DGKD | chr2:234299129 | chr2:240961757 | ENST00000409813 | + | 2 | 29 | 53_146 | 72 | 1171.0 | Domain | PH |
| Hgene | DGKD | chr2:234299129 | chr2:240961757 | ENST00000264057 | + | 3 | 30 | 163_213 | 116 | 1215.0 | Zinc finger | Phorbol-ester/DAG-type 1 |
| Hgene | DGKD | chr2:234299129 | chr2:240961757 | ENST00000264057 | + | 3 | 30 | 235_286 | 116 | 1215.0 | Zinc finger | Phorbol-ester/DAG-type 2 |
| Hgene | DGKD | chr2:234299129 | chr2:240961757 | ENST00000409813 | + | 2 | 29 | 163_213 | 72 | 1171.0 | Zinc finger | Phorbol-ester/DAG-type 1 |
| Hgene | DGKD | chr2:234299129 | chr2:240961757 | ENST00000409813 | + | 2 | 29 | 235_286 | 72 | 1171.0 | Zinc finger | Phorbol-ester/DAG-type 2 |
Top |
Fusion Gene Sequence for DGKD-NDUFA10 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >22450_22450_1_DGKD-NDUFA10_DGKD_chr2_234299129_ENST00000264057_NDUFA10_chr2_240961757_ENST00000252711_length(transcript)=5099nt_BP=360nt TGGCCCGGCAGCATGGCGGCGGCGGCGGGCGCCCCTCCGCCGGGTCCCCCGCAACCGCCTCCGCCGCCGCCGCCCGAGGAGTCGTCCGAC AGCGAGCCCGAGGCGGAGCCCGGCTCCCCACAGAAGCTCATCCGCAAGGTGTCCACGTCGGGTCAGATCCGACAGAAGACCATCATCAAA GAGGGGATGCTGACCAAACAGAACAATTCATTCCAGCGATCAAAAAGGAGATACTTTAAGCTTCGAGGGCGAACGCTTTACTATGCCAAA ACGGCAAAGTCAATCATATTTGATGAGGTGGATCTGACAGATGCCAGCGTAGCTGAATCCAGTACCAAAAACGTCAACAACAGTTTTACG AGAGGAATTCATAGCAGTGTGCAGTGCAAACTGCGCTATGGAATGTGGCATTTCCTACTTGGGGATAAAGCAAGCAAAAGACTGACAGAA CGCAGCAGAGTGATAACTGTAGATGGCAATATATGTACTGGAAAAGGCAAACTTGCAAAAGAAATAGCAGAGAAACTAGGCTTCAAGCAC TTTCCTGAAGCGGGGATTCATTATCCAGACAGTACCACAGGAGATGGGAAGCCCCTCGCCACCGACTATAATGGCAACTGTAGTTTGGAG AAATTTTACGATGATCCGAGAAGCAATGATGGCAACAGTTACCGCCTGCAGTCCTGGTTGTACAGCAGTCGCCTGCTGCAGTACTCAGAT GCCTTGGAGCACTTGCTGACCACAGGACAAGGTGTTGTGTTGGAGCGCTCCATCTTCAGTGACTTTGTGTTCCTGGAGGCGATGTACAAC CAGGGATTCATCCGAAAGCAGTGTGTGGACCACTACAACGAGGTGAAGAGCGTCACCATCTGCGATTACCTGCCCCCCCACCTGGTGATT TACATCGATGTGCCCGTTCCAGAGGTCCAGAGGCGGATTCAGAAGAAAGGAGATCCACATGAAATGAAGATCACCTCTGCCTATCTACAG GACATTGAGAATGCCTATAAGAAAACCTTTCTCCCTGAGATGAGTGAAAAATGTGAGGTTTTACAATATTCTGCAAGGGAAGCTCAAGAT TCAAAAAAGGTGGTAGAGGACATTGAATACCTGAAGTTCGATAAAGGGCCGTGGCTCAAGCAGGACAATCGCACTTTATACCACCTGCGA