
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:DHX9-XPR1 (FusionGDB2 ID:22735) |
Fusion Gene Summary for DHX9-XPR1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: DHX9-XPR1 | Fusion gene ID: 22735 | Hgene | Tgene | Gene symbol | DHX9 | XPR1 | Gene ID | 1660 | 9213 |
| Gene name | DExH-box helicase 9 | xenotropic and polytropic retrovirus receptor 1 | |
| Synonyms | DDX9|LKP|NDH2|NDHII|RHA | IBGC6|SLC53A1|SYG1|X3 | |
| Cytomap | 1q25.3 | 1q25.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | ATP-dependent RNA helicase ADEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 9DEAH (Asp-Glu-Ala-His) box helicase 9DEAH (Asp-Glu-Ala-His) box polypeptide 9DEAH box protein 9DEAH-box helicase 9RNA helicase Aleukophysinnuclear DNA helicase II | xenotropic and polytropic retrovirus receptor 1X-receptorprotein SYG1 homologsolute carrier family 53 (phosphate exporter), member 1xenotropic and polytropic murine leukemia virus receptor X3 | |
| Modification date | 20200322 | 20200313 | |
| UniProtAcc | Q08211 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000367549, ENST00000485081, | ENST00000467345, ENST00000367589, ENST00000367590, | |
| Fusion gene scores | * DoF score | 14 X 10 X 7=980 | 8 X 9 X 5=360 |
| # samples | 15 | 9 | |
| ** MAII score | log2(15/980*10)=-2.70781924850669 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/360*10)=-2 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: DHX9 [Title/Abstract] AND XPR1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | DHX9(182829319)-XPR1(180651495), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | DHX9 | GO:0010501 | RNA secondary structure unwinding | 1537828|9111062 |
| Hgene | DHX9 | GO:0032508 | DNA duplex unwinding | 9111062|21561811 |
| Hgene | DHX9 | GO:0034622 | cellular protein-containing complex assembly | 23361462 |
| Hgene | DHX9 | GO:0039695 | DNA-templated viral transcription | 11096080 |
| Hgene | DHX9 | GO:0044806 | G-quadruplex DNA unwinding | 21561811 |
| Hgene | DHX9 | GO:0046833 | positive regulation of RNA export from nucleus | 11402034 |
| Hgene | DHX9 | GO:0050434 | positive regulation of viral transcription | 11096080 |
| Hgene | DHX9 | GO:0050684 | regulation of mRNA processing | 28355180 |
| Hgene | DHX9 | GO:2000765 | regulation of cytoplasmic translation | 28355180 |
 Fusion gene breakpoints across DHX9 (5'-gene) Fusion gene breakpoints across DHX9 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
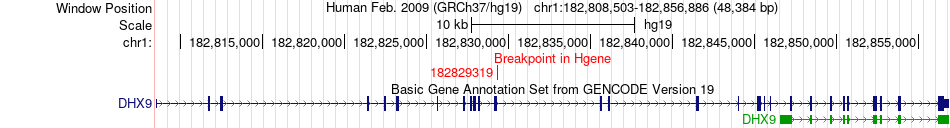 |
 Fusion gene breakpoints across XPR1 (3'-gene) Fusion gene breakpoints across XPR1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
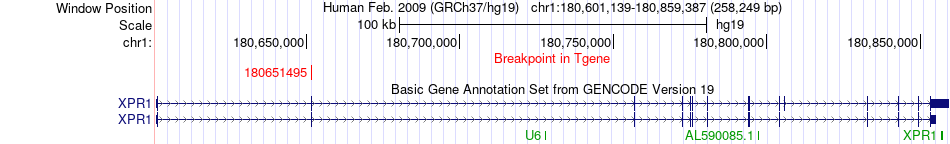 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-VR-A8EW | DHX9 | chr1 | 182829319 | + | XPR1 | chr1 | 180651495 | + |
Top |
Fusion Gene ORF analysis for DHX9-XPR1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000367549 | ENST00000467345 | DHX9 | chr1 | 182829319 | + | XPR1 | chr1 | 180651495 | + |
| In-frame | ENST00000367549 | ENST00000367589 | DHX9 | chr1 | 182829319 | + | XPR1 | chr1 | 180651495 | + |
| In-frame | ENST00000367549 | ENST00000367590 | DHX9 | chr1 | 182829319 | + | XPR1 | chr1 | 180651495 | + |
| intron-3CDS | ENST00000485081 | ENST00000367589 | DHX9 | chr1 | 182829319 | + | XPR1 | chr1 | 180651495 | + |
| intron-3CDS | ENST00000485081 | ENST00000367590 | DHX9 | chr1 | 182829319 | + | XPR1 | chr1 | 180651495 | + |
| intron-intron | ENST00000485081 | ENST00000467345 | DHX9 | chr1 | 182829319 | + | XPR1 | chr1 | 180651495 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000367549 | DHX9 | chr1 | 182829319 | + | ENST00000367590 | XPR1 | chr1 | 180651495 | + | 9649 | 1442 | 110 | 3463 | 1117 |
| ENST00000367549 | DHX9 | chr1 | 182829319 | + | ENST00000367589 | XPR1 | chr1 | 180651495 | + | 5329 | 1442 | 110 | 3268 | 1052 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000367549 | ENST00000367590 | DHX9 | chr1 | 182829319 | + | XPR1 | chr1 | 180651495 | + | 9.56E-05 | 0.9999044 |
| ENST00000367549 | ENST00000367589 | DHX9 | chr1 | 182829319 | + | XPR1 | chr1 | 180651495 | + | 0.000236656 | 0.9997633 |
Top |
Fusion Genomic Features for DHX9-XPR1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| DHX9 | chr1 | 182829319 | + | XPR1 | chr1 | 180651495 | + | 7.09E-10 | 1 |
| DHX9 | chr1 | 182829319 | + | XPR1 | chr1 | 180651495 | + | 7.09E-10 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
