
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:DIAPH1-BRIX1 (FusionGDB2 ID:22739) |
Fusion Gene Summary for DIAPH1-BRIX1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: DIAPH1-BRIX1 | Fusion gene ID: 22739 | Hgene | Tgene | Gene symbol | DIAPH1 | BRIX1 | Gene ID | 1729 | 55299 |
| Gene name | diaphanous related formin 1 | biogenesis of ribosomes BRX1 | |
| Synonyms | DFNA1|DIA1|DRF1|LFHL1|SCBMS|hDIA1 | BRIX|BXDC2 | |
| Cytomap | 5q31.3 | 5p13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein diaphanous homolog 1 | ribosome biogenesis protein BRX1 homologBRX1, biogenesis of ribosomes, homologbrix domain containing 2brix domain-containing protein 2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | O60610 | Q8TDN6 | |
| Ensembl transtripts involved in fusion gene | ENST00000253811, ENST00000389054, ENST00000389057, ENST00000398557, ENST00000398562, ENST00000398566, ENST00000518047, ENST00000520569, ENST00000494967, | ENST00000506023, ENST00000336767, | |
| Fusion gene scores | * DoF score | 6 X 6 X 5=180 | 2 X 2 X 2=8 |
| # samples | 7 | 2 | |
| ** MAII score | log2(7/180*10)=-1.36257007938471 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(2/8*10)=1.32192809488736 | |
| Context | PubMed: DIAPH1 [Title/Abstract] AND BRIX1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | DIAPH1(140963691)-BRIX1(34924952), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across DIAPH1 (5'-gene) Fusion gene breakpoints across DIAPH1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
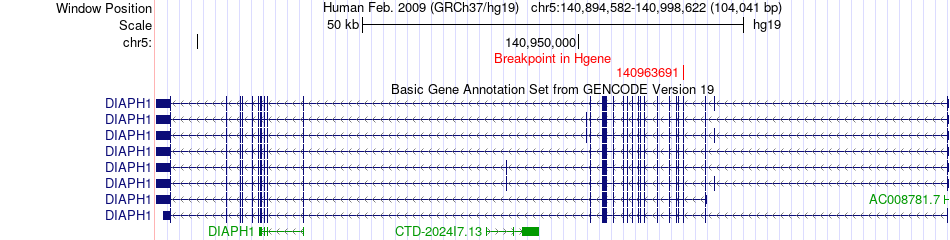 |
 Fusion gene breakpoints across BRIX1 (3'-gene) Fusion gene breakpoints across BRIX1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
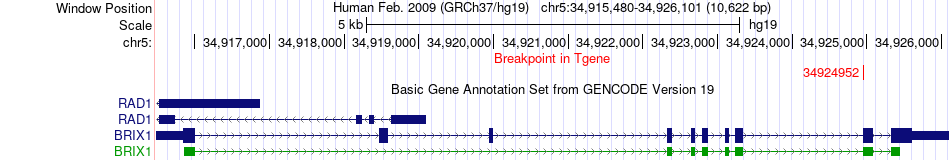 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-EW-A1P4-01A | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
Top |
Fusion Gene ORF analysis for DIAPH1-BRIX1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000253811 | ENST00000506023 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
| 5CDS-3UTR | ENST00000389054 | ENST00000506023 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
| 5CDS-3UTR | ENST00000389057 | ENST00000506023 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
| 5CDS-3UTR | ENST00000398557 | ENST00000506023 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
| 5CDS-3UTR | ENST00000398562 | ENST00000506023 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
| 5CDS-3UTR | ENST00000398566 | ENST00000506023 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
| 5CDS-3UTR | ENST00000518047 | ENST00000506023 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
| 5CDS-3UTR | ENST00000520569 | ENST00000506023 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
| In-frame | ENST00000253811 | ENST00000336767 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
| In-frame | ENST00000389054 | ENST00000336767 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
| In-frame | ENST00000389057 | ENST00000336767 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
| In-frame | ENST00000398557 | ENST00000336767 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
| In-frame | ENST00000398562 | ENST00000336767 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