TTACTGGTTCAGGATAAGTTTGAGGTGCTGAATTACACAAGCATTCCTATCTTTCTCCCGGAAGTCACCATTGGAGCTCATCAGACTGAC CGTGTCTTACATCAGTTCAGAGAGCTGCCGGGCCGCAAGTACAGCCCTGGGTACAACACCGAGGTGGGAGACAAGTGGATCTGGCTGAAG TGAACGGGCCGCCTTCTGCTCCAGCTGCATCACAGTGATGGCCAAGCTGCATCAGCCGCACTCTCCTGGACGCCATATAGCTTTAAGATC GGGGGAGGGTAAATAATGCAAAAATTGCACAGTGGAAGAAGGGGTCTCACAAAAAGCAATCCATCCTGTAGTATAGGTAATGGAGTTGGG GGAAGCAGCTTCCATTCTGGATGTTTGGAACCCTTTAGCTTTGTTTTGGAATGGCCCACCATTCTCACTGGAAAACAGTGGTCTGCTGTG AAAGGCCAGCTCTCGGCAGCCCCTGTGGTTTCAGCGCTGCCGCTCTGTGTCATTCAGGTTGTGCACATTGTTTTTCTTCTGACTTCCAGA AATAAAAGTGTTTCCATGGGAATTTCCCCATCGTACGTCTATGTGCGTGCGGGCACGTCGAAACTGCACACGTGCACACACGCTGCTCTG GGCCCTGTTTGGTGTGGTGTGGTTTAATTGAGGCTTACATTATGCTGAAGAGTTTTTCTAAAGGCCTACTGTATTTATTTTAGTAATGTG CTACATTAGAGTAGAAAAAATAAGAACGTGAAGAAGAAATAGCAAAAGGAGAAAATTACCACAGATTTACAGAACAATATGTTTTAAGAG AGACGTTGAGAAAGATTCACGTGGCCGTCCTCAGCGTCTCAAACTCCTTTGAAGGGAGATTTGAGCTTAGGCTGTTGATGACATAAGATG CAAAAGCAAAATAATATACTCTCTTTATTACCCAGAAAATCAAGTTGCCAGTCAAAACTGAGAAATGAGAAAGAAAATCAGATTTCTTTA TTCATCTCTGTTGAAGCCTAGAAAACCAAAACAAGCTGGTTGTGGTTCAGCATCATGGTTTAGGAATCAAGCTGTCAGTTGTGAGTCCTA CTGAACTGTGGGTTTTCCACTGTAAAGCTCCACTGTGTCCATTAGGAATCAGGCTGCCAGTTGTGAGTCCTACTGAACTGTGGGTTTTCC ACTGTAAAGCTCCACTGTGTCCAATATGGTAGCTCCTAGCCACATGTGGGTATTGAAATTTAAATGAGCTAAAATGTAATAGAGTTTAAA ATTGGGTTCCTTGGTTGCACTGGCCATAGTTCAGGCACTCCACAGCCGCCTGTGGGCAGTAGTTCCAGTGCTGGTGCAGAGGACGATGCT CTGCTCCCCGCGGAAGCTCTACCAGGCCTCGCTGCTGCGGAGCATGCGAAGAAGATGCACACTCCTTCCCTCCTTGCGTCCGCCTGCCTT TTTCCCTCTCCCCTTCCTTCCCTCCTTCTTTCCTCCTCCTTCCCTCCCTTCCTTTCTTCCTTCCATCTGTCCTTGTTTTCCTCCCTTCCC TTCCTCCCTGCCTCTTTCCTTCCCTCCTTCTCTTCCTTCCTTCCGTCTGTCCTTGTTTTCCTCCCTGCCTCTTTCCTTCCCTCCTTCTCT TCCTTCCTTCCTTCCGTCTGTCCTTGTTTTCCTCCCTTCCCTTCCTCCCTGCCTCCTTCCTTGTTTTAAGAATTTGAAAATAAAGTTTCC CAGGAGTTTTGAATTGGAGTCGAGATTTAAGTGGCTGAAGTTTTGCATTTCACTAGGAAACCAACAGCTTTAATCATGAAATGAAAGCAC AGCTGGGCATTGGGCCAGTCACAGAGTGTCTTTTGCTCTGAGCACTTCACAAACCCATTGTGGAGGGGAGGAAAGAAAAAGAAAGCCATC TTTAATTATCTTAGCTGTTCCTAATGTCTTTGGCTGCAAAGAGCTAGCTGTTTCCTTCAACCTTGGTGGTTGCACGGGTAGTGTCCTCAG CAGTGAGAGGGCAGGTCTGCCCTGTGGTCAGGATTGCCCTTGTGAGGGGCAGAGCTCTCCAACCATGCAGGGCTCCGTGTCGGCATGGTG