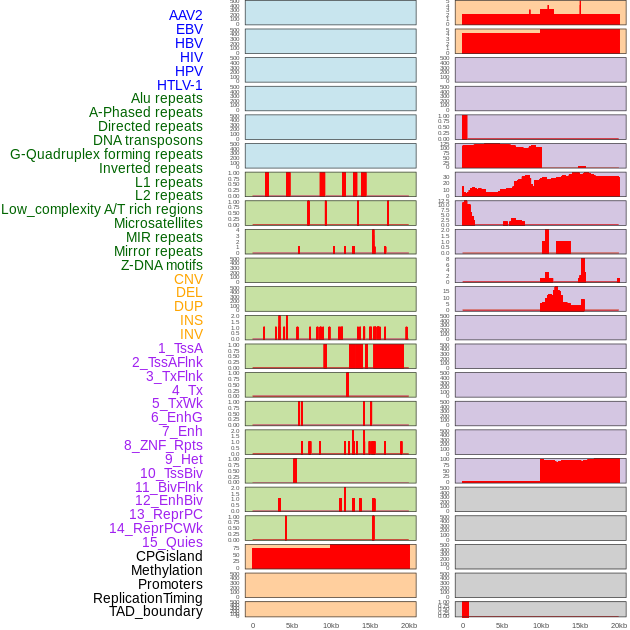 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
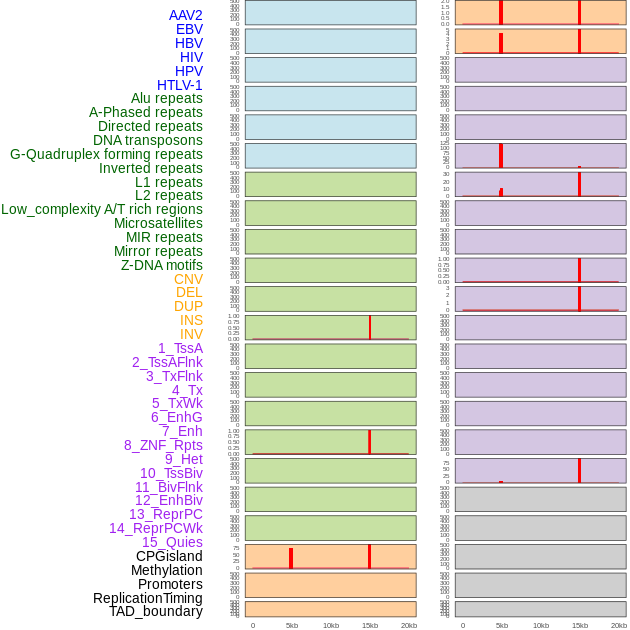 |
Top |
Fusion Protein Features for DHX9-XPR1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:182829319/chr1:180651495) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| DHX9 | . |
| FUNCTION: Multifunctional ATP-dependent nucleic acid helicase that unwinds DNA and RNA in a 3' to 5' direction and that plays important roles in many processes, such as DNA replication, transcriptional activation, post-transcriptional RNA regulation, mRNA translation and RNA-mediated gene silencing (PubMed:9111062, PubMed:11416126, PubMed:12711669, PubMed:15355351, PubMed:16680162, PubMed:17531811, PubMed:20669935, PubMed:21561811, PubMed:24049074, PubMed:25062910, PubMed:24990949, PubMed:28221134). Requires a 3'-single-stranded tail as entry site for acid nuclei unwinding activities as well as the binding and hydrolyzing of any of the four ribo- or deoxyribo-nucleotide triphosphates (NTPs) (PubMed:1537828). Unwinds numerous nucleic acid substrates such as double-stranded (ds) DNA and RNA, DNA:RNA hybrids, DNA and RNA forks composed of either partially complementary DNA duplexes or DNA:RNA hybrids, respectively, and also DNA and RNA displacement loops (D- and R-loops), triplex-helical DNA (H-DNA) structure and DNA and RNA-based G-quadruplexes (PubMed:20669935, PubMed:21561811, PubMed:24049074). Binds dsDNA, single-stranded DNA (ssDNA), dsRNA, ssRNA and poly(A)-containing RNA (PubMed:9111062, PubMed:10198287). Binds also to circular dsDNA or dsRNA of either linear and/or circular forms and stimulates the relaxation of supercoiled DNAs catalyzed by topoisomerase TOP2A (PubMed:12711669). Plays a role in DNA replication at origins of replication and cell cycle progression (PubMed:24990949). Plays a role as a transcriptional coactivator acting as a bridging factor between polymerase II holoenzyme and transcription factors or cofactors, such as BRCA1, CREBBP, RELA and SMN1 (PubMed:11149922, PubMed:9323138, PubMed:9662397, PubMed:11038348, PubMed:11416126, PubMed:15355351, PubMed:28221134). Binds to the CDKN2A promoter (PubMed:11038348). Plays several roles in post-transcriptional regulation of gene expression (PubMed:28221134, PubMed:28355180). In cooperation with NUP98, promotes pre-mRNA alternative splicing activities of a subset of genes (PubMed:11402034, PubMed:16680162, PubMed:28221134, PubMed:28355180). As component of a large PER complex, is involved in the negative regulation of 3' transcriptional termination of circadian target genes such as PER1 and NR1D1 and the control of the circadian rhythms (By similarity). Acts also as a nuclear resolvase that is able to bind and neutralize harmful massive secondary double-stranded RNA structures formed by inverted-repeat Alu retrotransposon elements that are inserted and transcribed as parts of genes during the process of gene transposition (PubMed:28355180). Involved in the positive regulation of nuclear export of constitutive transport element (CTE)-containing unspliced mRNA (PubMed:9162007, PubMed:10924507, PubMed:11402034). Component of the coding region determinant (CRD)-mediated complex that promotes cytoplasmic MYC mRNA stability (PubMed:19029303). Plays a role in mRNA translation (PubMed:28355180). Positively regulates translation of selected mRNAs through its binding to post-transcriptional control element (PCE) in the 5'-untranslated region (UTR) (PubMed:16680162). Involved with LARP6 in the translation stimulation of type I collagen mRNAs for CO1A1 and CO1A2 through binding of a specific stem-loop structure in their 5'-UTRs (PubMed:22190748). Stimulates LIN28A-dependent mRNA translation probably by facilitating ribonucleoprotein remodeling during the process of translation (PubMed:21247876). Plays also a role as a small interfering (siRNA)-loading factor involved in the RNA-induced silencing complex (RISC) loading complex (RLC) assembly, and hence functions in the RISC-mediated gene silencing process (PubMed:17531811). Binds preferentially to short double-stranded RNA, such as those produced during rotavirus intestinal infection (PubMed:28636595). This interaction may mediate NLRP9 inflammasome activation and trigger inflammatory response, including IL18 release and pyroptosis (PubMed:28636595). Finally, mediates the attachment of heterogeneous nuclear ribonucleoproteins (hnRNPs) to actin filaments in the nucleus (PubMed:11687588). {ECO:0000250|UniProtKB:O70133, ECO:0000269|PubMed:10198287, ECO:0000269|PubMed:10924507, ECO:0000269|PubMed:11038348, ECO:0000269|PubMed:11149922, ECO:0000269|PubMed:11402034, ECO:0000269|PubMed:11416126, ECO:0000269|PubMed:11687588, ECO:0000269|PubMed:12711669, ECO:0000269|PubMed:15355351, ECO:0000269|PubMed:1537828, ECO:0000269|PubMed:16680162, ECO:0000269|PubMed:17531811, ECO:0000269|PubMed:19029303, ECO:0000269|PubMed:20669935, ECO:0000269|PubMed:21247876, ECO:0000269|PubMed:21561811, ECO:0000269|PubMed:22190748, ECO:0000269|PubMed:24049074, ECO:0000269|PubMed:24990949, ECO:0000269|PubMed:25062910, ECO:0000269|PubMed:28221134, ECO:0000269|PubMed:28355180, ECO:0000269|PubMed:28636595, ECO:0000269|PubMed:9111062, ECO:0000269|PubMed:9162007, ECO:0000269|PubMed:9323138, ECO:0000269|PubMed:9662397}.; FUNCTION: (Microbial infection) Plays a role in HIV-1 replication and virion infectivity (PubMed:11096080, PubMed:19229320, PubMed:25149208, PubMed:27107641). Enhances HIV-1 transcription by facilitating the binding of RNA polymerase II holoenzyme to the proviral DNA (PubMed:11096080, PubMed:25149208). Binds (via DRBM domain 2) to the HIV-1 TAR RNA and stimulates HIV-1 transcription of transactivation response element (TAR)-containing mRNAs (PubMed:9892698, PubMed:11096080). Involved also in HIV-1 mRNA splicing and transport (PubMed:25149208). Positively regulates HIV-1 gag mRNA translation, through its binding to post-transcriptional control element (PCE) in the 5'-untranslated region (UTR) (PubMed:16680162). Binds (via DRBM domains) to a HIV-1 double-stranded RNA region of the primer binding site (PBS)-segment of the 5'-UTR, and hence stimulates DHX9 incorporation into virions and virion infectivity (PubMed:27107641). Plays also a role as a cytosolic viral MyD88-dependent DNA and RNA sensors in plasmacytoid dendritic cells (pDCs), and hence induce antiviral innate immune responses (PubMed:20696886, PubMed:21957149). Binds (via the OB-fold region) to viral single-stranded DNA unmethylated C-phosphate-G (CpG) oligonucleotide (PubMed:20696886). {ECO:0000269|PubMed:11096080, ECO:0000269|PubMed:16680162, ECO:0000269|PubMed:19229320, ECO:0000269|PubMed:20696886, ECO:0000269|PubMed:21957149, ECO:0000269|PubMed:25149208, ECO:0000269|PubMed:27107641, ECO:0000269|PubMed:9892698}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 180_252 | 444 | 1271.0 | Domain | DRBM 2 |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 3_71 | 444 | 1271.0 | Domain | DRBM 1 |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 411_419 | 444 | 1271.0 | Nucleotide binding | ATP |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 182_186 | 444 | 1271.0 | Region | siRNA-binding |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 234_236 | 444 | 1271.0 | Region | siRNA-binding |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 331_380 | 444 | 1271.0 | Region | MTAD |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 53_55 | 444 | 1271.0 | Region | siRNA-binding |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 5_9 | 444 | 1271.0 | Region | siRNA-binding |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 439_643 | 23 | 632.0 | Domain | EXS | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 439_643 | 23 | 697.0 | Domain | EXS | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 158_165 | 23 | 632.0 | Region | Important for inositol polyphosphate binding | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 158_165 | 23 | 697.0 | Region | Important for inositol polyphosphate binding | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 258_264 | 23 | 632.0 | Topological domain | Extracellular | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 286_318 | 23 | 632.0 | Topological domain | Cytoplasmic | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 340_345 | 23 | 632.0 | Topological domain | Extracellular | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 369_376 | 23 | 632.0 | Topological domain | Cytoplasmic | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 398_402 | 23 | 632.0 | Topological domain | Extracellular | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 424_473 | 23 | 632.0 | Topological domain | Cytoplasmic | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 497_507 | 23 | 632.0 | Topological domain | Extracellular | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 529_696 | 23 | 632.0 | Topological domain | Cytoplasmic | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 258_264 | 23 | 697.0 | Topological domain | Extracellular | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 286_318 | 23 | 697.0 | Topological domain | Cytoplasmic | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 340_345 | 23 | 697.0 | Topological domain | Extracellular | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 369_376 | 23 | 697.0 | Topological domain | Cytoplasmic | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 398_402 | 23 | 697.0 | Topological domain | Extracellular | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 424_473 | 23 | 697.0 | Topological domain | Cytoplasmic | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 497_507 | 23 | 697.0 | Topological domain | Extracellular | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 529_696 | 23 | 697.0 | Topological domain | Cytoplasmic | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 237_257 | 23 | 632.0 | Transmembrane | Helical | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 265_285 | 23 | 632.0 | Transmembrane | Helical | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 319_339 | 23 | 632.0 | Transmembrane | Helical | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 346_368 | 23 | 632.0 | Transmembrane | Helical | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 377_397 | 23 | 632.0 | Transmembrane | Helical | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 403_423 | 23 | 632.0 | Transmembrane | Helical | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 474_496 | 23 | 632.0 | Transmembrane | Helical | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 508_528 | 23 | 632.0 | Transmembrane | Helical | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 237_257 | 23 | 697.0 | Transmembrane | Helical | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 265_285 | 23 | 697.0 | Transmembrane | Helical | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 319_339 | 23 | 697.0 | Transmembrane | Helical | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 346_368 | 23 | 697.0 | Transmembrane | Helical | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 377_397 | 23 | 697.0 | Transmembrane | Helical | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 403_423 | 23 | 697.0 | Transmembrane | Helical | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 474_496 | 23 | 697.0 | Transmembrane | Helical | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 508_528 | 23 | 697.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 398_564 | 444 | 1271.0 | Domain | Helicase ATP-binding |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 636_809 | 444 | 1271.0 | Domain | Helicase C-terminal |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 1155_1173 | 444 | 1271.0 | Motif | Nuclear localization signal (NLS2) |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 511_514 | 444 | 1271.0 | Motif | Note=DEIH box |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 586_595 | 444 | 1271.0 | Motif | Nuclear localization signal (NLS1) |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 1150_1270 | 444 | 1271.0 | Region | RGG |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 398_809 | 444 | 1271.0 | Region | Core helicase |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 831_919 | 444 | 1271.0 | Region | HA2 |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 958_1074 | 444 | 1271.0 | Region | OB-fold |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 1_177 | 23 | 632.0 | Domain | SPX | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 1_177 | 23 | 697.0 | Domain | SPX | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367589 | 0 | 14 | 1_236 | 23 | 632.0 | Topological domain | Cytoplasmic | |
| Tgene | XPR1 | chr1:182829319 | chr1:180651495 | ENST00000367590 | 0 | 15 | 1_236 | 23 | 697.0 | Topological domain | Cytoplasmic |
Top |