| In-frame | ENST00000398566 | ENST00000336767 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
| In-frame | ENST00000518047 | ENST00000336767 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
| In-frame | ENST00000520569 | ENST00000336767 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
| intron-3CDS | ENST00000494967 | ENST00000336767 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
| intron-3UTR | ENST00000494967 | ENST00000506023 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000389054 | DIAPH1 | chr5 | 140963691 | - | ENST00000336767 | BRIX1 | chr5 | 34924952 | + | 1443 | 543 | 87 | 941 | 284 |
| ENST00000520569 | DIAPH1 | chr5 | 140963691 | - | ENST00000336767 | BRIX1 | chr5 | 34924952 | + | 1158 | 258 | 0 | 656 | 218 |
| ENST00000253811 | DIAPH1 | chr5 | 140963691 | - | ENST00000336767 | BRIX1 | chr5 | 34924952 | + | 1443 | 543 | 87 | 941 | 284 |
| ENST00000398566 | DIAPH1 | chr5 | 140963691 | - | ENST00000336767 | BRIX1 | chr5 | 34924952 | + | 1416 | 516 | 87 | 914 | 275 |
| ENST00000398562 | DIAPH1 | chr5 | 140963691 | - | ENST00000336767 | BRIX1 | chr5 | 34924952 | + | 1416 | 516 | 87 | 914 | 275 |
| ENST00000398557 | DIAPH1 | chr5 | 140963691 | - | ENST00000336767 | BRIX1 | chr5 | 34924952 | + | 1443 | 543 | 87 | 941 | 284 |
| ENST00000389057 | DIAPH1 | chr5 | 140963691 | - | ENST00000336767 | BRIX1 | chr5 | 34924952 | + | 1416 | 516 | 87 | 914 | 275 |
| ENST00000518047 | DIAPH1 | chr5 | 140963691 | - | ENST00000336767 | BRIX1 | chr5 | 34924952 | + | 1275 | 375 | 0 | 773 | 257 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000389054 | ENST00000336767 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + | 0.003282598 | 0.9967174 |
| ENST00000520569 | ENST00000336767 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + | 0.004748164 | 0.9952518 |
| ENST00000253811 | ENST00000336767 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + | 0.003282598 | 0.9967174 |
| ENST00000398566 | ENST00000336767 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + | 0.002986233 | 0.9970138 |
| ENST00000398562 | ENST00000336767 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + | 0.002986233 | 0.9970138 |
| ENST00000398557 | ENST00000336767 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + | 0.003282598 | 0.9967174 |
| ENST00000389057 | ENST00000336767 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + | 0.002986233 | 0.9970138 |
| ENST00000518047 | ENST00000336767 | DIAPH1 | chr5 | 140963691 | - | BRIX1 | chr5 | 34924952 | + | 0.002243735 | 0.9977563 |
Top |
Fusion Genomic Features for DIAPH1-BRIX1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| DIAPH1 | chr5 | 140963690 | - | BRIX1 | chr5 | 34924951 | + | 8.62E-07 | 0.99999917 |
| DIAPH1 | chr5 | 140963690 | - | BRIX1 | chr5 | 34924951 | + | 8.62E-07 | 0.99999917 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
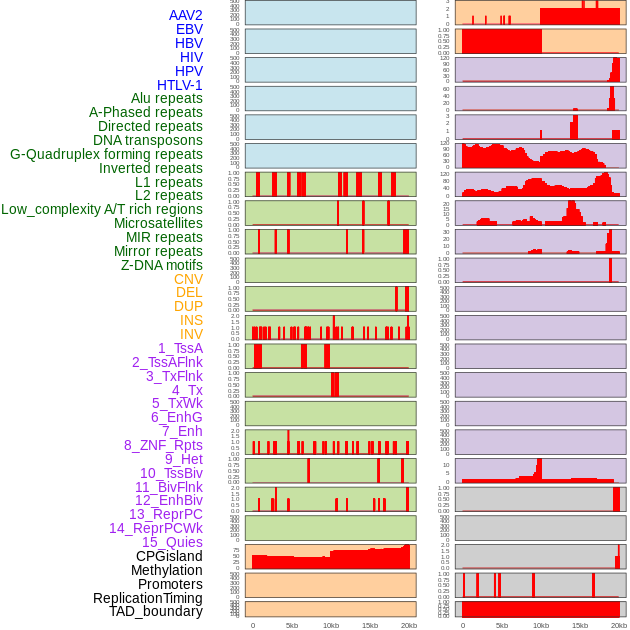 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
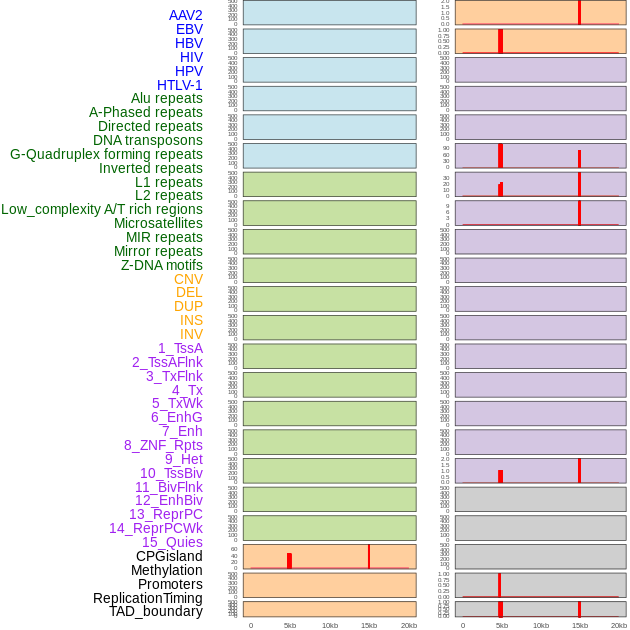 |
Top |
Fusion Protein Features for DIAPH1-BRIX1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:140963691/chr5:34924952) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| DIAPH1 | BRIX1 |
