
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:DIAPH3-DOK5 (FusionGDB2 ID:22769) |
Fusion Gene Summary for DIAPH3-DOK5 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: DIAPH3-DOK5 | Fusion gene ID: 22769 | Hgene | Tgene | Gene symbol | DIAPH3 | DOK5 | Gene ID | 81624 | 55816 |
| Gene name | diaphanous related formin 3 | docking protein 5 | |
| Synonyms | AN|AUNA1|DIA2|DRF3|NSDAN|diap3|mDia2 | C20orf180|IRS-6|IRS6 | |
| Cytomap | 13q21.2 | 20q13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein diaphanous homolog 3diaphanous homolog 3 | docking protein 5downstream of tyrosine kinase 5insulin receptor substrate 6 | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | Q9NSV4 | Q9P104 | |
| Ensembl transtripts involved in fusion gene | ENST00000267215, ENST00000377908, ENST00000400319, ENST00000400320, ENST00000400324, ENST00000400330, ENST00000465066, | ENST00000491469, ENST00000262593, ENST00000395939, | |
| Fusion gene scores | * DoF score | 16 X 12 X 4=768 | 11 X 9 X 8=792 |
| # samples | 18 | 12 | |
| ** MAII score | log2(18/768*10)=-2.09310940439148 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/792*10)=-2.72246602447109 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: DIAPH3 [Title/Abstract] AND DOK5 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | DIAPH3(60582677)-DOK5(53266953), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | DIAPH3-DOK5 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across DIAPH3 (5'-gene) Fusion gene breakpoints across DIAPH3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
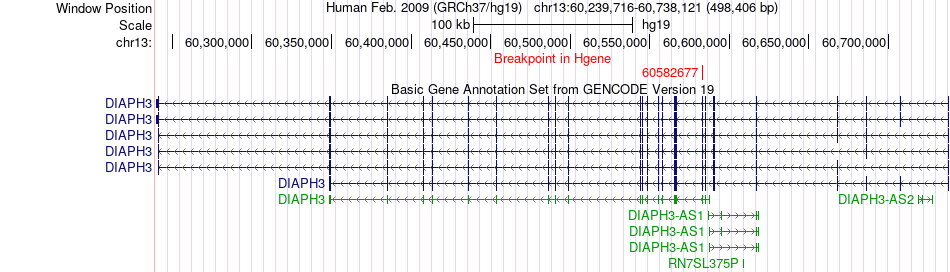 |
 Fusion gene breakpoints across DOK5 (3'-gene) Fusion gene breakpoints across DOK5 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
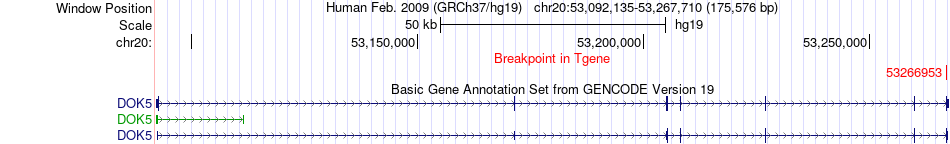 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-KK-A59V | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
Top |
Fusion Gene ORF analysis for DIAPH3-DOK5 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000267215 | ENST00000491469 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| 5CDS-intron | ENST00000377908 | ENST00000491469 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| 5CDS-intron | ENST00000400319 | ENST00000491469 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| 5CDS-intron | ENST00000400320 | ENST00000491469 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| 5CDS-intron | ENST00000400324 | ENST00000491469 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| 5CDS-intron | ENST00000400330 | ENST00000491469 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| 5UTR-3CDS | ENST00000465066 | ENST00000262593 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| 5UTR-3CDS | ENST00000465066 | ENST00000395939 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| 5UTR-intron | ENST00000465066 | ENST00000491469 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| Frame-shift | ENST00000267215 | ENST00000262593 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| Frame-shift | ENST00000377908 | ENST00000262593 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| Frame-shift | ENST00000400319 | ENST00000262593 