TGGACTCGGGTGCTCTGATCATCCCCTGTGTTTGCGTCTGACCCTGTGTGGTTGCATTCCCTTGATGGAGTGGGAAGTATGTAATTTGTA GTGCTGAGGACATTCGTCTGTTTGATTTTCTGGTGTTTTCAATGACATGCATGTTCAGCAGAGAAAGATGAATAAACACTGTCCAATTCT TCTCTCCACATAGAAGCCTCAAGAAGAGGTGAGGAGATGCCCCGATTTCCTTATTTCAGAGCAGTACATGCGTTCGTCTCACGAAAGCTC TTACAACTAAAACAACAGCACCAACAAACCAGGATCAAAATAATCGTGCATTTTCCAGGATTCCATGTTGGACTTCTCAGTGCTTTGGAG ATCAGGAAATAATCATGCTGTTGCAGCAGTATGGTTTTGTGTCTGGATTTTATTCATACAGGCATGTGTTTACAGGCATGAGAATTCTGT ATTGAAAAGAAAGTCCTTGTTTTTTTCTAACGTATCTTTCTGTTCCATCAACTGTTAAAACCAAATTGTTAAGAACATTGAGCTTCTTAG GAGAGATTACATGCAGCTACTTAGGACCACCTCAGCAAAATGGAGTTGCCTGGGTAGATAGCAGCAGCAGCTCACCAGCCCCACGCTGCC AGGCTGCTGGCTGCTGTCCACCAGGCAGCTGTGGAACCTCCATGTTCTGCCTAGGTGTTGTGGCAAGAACAGCAGGGAAACAGGCGGCCG CCTCCAGGCCACAGGTGGACCCAGGGACCTTCAGTTACGCCGCTTCAATTGAATTCGTGGTGGTTTCTCTGCTACAGCAGAAGTGGAAGA AGCTCTTGTCTTGTTTGGCATTTGTCCCATTTTTTGGCTAATAATTGGCTTTGAATTTCTCTGTGCCCTGCTTACCAATTATAGCTTTCT GGATTGGCTGTTCTTAGCAGTGCAAATGTCAATGCAAAGAAGCAAGCAGGCGGCTACTTTCTGGGAAGAAGCATAGTCCGTGCTCACACT GGATATAAATCTGCAAGATTTGGGGGTTTGGCAAAATAGGGGTGGAGGGAAAGCTGCCATGGCTGTCAGAAATTGGGGCTTTTTGAAGAA AAGCTTGTGTAGGCATCAGCCACTGGCTGGCCAGAATTCCATGAATGGAAGCAGGTGCCCGAGTGATCTGGGGCTTTACATGTGGATATA AATAGAAGCCGGGAGCCTGAGGCTTTGCTTTAGGAAAATCATGAGCCAGAAAACAGAGGAGGTGATTGTCAGTAGTCACCTCACGCCGCT AGTGTTTTCTAATACTCTCTCGAATTGTCAAAGCAAAGGACCTTATAACAGCAACACGTGTGTGACTCGAAGCAGCGAGCTTCCCTTGGG AAAAGGAGCCTCTTCTCAGCAGAGGGTCCCAGCCTTTCTGTCCTTCCCCATCGGAATGCCAGGCTCTGTGAGTTTGGATGTTGGACATTG CCACCTGAATTCCAGGAGGCCGGGGAGCTCCAGTGATGGACTTGTCCCCAAAAGAGTGGTGCTGTTCTCAGAGCCATAAATGTTCCACTG AAAGTGTCTGTCCCCTGGCTCACGCTGGCTGCGCTTGCTTGAAGGAGGCTGATTTAAGGGTTCTAATAGGAAGACAAATGAAACTGTCAA ATTTTATCTTTAAGCATGTGAGTAAGTTTGCTCATTCGCTTTTACTATTGATGTATAGTTTTAAAATTCTTAATTTAAAAATTATTTTAA TAGTAAATTTTTCACAAGTGAACACTTTTTATCTTTAAATTAAAATTCTGAAGCTCCCTAGTTAGTTAGATCCAATTGCTGGTTACATTT >22450_22450_1_DGKD-NDUFA10_DGKD_chr2_234299129_ENST00000264057_NDUFA10_chr2_240961757_ENST00000252711_length(amino acids)=446AA_BP=114 MAAAAGAPPPGPPQPPPPPPPEESSDSEPEAEPGSPQKLIRKVSTSGQIRQKTIIKEGMLTKQNNSFQRSKRRYFKLRGRTLYYAKTAKS