| FUNCTION: Actin nucleation and elongation factor required for the assembly of F-actin structures, such as actin cables and stress fibers (By similarity). Binds to the barbed end of the actin filament and slows down actin polymerization and depolymerization (By similarity). Required for cytokinesis, and transcriptional activation of the serum response factor (By similarity). DFR proteins couple Rho and Src tyrosine kinase during signaling and the regulation of actin dynamics (By similarity). Functions as a scaffold protein for MAPRE1 and APC to stabilize microtubules and promote cell migration (By similarity). Has neurite outgrowth promoting activity. Acts in a Rho-dependent manner to recruit PFY1 to the membrane (By similarity). In hear cells, it may play a role in the regulation of actin polymerization in hair cells (PubMed:20937854, PubMed:21834987, PubMed:26912466). The MEMO1-RHOA-DIAPH1 signaling pathway plays an important role in ERBB2-dependent stabilization of microtubules at the cell cortex (PubMed:20937854, PubMed:21834987). It controls the localization of APC and CLASP2 to the cell membrane, via the regulation of GSK3B activity (PubMed:20937854, PubMed:21834987). In turn, membrane-bound APC allows the localization of the MACF1 to the cell membrane, which is required for microtubule capture and stabilization (PubMed:20937854, PubMed:21834987). Plays a role in the regulation of cell morphology and cytoskeletal organization. Required in the control of cell shape (PubMed:20937854, PubMed:21834987). Plays a role in brain development (PubMed:24781755). Also acts as an actin nucleation and elongation factor in the nucleus by promoting nuclear actin polymerization inside the nucleus to drive serum-dependent SRF-MRTFA activity (By similarity). {ECO:0000250|UniProtKB:O08808, ECO:0000269|PubMed:20937854, ECO:0000269|PubMed:21834987, ECO:0000269|PubMed:24781755, ECO:0000269|PubMed:26912466}. | FUNCTION: Required for biogenesis of the 60S ribosomal subunit. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DIAPH1 | chr5:140963691 | chr5:34924952 | ENST00000389057 | - | 3 | 27 | 1039_1196 | 125 | 1264.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DIAPH1 | chr5:140963691 | chr5:34924952 | ENST00000389057 | - | 3 | 27 | 468_572 | 125 | 1264.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DIAPH1 | chr5:140963691 | chr5:34924952 | ENST00000398557 | - | 4 | 28 | 1039_1196 | 134 | 1273.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DIAPH1 | chr5:140963691 | chr5:34924952 | ENST00000398557 | - | 4 | 28 | 468_572 | 134 | 1273.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DIAPH1 | chr5:140963691 | chr5:34924952 | ENST00000389057 | - | 3 | 27 | 1213_1216 | 125 | 1264.0 | Compositional bias | Note=Arg/Lys-rich (basic) |
| Hgene | DIAPH1 | chr5:140963691 | chr5:34924952 | ENST00000398557 | - | 4 | 28 | 1213_1216 | 134 | 1273.0 | Compositional bias | Note=Arg/Lys-rich (basic) |
| Hgene | DIAPH1 | chr5:140963691 | chr5:34924952 | ENST00000389057 | - | 3 | 27 | 1194_1222 | 125 | 1264.0 | Domain | DAD |
| Hgene | DIAPH1 | chr5:140963691 | chr5:34924952 | ENST00000389057 | - | 3 | 27 | 583_764 | 125 | 1264.0 | Domain | Note=FH1 |
| Hgene | DIAPH1 | chr5:140963691 | chr5:34924952 | ENST00000389057 | - | 3 | 27 | 769_1171 | 125 | 1264.0 | Domain | FH2 |
| Hgene | DIAPH1 | chr5:140963691 | chr5:34924952 | ENST00000389057 | - | 3 | 27 | 84_449 | 125 | 1264.0 | Domain | GBD/FH3 |