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| Frame-shift | ENST00000400320 | ENST00000262593 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| Frame-shift | ENST00000400324 | ENST00000262593 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| Frame-shift | ENST00000400330 | ENST00000262593 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| In-frame | ENST00000267215 | ENST00000395939 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| In-frame | ENST00000377908 | ENST00000395939 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| In-frame | ENST00000400319 | ENST00000395939 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| In-frame | ENST00000400320 | ENST00000395939 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| In-frame | ENST00000400324 | ENST00000395939 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
| In-frame | ENST00000400330 | ENST00000395939 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000400324 | DIAPH3 | chr13 | 60582677 | - | ENST00000395939 | DOK5 | chr20 | 53266953 | + | 1711 | 1235 | 110 | 1252 | 380 |
| ENST00000400330 | DIAPH3 | chr13 | 60582677 | - | ENST00000395939 | DOK5 | chr20 | 53266953 | + | 1709 | 1233 | 108 | 1250 | 380 |
| ENST00000400319 | DIAPH3 | chr13 | 60582677 | - | ENST00000395939 | DOK5 | chr20 | 53266953 | + | 1280 | 804 | 0 | 821 | 273 |
| ENST00000400320 | DIAPH3 | chr13 | 60582677 | - | ENST00000395939 | DOK5 | chr20 | 53266953 | + | 1352 | 876 | 0 | 893 | 297 |
| ENST00000377908 | DIAPH3 | chr13 | 60582677 | - | ENST00000395939 | DOK5 | chr20 | 53266953 | + | 1457 | 981 | 0 | 998 | 332 |
| ENST00000267215 | DIAPH3 | chr13 | 60582677 | - | ENST00000395939 | DOK5 | chr20 | 53266953 | + | 1490 | 1014 | 0 | 1031 | 343 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000400324 | ENST00000395939 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + | 0.000500145 | 0.9994998 |
| ENST00000400330 | ENST00000395939 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + | 0.000508233 | 0.9994917 |
| ENST00000400319 | ENST00000395939 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + | 0.000490045 | 0.99950993 |
| ENST00000400320 | ENST00000395939 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + | 0.002438845 | 0.9975611 |
| ENST00000377908 | ENST00000395939 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + | 0.000569891 | 0.9994301 |
| ENST00000267215 | ENST00000395939 | DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + | 0.000428373 | 0.9995716 |
Top |
Fusion Genomic Features for DIAPH3-DOK5 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + | 4.48E-07 | 0.9999995 |
| DIAPH3 | chr13 | 60582677 | - | DOK5 | chr20 | 53266953 | + | 4.48E-07 | 0.9999995 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
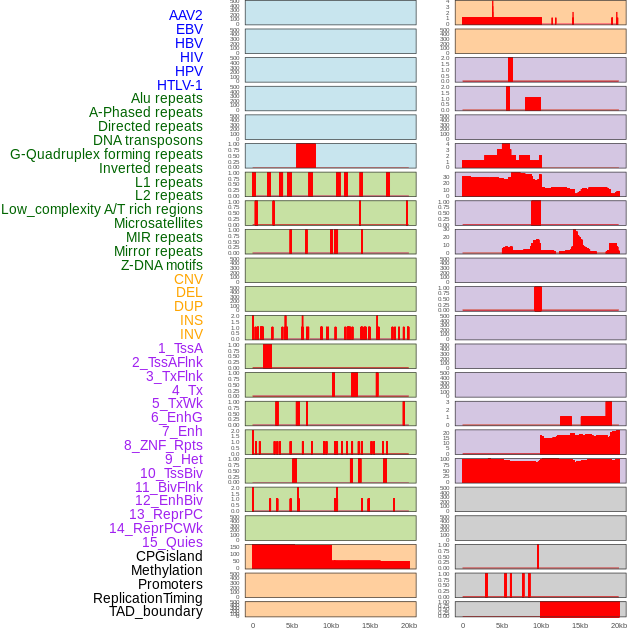 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
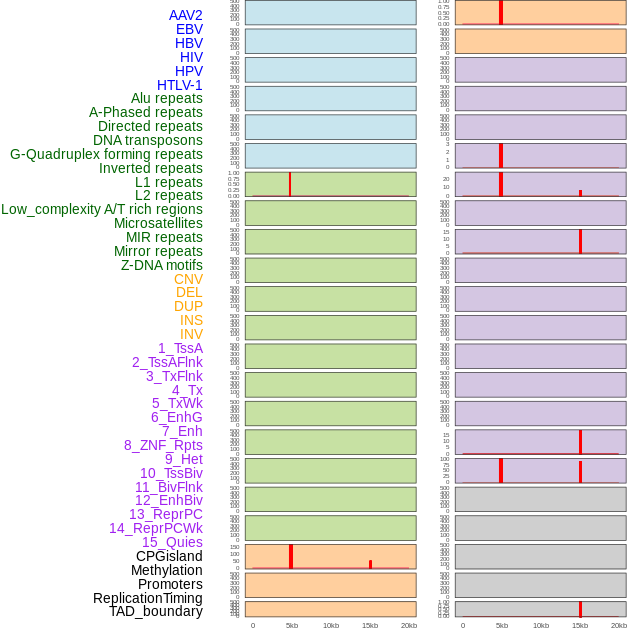 |
Top |
Fusion Protein Features for DIAPH3-DOK5 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr13:60582677/chr20:53266953) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| DIAPH3 | DOK5 |