Fusion Gene Sequence for DHX9-XPR1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >22735_22735_1_DHX9-XPR1_DHX9_chr1_182829319_ENST00000367549_XPR1_chr1_180651495_ENST00000367589_length(transcript)=5329nt_BP=1442nt GCGAGTTGCTGTGCGTTTCTCTGTTGTCTCGGTAGAAGGCCAGAGTCACACACGGTCCTAAGAGCTGGGCACCAGGAAGCGAAGGCTGAT CTGAAGAAGACACTTGAATCATGGGTGACGTTAAAAATTTTCTGTATGCCTGGTGTGGCAAAAGGAAGATGACCCCATCCTATGAAATTA GAGCAGTGGGGAACAAAAACAGGCAGAAATTCATGTGTGAGGTTCAGGTGGAAGGTTATAATTACACTGGCATGGGAAATTCCACCAATA AAAAAGATGCACAAAGCAATGCTGCCAGAGACTTTGTTAACTATTTGGTTCGAATAAATGAAATAAAGAGTGAAGAAGTTCCAGCTTTTG GGGTAGCATCTCCGCCCCCACTTACTGATACTCCTGACACTACAGCAAATGCTGAAGGAGATTTACCAACAACCATGGGAGGACCTCTTC CTCCACATCTGGCTCTCAAAGCAGAAAATAATTCTGAGGTAGGGGCCTCTGGCTATGGTGTTCCTGGGCCCACCTGGGACCGAGGAGCCA ACTTGAAGGATTACTACTCAAGAAAGGAAGAACAAGAAGTGCAAGCGACTCTAGAATCAGAAGAAGTGGATTTAAATGCTGGGCTTCATG GAAACTGGACCTTGGAAAATGCTAAAGCTCGTCTAAACCAATATTTTCAGAAAGAAAAGATCCAAGGAGAATATAAGTACACCCAAGTGG GTCCTGATCACAACAGGAGCTTTATTGCAGAAATGACCATTTATATCAAGCAGCTGGGCAGAAGGATTTTTGCACGAGAACATGGATCAA ATAAGAAATTGGCAGCACAGTCCTGTGCCCTGTCACTTGTCAGACAACTGTACCATCTTGGAGTGGTTGAAGCTTACTCCGGACTTACAA AGAAGAAGGAAGGAGAGACAGTGGAGCCTTACAAAGTAAACCTCTCTCAAGATTTAGAGCATCAGCTGCAAAACATCATTCAAGAGCTAA ATCTTGAGATTTTGCCCCCGCCTGAAGATCCTTCTGTGCCAGTTGCACTCAACATTGGCAAATTGGCTCAGTTCGAACCATCTCAGCGAC AAAACCAAGTGGGTGTGGTTCCTTGGTCACCTCCACAATCCAACTGGAATCCTTGGACTAGTAGCAACATTGATGAGGGGCCTCTGGCTT TTGCTACTCCAGAGCAAATAAGCATGGACCTCAAGAATGAATTGATGTACCAGTTGGAACAGGATCATGATTTGCAAGCAATCTTGCAGG AGAGAGAGTTACTGCCTGTGAAGAAATTTGAAAGTGAGATTCTGGAAGCAATCAGCCAAAATTCAGTTGTCATTATTAGAGGGGCTACTG GATGTGGGAAAACCACACAGGTTCCCCAGTTCATTCTAGATGACTTTATCCAGAATGACCGAGCAGCAGAGTGTAACATCGTAGTAACTC AGGCTTTCAAGGATATGCTGTATTCAGCTCAGGACCAGGCACCTTCTGTGGAAGTTACAGATGAGGACACAGTAAAGAGGTATTTTGCCA AGTTTGAAGAGAAGTTTTTCCAAACCTGTGAAAAAGAACTTGCCAAAATCAACACATTTTATTCAGAGAAGCTCGCAGAGGCTCAGCGCA GGTTTGCTACACTTCAGAATGAGCTTCAGTCATCACTGGATGCACAGAAAGAAAGCACTGGTGTTACTACGCTGCGACAACGCAGAAAGC CAGTCTTCCACTTGTCCCATGAGGAACGTGTCCAACATAGAAATATTAAAGACCTTAAACTGGCCTTCAGTGAGTTCTACCTCAGTCTAA TCCTGCTGCAGAACTATCAGAATCTGAATTTTACAGGGTTTCGAAAAATCCTGAAAAAGCATGACAAGATCCTGGAAACATCTCGTGGAG CAGATTGGCGAGTGGCTCACGTAGAGGTGGCCCCATTTTATACATGCAAGAAAATCAACCAGCTTATCTCTGAAACTGAGGCTGTAGTGA CCAATGAACTTGAAGATGGTGACAGACAAAAGGCTATGAAGCGTTTACGTGTCCCCCCTTTGGGAGCTGCTCAGCCTGCACCAGCATGGA CTACTTTTAGAGTTGGCCTATTTTGTGGAATATTCATTGTACTGAATATTACCCTTGTGCTTGCCGCTGTATTTAAACTTGAAACAGATA GAAGTATATGGCCCTTGATAAGAATCTATCGGGGTGGCTTTCTTCTGATTGAATTCCTTTTTCTACTGGGCATCAACACGTATGGTTGGA GACAGGCTGGAGTAAACCATGTACTCATCTTTGAACTTAATCCGAGAAGCAATTTGTCTCATCAACATCTCTTTGAGATTGCTGGATTCC TCGGGATATTGTGGTGCCTGAGCCTTCTGGCATGCTTCTTTGCTCCAATTAGTGTCATCCCCACATATGTGTATCCACTTGCCCTTTATG GATTTATGGTTTTCTTCCTTATCAACCCCACCAAAACTTTCTACTATAAATCCCGGTTTTGGCTGCTTAAACTGCTGTTTCGAGTATTTA CAGCCCCCTTCCATAAGGTAGGCTTTGCTGATTTCTGGCTGGCGGATCAGCTGAACAGCCTGTCAGTGATACTGATGGACCTGGAATATA TGATCTGCTTCTACAGTTTGGAGCTCAAATGGGATGAAAGTAAGGGCCTGTTGCCAAATAATTCAGAAGAACGAGGTCACTCGGACACTA TGGTGTTCTTTTACCTGTGGATTGTCTTTTATATCATCAGTTCCTGCTATACCCTCATCTGGGATCTCAAGATGGACTGGGGTCTCTTCG ATAAGAATGCTGGAGAGAACACTTTCCTCCGGGAAGAGATTGTATACCCCCAAAAAGCCTACTACTACTGTGCCATAATAGAGGATGTGA TTCTGCGCTTTGCTTGGACTATCCAAATCTCGATTACCTCTACAACTTTGTTGCCTCATTCTGGGGACATCATTGCTACTGTCTTTGCCC CACTTGAGGTTTTCCGGCGATTTGTGTGGAACTTCTTCCGCCTGGAGAATGAACATCTGAATAACTGTGGTGAATTCCGTGCTGTGCGGG ACATCTCTGTGGCCCCCCTGAACGCAGATGATCAGACTCTCCTAGAACAGATGATGGACCAGGATGATGGGGTACGAAACCGCCAGAAGA ATCGGTCATGGAAGTACAACCAGAGCATATCCCTGCGCCGGCCTCGCCTCGCTTCTCAATCCAAGGCTCGTGACACTAAGGTATTGATAG AAGACACAGATGATGAAGCTAACACTTGAATTTTCTGAAGTCTAGCTTAACATCTTTGGTTTTCCTACTCTACAATCCTTTCCTCGACCA ACGCAACCTCTAGTACCTTTCCAGCCGAAAACAGGAGAAAACACATAACACATTTTCCGAGCTCTTCCGGATCGGATCCTATGGACTCCA AACAAGCTCACTGTGTTTCTTTTCTTTTCTTCTGGTTTAATTTTAATTTTCTATTTTCAAAACAAATATTTACTTCATTTGCCAATCAGA GGATGTTTTAAGAAACAAAACATAGTATCTTATGGATTGTTTACAATCACAAGGACATAGATACCTATCAGGATGAAGAACAGGCATTGC AAGGACCCTCTGATGGGACGGTACTGAGATATCTCGGCTTCCGCTCAGCCCGGTTTTGACTGGTTGAAACCGGACATTGGTTTTTAAATT TTTTGTCAGTTTATGTGGAGAATTTTTTTCTTTCCTTCATACCCAGCGCAAAGGCACTGGCCGCACTTGCAGGAAAAGTGCAACTTAAAG CAGTACCTTCATTCATGAAGCTACTTTTTAATTTGATGTAACTTTTCTTATTTTGGGAAGGGTTGCTGGGTGGGTGGGAAATATGATGTA TTTGTTACACATAGTTTTCTCATTATTTATGAAACTTAACCATACAGAATGATATAACTCCTGTGCAATGAAGGTGATAACAGTAAAAGA AGGCAGGGGAAACTTACGTTGGATGACATTTATGAGGGTCAGTCCCACATACCTCTTTCAGGAGACAACTTGCACCAGTTTGACCTTTTC TTTTCTTTGTTTTTATTTTAAGCCAAAGTTTCATTGCTAACTTCTTAAGTTGCTGCTGCTTTAGAGTCCTGAGCATATCTCTCATAACAA GGAATCCCACACTTCACACCACCGGCTGAATTTCATGGAAGAGGTTCTGATAATTTTTTTAACTTTTTAAGGAACAGATGTGGAATACAC TGGCCCATATTTCAACCTTAACAGCTGAAGCTATGCCTTATTATGCATCCACATGTATGGTCCCTGTAGCGTGACCTTTACTAGCTCTGA ATCAGAAGACAGAGCTATTTCAGAGGCTCTGTGTGCCCTCACTAGATAGTTTTTCTTCTGGGTTCAACCACTTTAGCCAGAATTTGATCA AATTAAAAGTCTGTCATGGGGAAACTATATTTTTGAGCACATGGAACAAATTATACTTCCTCATTCATATTATGTTGATACAAAAGACCT TGGCAGCCATTTCTCCCAGCAGTTTTAAAGGATGAACATTGGATTTCATGCCATCCCATAGAAAACCTGTTTTAAAATTTTAGGGATCTT TACTTGGTCATACATGAAAAGTACACTGCTTAGAAATTATAGACTATTATGATCTGTCCACAGTGCCCATTGTCACTTCTTTGTCTCATT