| Hgene | DIAPH1 | chr5:140963691 | chr5:34924952 | ENST00000398557 | - | 4 | 28 | 1194_1222 | 134 | 1273.0 | Domain | DAD |
| Hgene | DIAPH1 | chr5:140963691 | chr5:34924952 | ENST00000398557 | - | 4 | 28 | 583_764 | 134 | 1273.0 | Domain | Note=FH1 |
| Hgene | DIAPH1 | chr5:140963691 | chr5:34924952 | ENST00000398557 | - | 4 | 28 | 769_1171 | 134 | 1273.0 | Domain | FH2 |
| Hgene | DIAPH1 | chr5:140963691 | chr5:34924952 | ENST00000398557 | - | 4 | 28 | 84_449 | 134 | 1273.0 | Domain | GBD/FH3 |
| Tgene | BRIX1 | chr5:140963691 | chr5:34924952 | ENST00000336767 | 7 | 10 | 60_249 | 221 | 354.0 | Domain | Brix |
Top |
Fusion Gene Sequence for DIAPH1-BRIX1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >22739_22739_1_DIAPH1-BRIX1_DIAPH1_chr5_140963691_ENST00000253811_BRIX1_chr5_34924952_ENST00000336767_length(transcript)=1443nt_BP=543nt ATTCATGAGGCGCGCGCAGCCCGGCATGCTAATGAGGCGGGGCGCGGCGGCTGGCTAAAGAGCGACTGGGCGGCGGCGGGCGCGGAGCTG CCAGGCGGGAGCGGCGTAGGCGCGGGGTCGCCGGCCAGCGTGAACCGGGACATGGAGCCGCCCGGCGGGAGCCTGGGGCCCGGCCGCGGG ACCCGGGACAAGAAGAAGGGCCGGAGCCCAGATGAGCTGCCCTCGGCGGGCGGCGACGGCGGCAAATCTAAGAAATTTACTCTGAAGCGG CTCATGGCAGATGAGCTGGAGAGATTTACCAGCATGAGAATTAAGAAGGAGAAGGAAAAGCCCAATTCTGCTCATAGAAATTCTTCTGCA TCATATGGGGATGATCCCACAGCACAGTCATTGCAAGATGTTTCAGATGAACAAGTGCTGGTTCTCTTTGAACAGATGCTGCTGGATATG AACCTGAATGAGGAGAAACAGCAACCTTTGAGGGAGAAGGACATCATCATCAAGAGGGAGATGGTGTCCCAATACTTGTACACCTCCAAG GCTATCATAGAAGAAGATGCTGCTCTTGTAGAAATAGGACCTCGTTTTGTCTTAAATCTCATAAAGATTTTCCAGGGAAGTTTTGGAGGA CCAACTTTATATGAAAATCCTCACTACCAGTCACCAAACATGCATCGGCGTGTCATAAGATCCATCACAGCTGCAAAATACAGAGAGAAA CAGCAAGTGAAAGATGTGCAAAAACTGAGAAAGAAAGAGCCGAAGACTCTTCTTCCACATGATCCCACTGCAGATGTTTTTGTAACACCA GCTGAGGAGAAACCAATAGAAATACAGTGGGTAAAACCAGAGCCAAAAGTTGATTTGAAAGCAAGAAAGAAACGGATTTACAAAAGGCAA AGAAAAATGAAACAGAGGATGGACAGTGGGAAAACAAAATAAGTCAATGGAAACCTGATTTGTTTTTCAGTTACTTTATATTTATTTTGT ATTCAATGTGTAAATACTTTTATTATCTAATACTATCTTACGTCTAATTAGTGTAGCATTTACAAGAAAGAAAAATTAAGATCTTAAAAT CAGTGATTATCTTTTTCTAAATAAAATATCACCAGAATTCATCAGTTAATTTCTGATTTCTTTTTGAAGTTTGTGTTGCTAAAAATGTAG CACACTTAATGTAGCCTGTTCTCTTGGGTTGGAATTTTTGGTTTAGCAAAGCTGAAATTCAGACATTTATTAGGTCATATTATTTGTAAA AGCATTCAACACTTCAAGAGCATCGGTTGTGGATGGTAAAGTAGGAGTAGACTGGTAGAAAGGAAGGCATCTCACTGAAGTTTACTCAGT CATCAGTTAATGAAGCCATGGAAGAACCAACCCCTGATCTGTCACTTCAAAAAAGAGTATATTAAAATGTTGAGGTTTTGAGAATCAAAT >22739_22739_1_DIAPH1-BRIX1_DIAPH1_chr5_140963691_ENST00000253811_BRIX1_chr5_34924952_ENST00000336767_length(amino acids)=284AA_BP=152 MPGGSGVGAGSPASVNRDMEPPGGSLGPGRGTRDKKKGRSPDELPSAGGDGGKSKKFTLKRLMADELERFTSMRIKKEKEKPNSAHRNSS ASYGDDPTAQSLQDVSDEQVLVLFEQMLLDMNLNEEKQQPLREKDIIIKREMVSQYLYTSKAIIEEDAALVEIGPRFVLNLIKIFQGSFG GPTLYENPHYQSPNMHRRVIRSITAAKYREKQQVKDVQKLRKKEPKTLLPHDPTADVFVTPAEEKPIEIQWVKPEPKVDLKARKKRIYKR -------------------------------------------------------------- >22739_22739_2_DIAPH1-BRIX1_DIAPH1_chr5_140963691_ENST00000389054_BRIX1_chr5_34924952_ENST00000336767_length(transcript)=1443nt_BP=543nt ATTCATGAGGCGCGCGCAGCCCGGCATGCTAATGAGGCGGGGCGCGGCGGCTGGCTAAAGAGCGACTGGGCGGCGGCGGGCGCGGAGCTG CCAGGCGGGAGCGGCGTAGGCGCGGGGTCGCCGGCCAGCGTGAACCGGGACATGGAGCCGCCCGGCGGGAGCCTGGGGCCCGGCCGCGGG ACCCGGGACAAGAAGAAGGGCCGGAGCCCAGATGAGCTGCCCTCGGCGGGCGGCGACGGCGGCAAATCTAAGAAATTTACTCTGAAGCGG CTCATGGCAGATGAGCTGGAGAGATTTACCAGCATGAGAATTAAGAAGGAGAAGGAAAAGCCCAATTCTGCTCATAGAAATTCTTCTGCA