IIFDEVDLTDASVAESSTKNVNNSFTRGIHSSVQCKLRYGMWHFLLGDKASKRLTERSRVITVDGNICTGKGKLAKEIAEKLGFKHFPEA GIHYPDSTTGDGKPLATDYNGNCSLEKFYDDPRSNDGNSYRLQSWLYSSRLLQYSDALEHLLTTGQGVVLERSIFSDFVFLEAMYNQGFI RKQCVDHYNEVKSVTICDYLPPHLVIYIDVPVPEVQRRIQKKGDPHEMKITSAYLQDIENAYKKTFLPEMSEKCEVLQYSAREAQDSKKV -------------------------------------------------------------- >22450_22450_2_DGKD-NDUFA10_DGKD_chr2_234299129_ENST00000409813_NDUFA10_chr2_240961757_ENST00000252711_length(transcript)=5034nt_BP=295nt AGAGACACGAATATGTTTCAGCCGCAACAGGCTGCGTTTCAGCCGGAAGAGTGAAAGGGCACCTTGAAAACGCAAGTTTATGAATATGTT TCTGTACTTTCAGACCATCATCAAAGAGGGGATGCTGACCAAACAGAACAATTCATTCCAGCGATCAAAAAGGAGATACTTTAAGCTTCG AGGGCGAACGCTTTACTATGCCAAAACGGCAAAGTCAATCATATTTGATGAGGTGGATCTGACAGATGCCAGCGTAGCTGAATCCAGTAC CAAAAACGTCAACAACAGTTTTACGAGAGGAATTCATAGCAGTGTGCAGTGCAAACTGCGCTATGGAATGTGGCATTTCCTACTTGGGGA TAAAGCAAGCAAAAGACTGACAGAACGCAGCAGAGTGATAACTGTAGATGGCAATATATGTACTGGAAAAGGCAAACTTGCAAAAGAAAT AGCAGAGAAACTAGGCTTCAAGCACTTTCCTGAAGCGGGGATTCATTATCCAGACAGTACCACAGGAGATGGGAAGCCCCTCGCCACCGA CTATAATGGCAACTGTAGTTTGGAGAAATTTTACGATGATCCGAGAAGCAATGATGGCAACAGTTACCGCCTGCAGTCCTGGTTGTACAG CAGTCGCCTGCTGCAGTACTCAGATGCCTTGGAGCACTTGCTGACCACAGGACAAGGTGTTGTGTTGGAGCGCTCCATCTTCAGTGACTT TGTGTTCCTGGAGGCGATGTACAACCAGGGATTCATCCGAAAGCAGTGTGTGGACCACTACAACGAGGTGAAGAGCGTCACCATCTGCGA TTACCTGCCCCCCCACCTGGTGATTTACATCGATGTGCCCGTTCCAGAGGTCCAGAGGCGGATTCAGAAGAAAGGAGATCCACATGAAAT GAAGATCACCTCTGCCTATCTACAGGACATTGAGAATGCCTATAAGAAAACCTTTCTCCCTGAGATGAGTGAAAAATGTGAGGTTTTACA ATATTCTGCAAGGGAAGCTCAAGATTCAAAAAAGGTGGTAGAGGACATTGAATACCTGAAGTTCGATAAAGGGCCGTGGCTCAAGCAGGA CAATCGCACTTTATACCACCTGCGATTACTGGTTCAGGATAAGTTTGAGGTGCTGAATTACACAAGCATTCCTATCTTTCTCCCGGAAGT CACCATTGGAGCTCATCAGACTGACCGTGTCTTACATCAGTTCAGAGAGCTGCCGGGCCGCAAGTACAGCCCTGGGTACAACACCGAGGT GGGAGACAAGTGGATCTGGCTGAAGTGAACGGGCCGCCTTCTGCTCCAGCTGCATCACAGTGATGGCCAAGCTGCATCAGCCGCACTCTC CTGGACGCCATATAGCTTTAAGATCGGGGGAGGGTAAATAATGCAAAAATTGCACAGTGGAAGAAGGGGTCTCACAAAAAGCAATCCATC CTGTAGTATAGGTAATGGAGTTGGGGGAAGCAGCTTCCATTCTGGATGTTTGGAACCCTTTAGCTTTGTTTTGGAATGGCCCACCATTCT CACTGGAAAACAGTGGTCTGCTGTGAAAGGCCAGCTCTCGGCAGCCCCTGTGGTTTCAGCGCTGCCGCTCTGTGTCATTCAGGTTGTGCA