TCTTCCCTTTGTTCCTTAGTCATCCAAATAAGCCTGAAAACCATAAGAGATATTACTTTATTGAATATGGTTGGCATTAAATTTAGCATT TCATTATCTAACAAAATTAATATAAATTCCAGGACATGGTAAAATGTGTTTTAATAACCCCCAGACCCAAATGAAAATTTCAAAGTCAAT ACCAGCAGATTCATGAAAGTAAATTTAGTCCTATAATTTTCAGCTTAATTATAAACAAAGGAACAAATAAGTGGAAGGGCAGCTATTACC ATTCGCTTAGTCAAAACATTCGGTTACTGCCCTTTAATACACTCCTATCATCAGCACTTCCACCATGTATTACAAGTCTTGACCCATCCC TGTCGTAACTCCAGTAAAAGTTACTGTTACTAGAAAATTTTTATCAATTAACTGACAAATAGTTTCTTTTTAAAGTAGTTTCTTCCATCT TTATTCTGACTAGCTTCCAAAATGTGTTCCCTTTTTGAATCGAGGTTTTTTTGTTTTGTTTTGTTTTCTGAAAAAATCATACAACTTTGT GCTTCTATTGCTTTTTTGTGTTTTGTTAAGCATGTCCCTTGGCCCAAATGGAAGAGGAAATGTTTAATTAATGCTTTTTAGTTTAAATAA >22735_22735_1_DHX9-XPR1_DHX9_chr1_182829319_ENST00000367549_XPR1_chr1_180651495_ENST00000367589_length(amino acids)=1052AA_BP=444 MGDVKNFLYAWCGKRKMTPSYEIRAVGNKNRQKFMCEVQVEGYNYTGMGNSTNKKDAQSNAARDFVNYLVRINEIKSEEVPAFGVASPPP LTDTPDTTANAEGDLPTTMGGPLPPHLALKAENNSEVGASGYGVPGPTWDRGANLKDYYSRKEEQEVQATLESEEVDLNAGLHGNWTLEN AKARLNQYFQKEKIQGEYKYTQVGPDHNRSFIAEMTIYIKQLGRRIFAREHGSNKKLAAQSCALSLVRQLYHLGVVEAYSGLTKKKEGET VEPYKVNLSQDLEHQLQNIIQELNLEILPPPEDPSVPVALNIGKLAQFEPSQRQNQVGVVPWSPPQSNWNPWTSSNIDEGPLAFATPEQI SMDLKNELMYQLEQDHDLQAILQERELLPVKKFESEILEAISQNSVVIIRGATGCGKTTQVPQFILDDFIQNDRAAECNIVVTQAFKDML YSAQDQAPSVEVTDEDTVKRYFAKFEEKFFQTCEKELAKINTFYSEKLAEAQRRFATLQNELQSSLDAQKESTGVTTLRQRRKPVFHLSH EERVQHRNIKDLKLAFSEFYLSLILLQNYQNLNFTGFRKILKKHDKILETSRGADWRVAHVEVAPFYTCKKINQLISETEAVVTNELEDG DRQKAMKRLRVPPLGAAQPAPAWTTFRVGLFCGIFIVLNITLVLAAVFKLETDRSIWPLIRIYRGGFLLIEFLFLLGINTYGWRQAGVNH VLIFELNPRSNLSHQHLFEIAGFLGILWCLSLLACFFAPISVIPTYVYPLALYGFMVFFLINPTKTFYYKSRFWLLKLLFRVFTAPFHKV GFADFWLADQLNSLSVILMDLEYMICFYSLELKWDESKGLLPNNSEERGHSDTMVFFYLWIVFYIISSCYTLIWDLKMDWGLFDKNAGEN TFLREEIVYPQKAYYYCAIIEDVILRFAWTIQISITSTTLLPHSGDIIATVFAPLEVFRRFVWNFFRLENEHLNNCGEFRAVRDISVAPL -------------------------------------------------------------- >22735_22735_2_DHX9-XPR1_DHX9_chr1_182829319_ENST00000367549_XPR1_chr1_180651495_ENST00000367590_length(transcript)=9649nt_BP=1442nt GCGAGTTGCTGTGCGTTTCTCTGTTGTCTCGGTAGAAGGCCAGAGTCACACACGGTCCTAAGAGCTGGGCACCAGGAAGCGAAGGCTGAT CTGAAGAAGACACTTGAATCATGGGTGACGTTAAAAATTTTCTGTATGCCTGGTGTGGCAAAAGGAAGATGACCCCATCCTATGAAATTA GAGCAGTGGGGAACAAAAACAGGCAGAAATTCATGTGTGAGGTTCAGGTGGAAGGTTATAATTACACTGGCATGGGAAATTCCACCAATA AAAAAGATGCACAAAGCAATGCTGCCAGAGACTTTGTTAACTATTTGGTTCGAATAAATGAAATAAAGAGTGAAGAAGTTCCAGCTTTTG GGGTAGCATCTCCGCCCCCACTTACTGATACTCCTGACACTACAGCAAATGCTGAAGGAGATTTACCAACAACCATGGGAGGACCTCTTC CTCCACATCTGGCTCTCAAAGCAGAAAATAATTCTGAGGTAGGGGCCTCTGGCTATGGTGTTCCTGGGCCCACCTGGGACCGAGGAGCCA ACTTGAAGGATTACTACTCAAGAAAGGAAGAACAAGAAGTGCAAGCGACTCTAGAATCAGAAGAAGTGGATTTAAATGCTGGGCTTCATG GAAACTGGACCTTGGAAAATGCTAAAGCTCGTCTAAACCAATATTTTCAGAAAGAAAAGATCCAAGGAGAATATAAGTACACCCAAGTGG GTCCTGATCACAACAGGAGCTTTATTGCAGAAATGACCATTTATATCAAGCAGCTGGGCAGAAGGATTTTTGCACGAGAACATGGATCAA ATAAGAAATTGGCAGCACAGTCCTGTGCCCTGTCACTTGTCAGACAACTGTACCATCTTGGAGTGGTTGAAGCTTACTCCGGACTTACAA AGAAGAAGGAAGGAGAGACAGTGGAGCCTTACAAAGTAAACCTCTCTCAAGATTTAGAGCATCAGCTGCAAAACATCATTCAAGAGCTAA ATCTTGAGATTTTGCCCCCGCCTGAAGATCCTTCTGTGCCAGTTGCACTCAACATTGGCAAATTGGCTCAGTTCGAACCATCTCAGCGAC AAAACCAAGTGGGTGTGGTTCCTTGGTCACCTCCACAATCCAACTGGAATCCTTGGACTAGTAGCAACATTGATGAGGGGCCTCTGGCTT TTGCTACTCCAGAGCAAATAAGCATGGACCTCAAGAATGAATTGATGTACCAGTTGGAACAGGATCATGATTTGCAAGCAATCTTGCAGG AGAGAGAGTTACTGCCTGTGAAGAAATTTGAAAGTGAGATTCTGGAAGCAATCAGCCAAAATTCAGTTGTCATTATTAGAGGGGCTACTG GATGTGGGAAAACCACACAGGTTCCCCAGTTCATTCTAGATGACTTTATCCAGAATGACCGAGCAGCAGAGTGTAACATCGTAGTAACTC AGGCTTTCAAGGATATGCTGTATTCAGCTCAGGACCAGGCACCTTCTGTGGAAGTTACAGATGAGGACACAGTAAAGAGGTATTTTGCCA AGTTTGAAGAGAAGTTTTTCCAAACCTGTGAAAAAGAACTTGCCAAAATCAACACATTTTATTCAGAGAAGCTCGCAGAGGCTCAGCGCA GGTTTGCTACACTTCAGAATGAGCTTCAGTCATCACTGGATGCACAGAAAGAAAGCACTGGTGTTACTACGCTGCGACAACGCAGAAAGC CAGTCTTCCACTTGTCCCATGAGGAACGTGTCCAACATAGAAATATTAAAGACCTTAAACTGGCCTTCAGTGAGTTCTACCTCAGTCTAA TCCTGCTGCAGAACTATCAGAATCTGAATTTTACAGGGTTTCGAAAAATCCTGAAAAAGCATGACAAGATCCTGGAAACATCTCGTGGAG CAGATTGGCGAGTGGCTCACGTAGAGGTGGCCCCATTTTATACATGCAAGAAAATCAACCAGCTTATCTCTGAAACTGAGGCTGTAGTGA CCAATGAACTTGAAGATGGTGACAGACAAAAGGCTATGAAGCGTTTACGTGTCCCCCCTTTGGGAGCTGCTCAGCCTGCACCAGCATGGA CTACTTTTAGAGTTGGCCTATTTTGTGGAATATTCATTGTACTGAATATTACCCTTGTGCTTGCCGCTGTATTTAAACTTGAAACAGATA GAAGTATATGGCCCTTGATAAGAATCTATCGGGGTGGCTTTCTTCTGATTGAATTCCTTTTTCTACTGGGCATCAACACGTATGGTTGGA GACAGGCTGGAGTAAACCATGTACTCATCTTTGAACTTAATCCGAGAAGCAATTTGTCTCATCAACATCTCTTTGAGATTGCTGGATTCC TCGGGATATTGTGGTGCCTGAGCCTTCTGGCATGCTTCTTTGCTCCAATTAGTGTCATCCCCACATATGTGTATCCACTTGCCCTTTATG GATTTATGGTTTTCTTCCTTATCAACCCCACCAAAACTTTCTACTATAAATCCCGGTTTTGGCTGCTTAAACTGCTGTTTCGAGTATTTA