TCATATGGGGATGATCCCACAGCACAGTCATTGCAAGATGTTTCAGATGAACAAGTGCTGGTTCTCTTTGAACAGATGCTGCTGGATATG AACCTGAATGAGGAGAAACAGCAACCTTTGAGGGAGAAGGACATCATCATCAAGAGGGAGATGGTGTCCCAATACTTGTACACCTCCAAG GCTATCATAGAAGAAGATGCTGCTCTTGTAGAAATAGGACCTCGTTTTGTCTTAAATCTCATAAAGATTTTCCAGGGAAGTTTTGGAGGA CCAACTTTATATGAAAATCCTCACTACCAGTCACCAAACATGCATCGGCGTGTCATAAGATCCATCACAGCTGCAAAATACAGAGAGAAA CAGCAAGTGAAAGATGTGCAAAAACTGAGAAAGAAAGAGCCGAAGACTCTTCTTCCACATGATCCCACTGCAGATGTTTTTGTAACACCA GCTGAGGAGAAACCAATAGAAATACAGTGGGTAAAACCAGAGCCAAAAGTTGATTTGAAAGCAAGAAAGAAACGGATTTACAAAAGGCAA AGAAAAATGAAACAGAGGATGGACAGTGGGAAAACAAAATAAGTCAATGGAAACCTGATTTGTTTTTCAGTTACTTTATATTTATTTTGT ATTCAATGTGTAAATACTTTTATTATCTAATACTATCTTACGTCTAATTAGTGTAGCATTTACAAGAAAGAAAAATTAAGATCTTAAAAT CAGTGATTATCTTTTTCTAAATAAAATATCACCAGAATTCATCAGTTAATTTCTGATTTCTTTTTGAAGTTTGTGTTGCTAAAAATGTAG CACACTTAATGTAGCCTGTTCTCTTGGGTTGGAATTTTTGGTTTAGCAAAGCTGAAATTCAGACATTTATTAGGTCATATTATTTGTAAA AGCATTCAACACTTCAAGAGCATCGGTTGTGGATGGTAAAGTAGGAGTAGACTGGTAGAAAGGAAGGCATCTCACTGAAGTTTACTCAGT CATCAGTTAATGAAGCCATGGAAGAACCAACCCCTGATCTGTCACTTCAAAAAAGAGTATATTAAAATGTTGAGGTTTTGAGAATCAAAT >22739_22739_2_DIAPH1-BRIX1_DIAPH1_chr5_140963691_ENST00000389054_BRIX1_chr5_34924952_ENST00000336767_length(amino acids)=284AA_BP=152 MPGGSGVGAGSPASVNRDMEPPGGSLGPGRGTRDKKKGRSPDELPSAGGDGGKSKKFTLKRLMADELERFTSMRIKKEKEKPNSAHRNSS ASYGDDPTAQSLQDVSDEQVLVLFEQMLLDMNLNEEKQQPLREKDIIIKREMVSQYLYTSKAIIEEDAALVEIGPRFVLNLIKIFQGSFG GPTLYENPHYQSPNMHRRVIRSITAAKYREKQQVKDVQKLRKKEPKTLLPHDPTADVFVTPAEEKPIEIQWVKPEPKVDLKARKKRIYKR -------------------------------------------------------------- >22739_22739_3_DIAPH1-BRIX1_DIAPH1_chr5_140963691_ENST00000389057_BRIX1_chr5_34924952_ENST00000336767_length(transcript)=1416nt_BP=516nt ATTCATGAGGCGCGCGCAGCCCGGCATGCTAATGAGGCGGGGCGCGGCGGCTGGCTAAAGAGCGACTGGGCGGCGGCGGGCGCGGAGCTG CCAGGCGGGAGCGGCGTAGGCGCGGGGTCGCCGGCCAGCGTGAACCGGGACATGGAGCCGCCCGGCGGGAGCCTGGGGCCCGGCCGCGGG ACCCGGGACAAGAAGAAGGGCCGGAGCCCAGATGAGCTGCCCTCGGCGGGCGGCGACGGCGGCAAATCTAAGAAATTTCTGGAGAGATTT ACCAGCATGAGAATTAAGAAGGAGAAGGAAAAGCCCAATTCTGCTCATAGAAATTCTTCTGCATCATATGGGGATGATCCCACAGCACAG TCATTGCAAGATGTTTCAGATGAACAAGTGCTGGTTCTCTTTGAACAGATGCTGCTGGATATGAACCTGAATGAGGAGAAACAGCAACCT TTGAGGGAGAAGGACATCATCATCAAGAGGGAGATGGTGTCCCAATACTTGTACACCTCCAAGGCTATCATAGAAGAAGATGCTGCTCTT GTAGAAATAGGACCTCGTTTTGTCTTAAATCTCATAAAGATTTTCCAGGGAAGTTTTGGAGGACCAACTTTATATGAAAATCCTCACTAC CAGTCACCAAACATGCATCGGCGTGTCATAAGATCCATCACAGCTGCAAAATACAGAGAGAAACAGCAAGTGAAAGATGTGCAAAAACTG AGAAAGAAAGAGCCGAAGACTCTTCTTCCACATGATCCCACTGCAGATGTTTTTGTAACACCAGCTGAGGAGAAACCAATAGAAATACAG TGGGTAAAACCAGAGCCAAAAGTTGATTTGAAAGCAAGAAAGAAACGGATTTACAAAAGGCAAAGAAAAATGAAACAGAGGATGGACAGT GGGAAAACAAAATAAGTCAATGGAAACCTGATTTGTTTTTCAGTTACTTTATATTTATTTTGTATTCAATGTGTAAATACTTTTATTATC TAATACTATCTTACGTCTAATTAGTGTAGCATTTACAAGAAAGAAAAATTAAGATCTTAAAATCAGTGATTATCTTTTTCTAAATAAAAT ATCACCAGAATTCATCAGTTAATTTCTGATTTCTTTTTGAAGTTTGTGTTGCTAAAAATGTAGCACACTTAATGTAGCCTGTTCTCTTGG GTTGGAATTTTTGGTTTAGCAAAGCTGAAATTCAGACATTTATTAGGTCATATTATTTGTAAAAGCATTCAACACTTCAAGAGCATCGGT TGTGGATGGTAAAGTAGGAGTAGACTGGTAGAAAGGAAGGCATCTCACTGAAGTTTACTCAGTCATCAGTTAATGAAGCCATGGAAGAAC >22739_22739_3_DIAPH1-BRIX1_DIAPH1_chr5_140963691_ENST00000389057_BRIX1_chr5_34924952_ENST00000336767_length(amino acids)=275AA_BP=143 MPGGSGVGAGSPASVNRDMEPPGGSLGPGRGTRDKKKGRSPDELPSAGGDGGKSKKFLERFTSMRIKKEKEKPNSAHRNSSASYGDDPTA