CATTGTTTTTCTTCTGACTTCCAGAAATAAAAGTGTTTCCATGGGAATTTCCCCATCGTACGTCTATGTGCGTGCGGGCACGTCGAAACT GCACACGTGCACACACGCTGCTCTGGGCCCTGTTTGGTGTGGTGTGGTTTAATTGAGGCTTACATTATGCTGAAGAGTTTTTCTAAAGGC CTACTGTATTTATTTTAGTAATGTGCTACATTAGAGTAGAAAAAATAAGAACGTGAAGAAGAAATAGCAAAAGGAGAAAATTACCACAGA TTTACAGAACAATATGTTTTAAGAGAGACGTTGAGAAAGATTCACGTGGCCGTCCTCAGCGTCTCAAACTCCTTTGAAGGGAGATTTGAG CTTAGGCTGTTGATGACATAAGATGCAAAAGCAAAATAATATACTCTCTTTATTACCCAGAAAATCAAGTTGCCAGTCAAAACTGAGAAA TGAGAAAGAAAATCAGATTTCTTTATTCATCTCTGTTGAAGCCTAGAAAACCAAAACAAGCTGGTTGTGGTTCAGCATCATGGTTTAGGA ATCAAGCTGTCAGTTGTGAGTCCTACTGAACTGTGGGTTTTCCACTGTAAAGCTCCACTGTGTCCATTAGGAATCAGGCTGCCAGTTGTG AGTCCTACTGAACTGTGGGTTTTCCACTGTAAAGCTCCACTGTGTCCAATATGGTAGCTCCTAGCCACATGTGGGTATTGAAATTTAAAT GAGCTAAAATGTAATAGAGTTTAAAATTGGGTTCCTTGGTTGCACTGGCCATAGTTCAGGCACTCCACAGCCGCCTGTGGGCAGTAGTTC CAGTGCTGGTGCAGAGGACGATGCTCTGCTCCCCGCGGAAGCTCTACCAGGCCTCGCTGCTGCGGAGCATGCGAAGAAGATGCACACTCC TTCCCTCCTTGCGTCCGCCTGCCTTTTTCCCTCTCCCCTTCCTTCCCTCCTTCTTTCCTCCTCCTTCCCTCCCTTCCTTTCTTCCTTCCA TCTGTCCTTGTTTTCCTCCCTTCCCTTCCTCCCTGCCTCTTTCCTTCCCTCCTTCTCTTCCTTCCTTCCGTCTGTCCTTGTTTTCCTCCC TGCCTCTTTCCTTCCCTCCTTCTCTTCCTTCCTTCCTTCCGTCTGTCCTTGTTTTCCTCCCTTCCCTTCCTCCCTGCCTCCTTCCTTGTT TTAAGAATTTGAAAATAAAGTTTCCCAGGAGTTTTGAATTGGAGTCGAGATTTAAGTGGCTGAAGTTTTGCATTTCACTAGGAAACCAAC AGCTTTAATCATGAAATGAAAGCACAGCTGGGCATTGGGCCAGTCACAGAGTGTCTTTTGCTCTGAGCACTTCACAAACCCATTGTGGAG GGGAGGAAAGAAAAAGAAAGCCATCTTTAATTATCTTAGCTGTTCCTAATGTCTTTGGCTGCAAAGAGCTAGCTGTTTCCTTCAACCTTG GTGGTTGCACGGGTAGTGTCCTCAGCAGTGAGAGGGCAGGTCTGCCCTGTGGTCAGGATTGCCCTTGTGAGGGGCAGAGCTCTCCAACCA TGCAGGGCTCCGTGTCGGCATGGTGTGGACTCGGGTGCTCTGATCATCCCCTGTGTTTGCGTCTGACCCTGTGTGGTTGCATTCCCTTGA TGGAGTGGGAAGTATGTAATTTGTAGTGCTGAGGACATTCGTCTGTTTGATTTTCTGGTGTTTTCAATGACATGCATGTTCAGCAGAGAA AGATGAATAAACACTGTCCAATTCTTCTCTCCACATAGAAGCCTCAAGAAGAGGTGAGGAGATGCCCCGATTTCCTTATTTCAGAGCAGT ACATGCGTTCGTCTCACGAAAGCTCTTACAACTAAAACAACAGCACCAACAAACCAGGATCAAAATAATCGTGCATTTTCCAGGATTCCA TGTTGGACTTCTCAGTGCTTTGGAGATCAGGAAATAATCATGCTGTTGCAGCAGTATGGTTTTGTGTCTGGATTTTATTCATACAGGCAT GTGTTTACAGGCATGAGAATTCTGTATTGAAAAGAAAGTCCTTGTTTTTTTCTAACGTATCTTTCTGTTCCATCAACTGTTAAAACCAAA