CAGCCCCCTTCCATAAGGTAGGCTTTGCTGATTTCTGGCTGGCGGATCAGCTGAACAGCCTGTCAGTGATACTGATGGACCTGGAATATA TGATCTGCTTCTACAGTTTGGAGCTCAAATGGGATGAAAGTAAGGGCCTGTTGCCAAATAATTCAGAAGAATCAGGAATTTGCCACAAAT ATACATATGGTGTGCGGGCCATTGTTCAGTGCATTCCTGCTTGGCTTCGCTTCATCCAGTGCCTGCGCCGATATCGAGACACAAAAAGGG CCTTTCCTCATTTAGTTAATGCTGGCAAATACTCCACAACTTTCTTCATGGTGACGTTTGCAGCCCTTTACAGCACTCACAAAGAACGAG GTCACTCGGACACTATGGTGTTCTTTTACCTGTGGATTGTCTTTTATATCATCAGTTCCTGCTATACCCTCATCTGGGATCTCAAGATGG ACTGGGGTCTCTTCGATAAGAATGCTGGAGAGAACACTTTCCTCCGGGAAGAGATTGTATACCCCCAAAAAGCCTACTACTACTGTGCCA TAATAGAGGATGTGATTCTGCGCTTTGCTTGGACTATCCAAATCTCGATTACCTCTACAACTTTGTTGCCTCATTCTGGGGACATCATTG CTACTGTCTTTGCCCCACTTGAGGTTTTCCGGCGATTTGTGTGGAACTTCTTCCGCCTGGAGAATGAACATCTGAATAACTGTGGTGAAT TCCGTGCTGTGCGGGACATCTCTGTGGCCCCCCTGAACGCAGATGATCAGACTCTCCTAGAACAGATGATGGACCAGGATGATGGGGTAC GAAACCGCCAGAAGAATCGGTCATGGAAGTACAACCAGAGCATATCCCTGCGCCGGCCTCGCCTCGCTTCTCAATCCAAGGCTCGTGACA CTAAGGTATTGATAGAAGACACAGATGATGAAGCTAACACTTGAATTTTCTGAAGTCTAGCTTAACATCTTTGGTTTTCCTACTCTACAA TCCTTTCCTCGACCAACGCAACCTCTAGTACCTTTCCAGCCGAAAACAGGAGAAAACACATAACACATTTTCCGAGCTCTTCCGGATCGG ATCCTATGGACTCCAAACAAGCTCACTGTGTTTCTTTTCTTTTCTTCTGGTTTAATTTTAATTTTCTATTTTCAAAACAAATATTTACTT CATTTGCCAATCAGAGGATGTTTTAAGAAACAAAACATAGTATCTTATGGATTGTTTACAATCACAAGGACATAGATACCTATCAGGATG AAGAACAGGCATTGCAAGGACCCTCTGATGGGACGGTACTGAGATATCTCGGCTTCCGCTCAGCCCGGTTTTGACTGGTTGAAACCGGAC ATTGGTTTTTAAATTTTTTGTCAGTTTATGTGGAGAATTTTTTTCTTTCCTTCATACCCAGCGCAAAGGCACTGGCCGCACTTGCAGGAA AAGTGCAACTTAAAGCAGTACCTTCATTCATGAAGCTACTTTTTAATTTGATGTAACTTTTCTTATTTTGGGAAGGGTTGCTGGGTGGGT GGGAAATATGATGTATTTGTTACACATAGTTTTCTCATTATTTATGAAACTTAACCATACAGAATGATATAACTCCTGTGCAATGAAGGT GATAACAGTAAAAGAAGGCAGGGGAAACTTACGTTGGATGACATTTATGAGGGTCAGTCCCACATACCTCTTTCAGGAGACAACTTGCAC CAGTTTGACCTTTTCTTTTCTTTGTTTTTATTTTAAGCCAAAGTTTCATTGCTAACTTCTTAAGTTGCTGCTGCTTTAGAGTCCTGAGCA TATCTCTCATAACAAGGAATCCCACACTTCACACCACCGGCTGAATTTCATGGAAGAGGTTCTGATAATTTTTTTAACTTTTTAAGGAAC AGATGTGGAATACACTGGCCCATATTTCAACCTTAACAGCTGAAGCTATGCCTTATTATGCATCCACATGTATGGTCCCTGTAGCGTGAC CTTTACTAGCTCTGAATCAGAAGACAGAGCTATTTCAGAGGCTCTGTGTGCCCTCACTAGATAGTTTTTCTTCTGGGTTCAACCACTTTA GCCAGAATTTGATCAAATTAAAAGTCTGTCATGGGGAAACTATATTTTTGAGCACATGGAACAAATTATACTTCCTCATTCATATTATGT TGATACAAAAGACCTTGGCAGCCATTTCTCCCAGCAGTTTTAAAGGATGAACATTGGATTTCATGCCATCCCATAGAAAACCTGTTTTAA AATTTTAGGGATCTTTACTTGGTCATACATGAAAAGTACACTGCTTAGAAATTATAGACTATTATGATCTGTCCACAGTGCCCATTGTCA CTTCTTTGTCTCATTTCTTCCCTTTGTTCCTTAGTCATCCAAATAAGCCTGAAAACCATAAGAGATATTACTTTATTGAATATGGTTGGC ATTAAATTTAGCATTTCATTATCTAACAAAATTAATATAAATTCCAGGACATGGTAAAATGTGTTTTAATAACCCCCAGACCCAAATGAA AATTTCAAAGTCAATACCAGCAGATTCATGAAAGTAAATTTAGTCCTATAATTTTCAGCTTAATTATAAACAAAGGAACAAATAAGTGGA AGGGCAGCTATTACCATTCGCTTAGTCAAAACATTCGGTTACTGCCCTTTAATACACTCCTATCATCAGCACTTCCACCATGTATTACAA GTCTTGACCCATCCCTGTCGTAACTCCAGTAAAAGTTACTGTTACTAGAAAATTTTTATCAATTAACTGACAAATAGTTTCTTTTTAAAG TAGTTTCTTCCATCTTTATTCTGACTAGCTTCCAAAATGTGTTCCCTTTTTGAATCGAGGTTTTTTTGTTTTGTTTTGTTTTCTGAAAAA ATCATACAACTTTGTGCTTCTATTGCTTTTTTGTGTTTTGTTAAGCATGTCCCTTGGCCCAAATGGAAGAGGAAATGTTTAATTAATGCT TTTTAGTTTAAATAAATTGAATCATTTATAATAATCAGTGTTAACAATTTAGTGACCCTTGGTAGGTTAAAGGTTGCATTATTTATACTT GAGATTTTTTTCCCCTAACTATTCTGTTTTTTGTACTTTAAAACTATGGGGGAAATATCACTGGTCTGTCAAGAAACAGCAGTAATTATT ACTGAGTTAAATTGAAAAGTCCAGTGGACCAGGCATTTCTTATATAAATAAAATTGGTGGTACTAATGTGTATCCAGAGACATTTGAATT ATTAACTTTATTCTTTATTAGTGAATCTCTATGATGGCACACATAAAATCTTACAGTTGACTTCTGCTGTTTGAATTCCCCAGTGTTTTT ACATTAATGTATCCTATGGCCAGTTTAGTCTTTCTTCAACTGTTACAGTTCTAGGCCCTAATCGGTCTTATTCTTTCACATATGATGCAA TGATAGATGTAATCCTTTTCTGAAGTGTTTGTCGGATTGGCATAGTCTATTCAAAAATAAGGCTTATGTAACTTTCAGCTTGCTATGTGA CAGAAGTTTAAATGTGTATTTAAGTACACAGCTTTATGTTCCTCTTAAAGCTGTAAAGAGAAAAGAGGACTTTTCCTCATCAAGAGAGCT TGGGAGACAAAGCATACATTTAATTTTTTTCTTTCAAAAAAGGCAATGTGCACTTTCCTCTCCTAAATTTTATATGCCCTAGCAGTTGCG AATGTTGTGTCCACCTTACAATTTGATTGGAGGCTCCCATACACTGAAACTCTCAATAGAATTCTTAAAAATGAACATCAGCTGGGCGCA GTGGCTCACGTCTGTAATCCCAGCACTTTGGGAGGCCAAGGTGGGCAGATCACGAGCTCAGGAGTTCGAGACCAGCCTGACCAACATGGT GAAACCCCGTCTCTACTAAAAATACAAAAATTAGCTGGGCGTGGTGGCGCATGCCTGTAATCCCAGCTACTCAGGAGACTGAGGCAGGAG AATTGCTTAAACCTGGGAAGCAGAGGTTGCAGTGAGACTAGATCATGCCACTGCACTCCACTGCCACTGCCTGGGCAACAGAGCAAGACT CCATCTCAAAAACAAACAAAAAAAATCACAGACTCATTTTAAAGGAGCAGCAACCTAACCAAATACAAACTTCATTTGATTTTAGGTTTA GAAATTCTAGTTTTATTTTGGAGAATCACTCAGTTTTTAATATCAATTAAAATAGTCCTCAAAGGAATGAAGAGGAAGTCTAAATAAATG AATAAAACTCATTTTTCATTTTATGGTAGTATAGAAAGCATTGATGTGTGTAGAGAAATTAAAAGACAAGAGTGGTTTACTGTCTATGCA CAAATGAGTGCCTATATTTAAGGCTGCCTATACCATTACAGTGGCGTAATTGGTGATTTCATAGCATACAGAGAAACAATGAAATCAAAG