QSLQDVSDEQVLVLFEQMLLDMNLNEEKQQPLREKDIIIKREMVSQYLYTSKAIIEEDAALVEIGPRFVLNLIKIFQGSFGGPTLYENPH YQSPNMHRRVIRSITAAKYREKQQVKDVQKLRKKEPKTLLPHDPTADVFVTPAEEKPIEIQWVKPEPKVDLKARKKRIYKRQRKMKQRMD -------------------------------------------------------------- >22739_22739_4_DIAPH1-BRIX1_DIAPH1_chr5_140963691_ENST00000398557_BRIX1_chr5_34924952_ENST00000336767_length(transcript)=1443nt_BP=543nt ATTCATGAGGCGCGCGCAGCCCGGCATGCTAATGAGGCGGGGCGCGGCGGCTGGCTAAAGAGCGACTGGGCGGCGGCGGGCGCGGAGCTG CCAGGCGGGAGCGGCGTAGGCGCGGGGTCGCCGGCCAGCGTGAACCGGGACATGGAGCCGCCCGGCGGGAGCCTGGGGCCCGGCCGCGGG ACCCGGGACAAGAAGAAGGGCCGGAGCCCAGATGAGCTGCCCTCGGCGGGCGGCGACGGCGGCAAATCTAAGAAATTTACTCTGAAGCGG CTCATGGCAGATGAGCTGGAGAGATTTACCAGCATGAGAATTAAGAAGGAGAAGGAAAAGCCCAATTCTGCTCATAGAAATTCTTCTGCA TCATATGGGGATGATCCCACAGCACAGTCATTGCAAGATGTTTCAGATGAACAAGTGCTGGTTCTCTTTGAACAGATGCTGCTGGATATG AACCTGAATGAGGAGAAACAGCAACCTTTGAGGGAGAAGGACATCATCATCAAGAGGGAGATGGTGTCCCAATACTTGTACACCTCCAAG GCTATCATAGAAGAAGATGCTGCTCTTGTAGAAATAGGACCTCGTTTTGTCTTAAATCTCATAAAGATTTTCCAGGGAAGTTTTGGAGGA CCAACTTTATATGAAAATCCTCACTACCAGTCACCAAACATGCATCGGCGTGTCATAAGATCCATCACAGCTGCAAAATACAGAGAGAAA CAGCAAGTGAAAGATGTGCAAAAACTGAGAAAGAAAGAGCCGAAGACTCTTCTTCCACATGATCCCACTGCAGATGTTTTTGTAACACCA GCTGAGGAGAAACCAATAGAAATACAGTGGGTAAAACCAGAGCCAAAAGTTGATTTGAAAGCAAGAAAGAAACGGATTTACAAAAGGCAA AGAAAAATGAAACAGAGGATGGACAGTGGGAAAACAAAATAAGTCAATGGAAACCTGATTTGTTTTTCAGTTACTTTATATTTATTTTGT ATTCAATGTGTAAATACTTTTATTATCTAATACTATCTTACGTCTAATTAGTGTAGCATTTACAAGAAAGAAAAATTAAGATCTTAAAAT CAGTGATTATCTTTTTCTAAATAAAATATCACCAGAATTCATCAGTTAATTTCTGATTTCTTTTTGAAGTTTGTGTTGCTAAAAATGTAG CACACTTAATGTAGCCTGTTCTCTTGGGTTGGAATTTTTGGTTTAGCAAAGCTGAAATTCAGACATTTATTAGGTCATATTATTTGTAAA AGCATTCAACACTTCAAGAGCATCGGTTGTGGATGGTAAAGTAGGAGTAGACTGGTAGAAAGGAAGGCATCTCACTGAAGTTTACTCAGT CATCAGTTAATGAAGCCATGGAAGAACCAACCCCTGATCTGTCACTTCAAAAAAGAGTATATTAAAATGTTGAGGTTTTGAGAATCAAAT >22739_22739_4_DIAPH1-BRIX1_DIAPH1_chr5_140963691_ENST00000398557_BRIX1_chr5_34924952_ENST00000336767_length(amino acids)=284AA_BP=152 MPGGSGVGAGSPASVNRDMEPPGGSLGPGRGTRDKKKGRSPDELPSAGGDGGKSKKFTLKRLMADELERFTSMRIKKEKEKPNSAHRNSS ASYGDDPTAQSLQDVSDEQVLVLFEQMLLDMNLNEEKQQPLREKDIIIKREMVSQYLYTSKAIIEEDAALVEIGPRFVLNLIKIFQGSFG GPTLYENPHYQSPNMHRRVIRSITAAKYREKQQVKDVQKLRKKEPKTLLPHDPTADVFVTPAEEKPIEIQWVKPEPKVDLKARKKRIYKR -------------------------------------------------------------- >22739_22739_5_DIAPH1-BRIX1_DIAPH1_chr5_140963691_ENST00000398562_BRIX1_chr5_34924952_ENST00000336767_length(transcript)=1416nt_BP=516nt ATTCATGAGGCGCGCGCAGCCCGGCATGCTAATGAGGCGGGGCGCGGCGGCTGGCTAAAGAGCGACTGGGCGGCGGCGGGCGCGGAGCTG CCAGGCGGGAGCGGCGTAGGCGCGGGGTCGCCGGCCAGCGTGAACCGGGACATGGAGCCGCCCGGCGGGAGCCTGGGGCCCGGCCGCGGG ACCCGGGACAAGAAGAAGGGCCGGAGCCCAGATGAGCTGCCCTCGGCGGGCGGCGACGGCGGCAAATCTAAGAAATTTCTGGAGAGATTT ACCAGCATGAGAATTAAGAAGGAGAAGGAAAAGCCCAATTCTGCTCATAGAAATTCTTCTGCATCATATGGGGATGATCCCACAGCACAG TCATTGCAAGATGTTTCAGATGAACAAGTGCTGGTTCTCTTTGAACAGATGCTGCTGGATATGAACCTGAATGAGGAGAAACAGCAACCT TTGAGGGAGAAGGACATCATCATCAAGAGGGAGATGGTGTCCCAATACTTGTACACCTCCAAGGCTATCATAGAAGAAGATGCTGCTCTT GTAGAAATAGGACCTCGTTTTGTCTTAAATCTCATAAAGATTTTCCAGGGAAGTTTTGGAGGACCAACTTTATATGAAAATCCTCACTAC CAGTCACCAAACATGCATCGGCGTGTCATAAGATCCATCACAGCTGCAAAATACAGAGAGAAACAGCAAGTGAAAGATGTGCAAAAACTG AGAAAGAAAGAGCCGAAGACTCTTCTTCCACATGATCCCACTGCAGATGTTTTTGTAACACCAGCTGAGGAGAAACCAATAGAAATACAG