| FUNCTION: Actin nucleation and elongation factor required for the assembly of F-actin structures, such as actin cables and stress fibers. Required for cytokinesis, stress fiber formation and transcriptional activation of the serum response factor. Binds to GTP-bound form of Rho and to profilin: acts in a Rho-dependent manner to recruit profilin to the membrane, where it promotes actin polymerization. DFR proteins couple Rho and Src tyrosine kinase during signaling and the regulation of actin dynamics. Also acts as an actin nucleation and elongation factor in the nucleus by promoting nuclear actin polymerization inside the nucleus to drive serum-dependent SRF-MRTFA activity. {ECO:0000250|UniProtKB:Q9Z207}. | FUNCTION: DOK proteins are enzymatically inert adaptor or scaffolding proteins. They provide a docking platform for the assembly of multimolecular signaling complexes. DOK5 functions in RET-mediated neurite outgrowth and plays a positive role in activation of the MAP kinase pathway. Putative link with downstream effectors of RET in neuronal differentiation. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000267215 | - | 9 | 27 | 36_60 | 338 | 1113.0 | Motif | Nuclear localization signal |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000377908 | - | 8 | 27 | 36_60 | 327 | 1183.0 | Motif | Nuclear localization signal |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400319 | - | 7 | 26 | 36_60 | 268 | 1124.0 | Motif | Nuclear localization signal |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400320 | - | 7 | 26 | 36_60 | 292 | 1148.0 | Motif | Nuclear localization signal |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400324 | - | 9 | 28 | 36_60 | 338 | 1194.0 | Motif | Nuclear localization signal |
| Tgene | DOK5 | chr13:60582677 | chr20:53266953 | ENST00000395939 | 6 | 8 | 263_273 | 177 | 199.0 | Motif | Note=DKFBH motif |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000267215 | - | 9 | 27 | 1013_1056 | 338 | 1113.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000267215 | - | 9 | 27 | 497_554 | 338 | 1113.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000377908 | - | 8 | 27 | 1013_1056 | 327 | 1183.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000377908 | - | 8 | 27 | 497_554 | 327 | 1183.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400319 | - | 7 | 26 | 1013_1056 | 268 | 1124.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400319 | - | 7 | 26 | 497_554 | 268 | 1124.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400320 | - | 7 | 26 | 1013_1056 | 292 | 1148.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400320 | - | 7 | 26 | 497_554 | 292 | 1148.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400324 | - | 9 | 28 | 1013_1056 | 338 | 1194.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400324 | - | 9 | 28 | 497_554 | 338 | 1194.