ATTAAATCATTAGGTTATGATGGTCATTTGTAAGATTGGAAAGTAGTCAAAAACCCATGTGCAACTTTTTTTCAGCACTCTACCATTAGG CTCCTTAGGCAAGCTGCTTAACTTCCGTCAGAAGCAAAGGTGTGGGAGAACACCATTCACCTCAGTAGTAACTTATAAAAACCGTTTAAA GAAGGAAAATGGGCCCCAGCCCCTAGGATCCCTGACTTCCTGATATTCTAAACATGAGAGTTCTGTGACTTTAACTGAGCTCTTTACCAG AAGTAACGTGTTGATGTATATTATCTTTCCACCATGCTTTGACCAAAGTACATTCACAGTAACTAGAGAAAGCAGATCATCCATTTCTGG ATAGGTGAACTACGGCAAGGTAAGAACAGAAAAAAGTAAAGTAGTATTGAGTCATGCAGGTATCAGATTAACAGACATTCATTGAAGATA AGTGAAATAACTTTGGAGATAGCAGAGAAGAGCAAGAAGATGTTGGACGTTGGCAGTAAGCCGCAGCAAGGCAGTTAGGAAAGTACGTGA GCCTGATGACGCGAAGGTGATGCCTCCATCTGTCCGCTTAGTGGTTATTAAATTGCTGGGAAAGACCTGCCTTTATTTTCCTATCATCTT TTCCCCATTACTTTGCATTATTCAGGAAATAATTATTTATTCCCAAATGCAGTTTTCTAAGCAGTGACCACAAAAATTTCTTATTCTAAG GAAAATAATTAAGAAAATAACCCTATCTGTGGCATTGTTTTAAAATGTGGGATACAAATCTGGTAAGCAGATTTCAGGTCATTATTGTTT TCTTTTAATGGTTTACATGAAATTTTCCTCTCAAAAAACAAGTACCTCTTACTGTATAACATTTGAGTTTTTAAAATTGAGATAGCTTCT GAGAGAAAAAAAGAAAGTTGAATTTGTAGCTCTCTCAACTACAAAATCACCACACTTTTCTCAAATCTAATAGGAATTCAGTGTCATTAC AGATACGGTCTCTAGTGACCAGGTGAAGACAATTGAAGCTTTGGTAGAACTTCTGCAAAAGTTAGGTAAAGAAGAATAAATCCAATTTAT CAGCTTTGTTCATATTCTTTTTCTGAGTGATCTAAATTTTTCTTGCTTGTGATCATTAGCTAAGACAAGGAGTGTTAAAGGTTTTCTAAA TGCAAACACATACGTTTCTGCATTTCAGAAGTCAGTCTTGATAGTAAATCCTACAAACTCTCAACTTTCTGCATTGTATTTTAAGTCTAT ACAATGTTAGAGCACATGGTAATGTGGGACAGAGTTGATTATATGATAGCATGAGAAATTTGCACTGGTTGGTGTTACAAACCAATGATC TAACCAGCATACTCTCCAAATATGAAAAAGTAAAAACATCTCATTAGGTTTGGAAGAGCCTAAGGTAGTCCATACAGGAAGTTTGCCTGA GAACAGAGAGGAGCCTGTTGATTGGCATGGCTCATGATGCCTTTCTTGTATGAATTATCAAAACTGATACTGAGTTGCCCCTAGAAAACA GCCTTCCAGAATATGTTTGCTTATTTATGGTGGAAGAGGAAGTGAGAACAAGATGAGAGTTTCCCTTAAAGTGAGGAGAATCATTATACA ACTTTGCAACTGAAGTATGTTTCCTTGTAGCAAACCTTTGGTTTATTTTATCGTCAAATCAAAGCCACCCTCAGTTTTTTGTATGGTGCC ATCAACCTAAGGAGGACAATCAAATAACCTATTGTTTGCCTTGGATTCAGTATATCTACTAAGTACACAGTCTCGTATACCTTCAAACCA GTTACTTGACTAAACTTCCACCATATTACCTTAGTTCTTCCTATGCCCTTTCCTCTGTGCCACCACAAATCAGAGGGAAAGACTGCAGTA CTTCTAGAAAGTTGTGGACTTCTAAGAAAGAGCCAGGCTTCCATCTCACTATCCCTTGATCATTATCTCTGAAGTCCCTACCTGCACTTC CCTGATTGCCCTGTAGCAACACCAGCATGGTGGGAATTGGAGGGAAAGATTGTGGAAAATCCCTAAAGGGTCATTGTTCATGACGTTATT TTGAATTGTAAAATTGTGAGCCACTCACATTTCAATTACAAAAAGAACAGAACTCTTAATCACGTTGTGAAGATTAATTTCTTGCCATTT TCTGGATTAGCACAGAAGCTTGTATCAATGAAGAGTATTTAGGAGGGGAATAGGGGGAAAATTGTATTTTTCTGTTCAAACTCTCTCCTT CTATTTGTGTTTCCTTCACCCCTCTCCCTCCACCACACTGTGTTGGTCAAATGATACTATTCTATGAAACCCTGAGCTATAGTTGTCAAA AGTAACTATGGATTGTGATTAGTCCTCTGTGGGTGCTTCCTTCCCTCTTTTTTCTCCCTCACTCTCTTTCTCAGTCTCCACTCTGGTAAA TGGAAAAGGTGATGTCGAAGAATTGATGCTTGCTTGGACTCATGTTGTATCTGTGACTATGCTATTAGCTGTCTCCTTTTTCCTTCCTAT ATGGGTAGAATTCTTATGACTTATGAAGTTCCCTTCTTGATAAGTAGGTTAAATGCACAATGTGGTACTGTTTTATAGTTTCTAGATGGT TGTATAAAGCAAAAAGTTAATGTGGTTATGTGTTTTTAAAGAAAATAAGTGATTGGTCACTAAACTCTGTCCTTTTTTCATTATTACAAT >22735_22735_2_DHX9-XPR1_DHX9_chr1_182829319_ENST00000367549_XPR1_chr1_180651495_ENST00000367590_length(amino acids)=1117AA_BP=444 MGDVKNFLYAWCGKRKMTPSYEIRAVGNKNRQKFMCEVQVEGYNYTGMGNSTNKKDAQSNAARDFVNYLVRINEIKSEEVPAFGVASPPP LTDTPDTTANAEGDLPTTMGGPLPPHLALKAENNSEVGASGYGVPGPTWDRGANLKDYYSRKEEQEVQATLESEEVDLNAGLHGNWTLEN AKARLNQYFQKEKIQGEYKYTQVGPDHNRSFIAEMTIYIKQLGRRIFAREHGSNKKLAAQSCALSLVRQLYHLGVVEAYSGLTKKKEGET VEPYKVNLSQDLEHQLQNIIQELNLEILPPPEDPSVPVALNIGKLAQFEPSQRQNQVGVVPWSPPQSNWNPWTSSNIDEGPLAFATPEQI SMDLKNELMYQLEQDHDLQAILQERELLPVKKFESEILEAISQNSVVIIRGATGCGKTTQVPQFILDDFIQNDRAAECNIVVTQAFKDML YSAQDQAPSVEVTDEDTVKRYFAKFEEKFFQTCEKELAKINTFYSEKLAEAQRRFATLQNELQSSLDAQKESTGVTTLRQRRKPVFHLSH EERVQHRNIKDLKLAFSEFYLSLILLQNYQNLNFTGFRKILKKHDKILETSRGADWRVAHVEVAPFYTCKKINQLISETEAVVTNELEDG DRQKAMKRLRVPPLGAAQPAPAWTTFRVGLFCGIFIVLNITLVLAAVFKLETDRSIWPLIRIYRGGFLLIEFLFLLGINTYGWRQAGVNH VLIFELNPRSNLSHQHLFEIAGFLGILWCLSLLACFFAPISVIPTYVYPLALYGFMVFFLINPTKTFYYKSRFWLLKLLFRVFTAPFHKV GFADFWLADQLNSLSVILMDLEYMICFYSLELKWDESKGLLPNNSEESGICHKYTYGVRAIVQCIPAWLRFIQCLRRYRDTKRAFPHLVN AGKYSTTFFMVTFAALYSTHKERGHSDTMVFFYLWIVFYIISSCYTLIWDLKMDWGLFDKNAGENTFLREEIVYPQKAYYYCAIIEDVIL RFAWTIQISITSTTLLPHSGDIIATVFAPLEVFRRFVWNFFRLENEHLNNCGEFRAVRDISVAPLNADDQTLLEQMMDQDDGVRNRQKNR -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for DHX9-XPR1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 230_325 | 444.0 | 1271.0 | BRCA1 |
| Hgene | DHX9 | chr1:182829319 | chr1:180651495 | ENST00000367549 | + | 12 | 28 | 1_250 | 444.0 | 1271.0 | CREBBP |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for DHX9-XPR1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for DHX9-XPR1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