TGGGTAAAACCAGAGCCAAAAGTTGATTTGAAAGCAAGAAAGAAACGGATTTACAAAAGGCAAAGAAAAATGAAACAGAGGATGGACAGT GGGAAAACAAAATAAGTCAATGGAAACCTGATTTGTTTTTCAGTTACTTTATATTTATTTTGTATTCAATGTGTAAATACTTTTATTATC TAATACTATCTTACGTCTAATTAGTGTAGCATTTACAAGAAAGAAAAATTAAGATCTTAAAATCAGTGATTATCTTTTTCTAAATAAAAT ATCACCAGAATTCATCAGTTAATTTCTGATTTCTTTTTGAAGTTTGTGTTGCTAAAAATGTAGCACACTTAATGTAGCCTGTTCTCTTGG GTTGGAATTTTTGGTTTAGCAAAGCTGAAATTCAGACATTTATTAGGTCATATTATTTGTAAAAGCATTCAACACTTCAAGAGCATCGGT TGTGGATGGTAAAGTAGGAGTAGACTGGTAGAAAGGAAGGCATCTCACTGAAGTTTACTCAGTCATCAGTTAATGAAGCCATGGAAGAAC >22739_22739_5_DIAPH1-BRIX1_DIAPH1_chr5_140963691_ENST00000398562_BRIX1_chr5_34924952_ENST00000336767_length(amino acids)=275AA_BP=143 MPGGSGVGAGSPASVNRDMEPPGGSLGPGRGTRDKKKGRSPDELPSAGGDGGKSKKFLERFTSMRIKKEKEKPNSAHRNSSASYGDDPTA QSLQDVSDEQVLVLFEQMLLDMNLNEEKQQPLREKDIIIKREMVSQYLYTSKAIIEEDAALVEIGPRFVLNLIKIFQGSFGGPTLYENPH YQSPNMHRRVIRSITAAKYREKQQVKDVQKLRKKEPKTLLPHDPTADVFVTPAEEKPIEIQWVKPEPKVDLKARKKRIYKRQRKMKQRMD -------------------------------------------------------------- >22739_22739_6_DIAPH1-BRIX1_DIAPH1_chr5_140963691_ENST00000398566_BRIX1_chr5_34924952_ENST00000336767_length(transcript)=1416nt_BP=516nt ATTCATGAGGCGCGCGCAGCCCGGCATGCTAATGAGGCGGGGCGCGGCGGCTGGCTAAAGAGCGACTGGGCGGCGGCGGGCGCGGAGCTG CCAGGCGGGAGCGGCGTAGGCGCGGGGTCGCCGGCCAGCGTGAACCGGGACATGGAGCCGCCCGGCGGGAGCCTGGGGCCCGGCCGCGGG ACCCGGGACAAGAAGAAGGGCCGGAGCCCAGATGAGCTGCCCTCGGCGGGCGGCGACGGCGGCAAATCTAAGAAATTTCTGGAGAGATTT ACCAGCATGAGAATTAAGAAGGAGAAGGAAAAGCCCAATTCTGCTCATAGAAATTCTTCTGCATCATATGGGGATGATCCCACAGCACAG TCATTGCAAGATGTTTCAGATGAACAAGTGCTGGTTCTCTTTGAACAGATGCTGCTGGATATGAACCTGAATGAGGAGAAACAGCAACCT TTGAGGGAGAAGGACATCATCATCAAGAGGGAGATGGTGTCCCAATACTTGTACACCTCCAAGGCTATCATAGAAGAAGATGCTGCTCTT GTAGAAATAGGACCTCGTTTTGTCTTAAATCTCATAAAGATTTTCCAGGGAAGTTTTGGAGGACCAACTTTATATGAAAATCCTCACTAC CAGTCACCAAACATGCATCGGCGTGTCATAAGATCCATCACAGCTGCAAAATACAGAGAGAAACAGCAAGTGAAAGATGTGCAAAAACTG AGAAAGAAAGAGCCGAAGACTCTTCTTCCACATGATCCCACTGCAGATGTTTTTGTAACACCAGCTGAGGAGAAACCAATAGAAATACAG TGGGTAAAACCAGAGCCAAAAGTTGATTTGAAAGCAAGAAAGAAACGGATTTACAAAAGGCAAAGAAAAATGAAACAGAGGATGGACAGT GGGAAAACAAAATAAGTCAATGGAAACCTGATTTGTTTTTCAGTTACTTTATATTTATTTTGTATTCAATGTGTAAATACTTTTATTATC TAATACTATCTTACGTCTAATTAGTGTAGCATTTACAAGAAAGAAAAATTAAGATCTTAAAATCAGTGATTATCTTTTTCTAAATAAAAT ATCACCAGAATTCATCAGTTAATTTCTGATTTCTTTTTGAAGTTTGTGTTGCTAAAAATGTAGCACACTTAATGTAGCCTGTTCTCTTGG GTTGGAATTTTTGGTTTAGCAAAGCTGAAATTCAGACATTTATTAGGTCATATTATTTGTAAAAGCATTCAACACTTCAAGAGCATCGGT TGTGGATGGTAAAGTAGGAGTAGACTGGTAGAAAGGAAGGCATCTCACTGAAGTTTACTCAGTCATCAGTTAATGAAGCCATGGAAGAAC >22739_22739_6_DIAPH1-BRIX1_DIAPH1_chr5_140963691_ENST00000398566_BRIX1_chr5_34924952_ENST00000336767_length(amino acids)=275AA_BP=143 MPGGSGVGAGSPASVNRDMEPPGGSLGPGRGTRDKKKGRSPDELPSAGGDGGKSKKFLERFTSMRIKKEKEKPNSAHRNSSASYGDDPTA QSLQDVSDEQVLVLFEQMLLDMNLNEEKQQPLREKDIIIKREMVSQYLYTSKAIIEEDAALVEIGPRFVLNLIKIFQGSFGGPTLYENPH YQSPNMHRRVIRSITAAKYREKQQVKDVQKLRKKEPKTLLPHDPTADVFVTPAEEKPIEIQWVKPEPKVDLKARKKRIYKRQRKMKQRMD -------------------------------------------------------------- >22739_22739_7_DIAPH1-BRIX1_DIAPH1_chr5_140963691_ENST00000518047_BRIX1_chr5_34924952_ENST00000336767_length(transcript)=1275nt_BP=375nt ATGGAGCCGCCCGGCGGGAGCCTGGGGCCCGGCCGCGGGACCCGGGACAAGAAGAAGGGCCGGAGCCCAGATGAGCTGCCCTCGGCGGGC GGCGACGGCGGCAAATCTAAGAAATTTCTGGAGAGATTTACCAGCATGAGAATTAAGAAGGAGAAGGAAAAGCCCAATTCTGCTCATAGA