TTGTTAAGAACATTGAGCTTCTTAGGAGAGATTACATGCAGCTACTTAGGACCACCTCAGCAAAATGGAGTTGCCTGGGTAGATAGCAGC AGCAGCTCACCAGCCCCACGCTGCCAGGCTGCTGGCTGCTGTCCACCAGGCAGCTGTGGAACCTCCATGTTCTGCCTAGGTGTTGTGGCA AGAACAGCAGGGAAACAGGCGGCCGCCTCCAGGCCACAGGTGGACCCAGGGACCTTCAGTTACGCCGCTTCAATTGAATTCGTGGTGGTT TCTCTGCTACAGCAGAAGTGGAAGAAGCTCTTGTCTTGTTTGGCATTTGTCCCATTTTTTGGCTAATAATTGGCTTTGAATTTCTCTGTG CCCTGCTTACCAATTATAGCTTTCTGGATTGGCTGTTCTTAGCAGTGCAAATGTCAATGCAAAGAAGCAAGCAGGCGGCTACTTTCTGGG AAGAAGCATAGTCCGTGCTCACACTGGATATAAATCTGCAAGATTTGGGGGTTTGGCAAAATAGGGGTGGAGGGAAAGCTGCCATGGCTG TCAGAAATTGGGGCTTTTTGAAGAAAAGCTTGTGTAGGCATCAGCCACTGGCTGGCCAGAATTCCATGAATGGAAGCAGGTGCCCGAGTG ATCTGGGGCTTTACATGTGGATATAAATAGAAGCCGGGAGCCTGAGGCTTTGCTTTAGGAAAATCATGAGCCAGAAAACAGAGGAGGTGA TTGTCAGTAGTCACCTCACGCCGCTAGTGTTTTCTAATACTCTCTCGAATTGTCAAAGCAAAGGACCTTATAACAGCAACACGTGTGTGA CTCGAAGCAGCGAGCTTCCCTTGGGAAAAGGAGCCTCTTCTCAGCAGAGGGTCCCAGCCTTTCTGTCCTTCCCCATCGGAATGCCAGGCT CTGTGAGTTTGGATGTTGGACATTGCCACCTGAATTCCAGGAGGCCGGGGAGCTCCAGTGATGGACTTGTCCCCAAAAGAGTGGTGCTGT TCTCAGAGCCATAAATGTTCCACTGAAAGTGTCTGTCCCCTGGCTCACGCTGGCTGCGCTTGCTTGAAGGAGGCTGATTTAAGGGTTCTA ATAGGAAGACAAATGAAACTGTCAAATTTTATCTTTAAGCATGTGAGTAAGTTTGCTCATTCGCTTTTACTATTGATGTATAGTTTTAAA ATTCTTAATTTAAAAATTATTTTAATAGTAAATTTTTCACAAGTGAACACTTTTTATCTTTAAATTAAAATTCTGAAGCTCCCTAGTTAG >22450_22450_2_DGKD-NDUFA10_DGKD_chr2_234299129_ENST00000409813_NDUFA10_chr2_240961757_ENST00000252711_length(amino acids)=402AA_BP=70 MNMFLYFQTIIKEGMLTKQNNSFQRSKRRYFKLRGRTLYYAKTAKSIIFDEVDLTDASVAESSTKNVNNSFTRGIHSSVQCKLRYGMWHF LLGDKASKRLTERSRVITVDGNICTGKGKLAKEIAEKLGFKHFPEAGIHYPDSTTGDGKPLATDYNGNCSLEKFYDDPRSNDGNSYRLQS WLYSSRLLQYSDALEHLLTTGQGVVLERSIFSDFVFLEAMYNQGFIRKQCVDHYNEVKSVTICDYLPPHLVIYIDVPVPEVQRRIQKKGD PHEMKITSAYLQDIENAYKKTFLPEMSEKCEVLQYSAREAQDSKKVVEDIEYLKFDKGPWLKQDNRTLYHLRLLVQDKFEVLNYTSIPIF -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for DGKD-NDUFA10 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for DGKD-NDUFA10 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for DGKD-NDUFA10 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