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000267215 | - | 9 | 27 | 561_636 | 338 | 1113.0 | Compositional bias | Note=Pro-rich |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000377908 | - | 8 | 27 | 561_636 | 327 | 1183.0 | Compositional bias | Note=Pro-rich |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400319 | - | 7 | 26 | 561_636 | 268 | 1124.0 | Compositional bias | Note=Pro-rich |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400320 | - | 7 | 26 | 561_636 | 292 | 1148.0 | Compositional bias | Note=Pro-rich |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400324 | - | 9 | 28 | 561_636 | 338 | 1194.0 | Compositional bias | Note=Pro-rich |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000267215 | - | 9 | 27 | 1057_1087 | 338 | 1113.0 | Domain | DAD |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000267215 | - | 9 | 27 | 114_476 | 338 | 1113.0 | Domain | GBD/FH3 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000267215 | - | 9 | 27 | 561_631 | 338 | 1113.0 | Domain | Note=FH1 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000267215 | - | 9 | 27 | 636_1034 | 338 | 1113.0 | Domain | FH2 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000377908 | - | 8 | 27 | 1057_1087 | 327 | 1183.0 | Domain | DAD |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000377908 | - | 8 | 27 | 114_476 | 327 | 1183.0 | Domain | GBD/FH3 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000377908 | - | 8 | 27 | 561_631 | 327 | 1183.0 | Domain | Note=FH1 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000377908 | - | 8 | 27 | 636_1034 | 327 | 1183.0 | Domain | FH2 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400319 | - | 7 | 26 | 1057_1087 | 268 | 1124.0 | Domain | DAD |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400319 | - | 7 | 26 | 114_476 | 268 | 1124.0 | Domain | GBD/FH3 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400319 | - | 7 | 26 | 561_631 | 268 | 1124.0 | Domain | Note=FH1 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400319 | - | 7 | 26 | 636_1034 | 268 | 1124.0 | Domain | FH2 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400320 | - | 7 | 26 | 1057_1087 | 292 | 1148.0 | Domain | DAD |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400320 | - | 7 | 26 | 114_476 | 292 | 1148.0 | Domain | GBD/FH3 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400320 | - | 7 | 26 | 561_631 | 292 | 1148.0 | Domain | Note=FH1 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400320 | - | 7 | 26 | 636_1034 | 292 | 1148.0 | Domain | FH2 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400324 | - | 9 | 28 | 1057_1087 | 338 | 1194.0 | Domain | DAD |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400324 | - | 9 | 28 | 114_476 | 338 | 1194.0 | Domain | GBD/FH3 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400324 | - | 9 | 28 | 561_631 | 338 | 1194.0 | Domain | Note=FH1 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400324 | - | 9 | 28 | 636_1034 | 338 | 1194.0 | Domain | FH2 |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000267215 | - | 9 | 27 | 1184_1193 | 338 | 1113.0 | Motif | Nuclear export signal |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000377908 | - | 8 | 27 | 1184_1193 | 327 | 1183.0 | Motif | Nuclear export signal |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400319 | - | 7 | 26 | 1184_1193 | 268 | 1124.0 | Motif | Nuclear export signal |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400320 | - | 7 | 26 | 1184_1193 | 292 | 1148.0 | Motif | Nuclear export signal |