AATTCTTCTGCATCATATGGGGATGATCCCACAGCACAGTCATTGCAAGATGTTTCAGATGAACAAGTGCTGGTTCTCTTTGAACAGATG CTGCTGGATATGAACCTGAATGAGGAGAAACAGCAACCTTTGAGGGAGAAGGACATCATCATCAAGAGGGAGATGGTGTCCCAATACTTG TACACCTCCAAGGCTATCATAGAAGAAGATGCTGCTCTTGTAGAAATAGGACCTCGTTTTGTCTTAAATCTCATAAAGATTTTCCAGGGA AGTTTTGGAGGACCAACTTTATATGAAAATCCTCACTACCAGTCACCAAACATGCATCGGCGTGTCATAAGATCCATCACAGCTGCAAAA TACAGAGAGAAACAGCAAGTGAAAGATGTGCAAAAACTGAGAAAGAAAGAGCCGAAGACTCTTCTTCCACATGATCCCACTGCAGATGTT TTTGTAACACCAGCTGAGGAGAAACCAATAGAAATACAGTGGGTAAAACCAGAGCCAAAAGTTGATTTGAAAGCAAGAAAGAAACGGATT TACAAAAGGCAAAGAAAAATGAAACAGAGGATGGACAGTGGGAAAACAAAATAAGTCAATGGAAACCTGATTTGTTTTTCAGTTACTTTA TATTTATTTTGTATTCAATGTGTAAATACTTTTATTATCTAATACTATCTTACGTCTAATTAGTGTAGCATTTACAAGAAAGAAAAATTA AGATCTTAAAATCAGTGATTATCTTTTTCTAAATAAAATATCACCAGAATTCATCAGTTAATTTCTGATTTCTTTTTGAAGTTTGTGTTG CTAAAAATGTAGCACACTTAATGTAGCCTGTTCTCTTGGGTTGGAATTTTTGGTTTAGCAAAGCTGAAATTCAGACATTTATTAGGTCAT ATTATTTGTAAAAGCATTCAACACTTCAAGAGCATCGGTTGTGGATGGTAAAGTAGGAGTAGACTGGTAGAAAGGAAGGCATCTCACTGA AGTTTACTCAGTCATCAGTTAATGAAGCCATGGAAGAACCAACCCCTGATCTGTCACTTCAAAAAAGAGTATATTAAAATGTTGAGGTTT >22739_22739_7_DIAPH1-BRIX1_DIAPH1_chr5_140963691_ENST00000518047_BRIX1_chr5_34924952_ENST00000336767_length(amino acids)=257AA_BP=125 MEPPGGSLGPGRGTRDKKKGRSPDELPSAGGDGGKSKKFLERFTSMRIKKEKEKPNSAHRNSSASYGDDPTAQSLQDVSDEQVLVLFEQM LLDMNLNEEKQQPLREKDIIIKREMVSQYLYTSKAIIEEDAALVEIGPRFVLNLIKIFQGSFGGPTLYENPHYQSPNMHRRVIRSITAAK -------------------------------------------------------------- >22739_22739_8_DIAPH1-BRIX1_DIAPH1_chr5_140963691_ENST00000520569_BRIX1_chr5_34924952_ENST00000336767_length(transcript)=1158nt_BP=258nt CTGGAGAGATTTACCAGCATGAGAATTAAGAAGGAGAAGGAAAAGCCCAATTCTGCTCATAGAAATTCTTCTGCATCATATGGGGATGAT CCCACAGCACAGTCATTGCAAGATGTTTCAGATGAACAAGTGCTGGTTCTCTTTGAACAGATGCTGCTGGATATGAACCTGAATGAGGAG AAACAGCAACCTTTGAGGGAGAAGGACATCATCATCAAGAGGGAGATGGTGTCCCAATACTTGTACACCTCCAAGGCTATCATAGAAGAA GATGCTGCTCTTGTAGAAATAGGACCTCGTTTTGTCTTAAATCTCATAAAGATTTTCCAGGGAAGTTTTGGAGGACCAACTTTATATGAA AATCCTCACTACCAGTCACCAAACATGCATCGGCGTGTCATAAGATCCATCACAGCTGCAAAATACAGAGAGAAACAGCAAGTGAAAGAT GTGCAAAAACTGAGAAAGAAAGAGCCGAAGACTCTTCTTCCACATGATCCCACTGCAGATGTTTTTGTAACACCAGCTGAGGAGAAACCA ATAGAAATACAGTGGGTAAAACCAGAGCCAAAAGTTGATTTGAAAGCAAGAAAGAAACGGATTTACAAAAGGCAAAGAAAAATGAAACAG AGGATGGACAGTGGGAAAACAAAATAAGTCAATGGAAACCTGATTTGTTTTTCAGTTACTTTATATTTATTTTGTATTCAATGTGTAAAT ACTTTTATTATCTAATACTATCTTACGTCTAATTAGTGTAGCATTTACAAGAAAGAAAAATTAAGATCTTAAAATCAGTGATTATCTTTT TCTAAATAAAATATCACCAGAATTCATCAGTTAATTTCTGATTTCTTTTTGAAGTTTGTGTTGCTAAAAATGTAGCACACTTAATGTAGC CTGTTCTCTTGGGTTGGAATTTTTGGTTTAGCAAAGCTGAAATTCAGACATTTATTAGGTCATATTATTTGTAAAAGCATTCAACACTTC AAGAGCATCGGTTGTGGATGGTAAAGTAGGAGTAGACTGGTAGAAAGGAAGGCATCTCACTGAAGTTTACTCAGTCATCAGTTAATGAAG >22739_22739_8_DIAPH1-BRIX1_DIAPH1_chr5_140963691_ENST00000520569_BRIX1_chr5_34924952_ENST00000336767_length(amino acids)=218AA_BP=86 LERFTSMRIKKEKEKPNSAHRNSSASYGDDPTAQSLQDVSDEQVLVLFEQMLLDMNLNEEKQQPLREKDIIIKREMVSQYLYTSKAIIEE DAALVEIGPRFVLNLIKIFQGSFGGPTLYENPHYQSPNMHRRVIRSITAAKYREKQQVKDVQKLRKKEPKTLLPHDPTADVFVTPAEEKP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for DIAPH1-BRIX1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for DIAPH1-BRIX1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for DIAPH1-BRIX1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