| Hgene | DIAPH3 | chr13:60582677 | chr20:53266953 | ENST00000400324 | - | 9 | 28 | 1184_1193 | 338 | 1194.0 | Motif | Nuclear export signal |
| Tgene | DOK5 | chr13:60582677 | chr20:53266953 | ENST00000262593 | 6 | 8 | 132_237 | 285 | 307.0 | Domain | IRS-type PTB | |
| Tgene | DOK5 | chr13:60582677 | chr20:53266953 | ENST00000262593 | 6 | 8 | 8_112 | 285 | 307.0 | Domain | Note=PH | |
| Tgene | DOK5 | chr13:60582677 | chr20:53266953 | ENST00000395939 | 6 | 8 | 132_237 | 177 | 199.0 | Domain | IRS-type PTB | |
| Tgene | DOK5 | chr13:60582677 | chr20:53266953 | ENST00000395939 | 6 | 8 | 8_112 | 177 | 199.0 | Domain | Note=PH | |
| Tgene | DOK5 | chr13:60582677 | chr20:53266953 | ENST00000262593 | 6 | 8 | 263_273 | 285 | 307.0 | Motif | Note=DKFBH motif |
Top |
Fusion Gene Sequence for DIAPH3-DOK5 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >22769_22769_1_DIAPH3-DOK5_DIAPH3_chr13_60582677_ENST00000267215_DOK5_chr20_53266953_ENST00000395939_length(transcript)=1490nt_BP=1014nt ATGGAACGGCACCAGCCGCGGCTGCACCACCCGGCCCAAGGCTCAGCCGCTGGGACTCCCTACCCTTCCTCAGCCTCTCTCCGCGGCTGC CGGGAAAGCAAGATGCCGCGCAGGAAGGGCCCCCAACACCCTCCGCCGCCCAGTGGCCCCGAGGAGCCTGGGGAGAAGCGCCCCAAGTTT CATTTAAATATTAGGACACTGACGGATGATATGCTGGACAAATTTGCCAGCATAAGAATTCCAGGGAGCAAGAAAGAGAGACCTCCACTT CCCAACCTGAAGACTGCATTTGCAAGCAGTGATTGCTCAGCAGCACCTTTAGAGATGATGGAGAACTTTCCAAAGCCACTGTCAGAGAAT GAACTCTTAGAACTTTTTGAAAAAATGATGGAAGATATGAATTTAAATGAAGATAAAAAGGCACCATTGCGGGAAAAGGACTTCAGTATC AAAAAAGAAATGGTGATGCAGTACATTAATACTGCTTCTAAGACAGGAAGTCTTAAGAGAAGCCGACAGATCTCACCTCAGGAATTCATT CATGAGCTGAAAATGGGGTCTGCAGATGAGAGACTTGTCACATGCCTGGAGTCTCTCCGAGTGTCTTTGACCAGCAATCCTGTGAGTTGG GTGGAAAGCTTTGGACATGAAGGGCTTGGATTATTATTAGACATTTTGGAAAAACTGATTAGTGGAAAAATCCAAGAAAAAGTTGTAAAG AAAAATCAACATAAAGTCATACAGTGTCTAAAAGCCCTGATGAATACGCAGTATGGCTTGGAAAGAATTATGAGTGAGGAGAGGAGCCTT TCCTTATTGGCCAAAGCCGTGGATCCCAGACACCCCAATATGATGACAGATGTGGTTAAACTTCTCTCTGCGGTATGCATTGTAGGGGAA GAAAGCATCCTTGAAGAAGTTTTAGAAGCTTTAACTTCAGCTGGTGAAGAAAAAAAAATTGACAGATTTTTTTGTATTGTGGAAGGCCTC CGGCACAATTCAGTTCAACTGCAAATGTTTCCAGCCCTCTGAAGCTTCATCGAACAGAGACTTTTCCAGCCTACAGATCTGAGCACTGAC AGTAACTGCCAAGAATTGTTAACACACTTGTGATGTGTCAGCCACAGATTCACCCAGGAGGTCACAGAATGACAGCAAGGGAAATGACGA CCAAGAGAAGAAGCTTAAAGTCCTGGCTAATTGTGTGGTCATTGGAAAACTCTGCAATACAATAATTTTCTTTATTTTCTTTTTCTTTTT TAAATTCTTAGTGTAATTGAAACGTGCTCTATAGATATTGACTCTGTGTTCCCTCTTTTACAGCTGGACAGAAAGAAGTCAATGTCACGA AATGATTTTCTATTGTAGATACTTTGTCCCTTGCACTTCTCTGAATCTGTCCTTTTGTGGATTCTTGTGATTTTCCTTCCAAGTGTTTCA >22769_22769_1_DIAPH3-DOK5_DIAPH3_chr13_60582677_ENST00000267215_DOK5_chr20_53266953_ENST00000395939_length(amino acids)=343AA_BP= MERHQPRLHHPAQGSAAGTPYPSSASLRGCRESKMPRRKGPQHPPPPSGPEEPGEKRPKFHLNIRTLTDDMLDKFASIRIPGSKKERPPL PNLKTAFASSDCSAAPLEMMENFPKPLSENELLELFEKMMEDMNLNEDKKAPLREKDFSIKKEMVMQYINTASKTGSLKRSRQISPQEFI HELKMGSADERLVTCLESLRVSLTSNPVSWVESFGHEGLGLLLDILEKLISGKIQEKVVKKNQHKVIQCLKALMNTQYGLERIMSEERSL -------------------------------------------------------------- >22769_22769_2_DIAPH3-DOK5_DIAPH3_chr13_60582677_ENST00000377908_DOK5_chr20_53266953_ENST00000395939_length(transcript)=1457nt_BP=981nt ATGGAACGGCACCAGCCGCGGCTGCACCACCCGGCCCAAGGCTCAGCCGCTGGGACTCCCTACCCTTCCTCAGCCTCTCTCCGCGGCTGC CGGGAAAGCAAGATGCCGCGCAGGAAGGGCCCCCAACACCCTCCGCCGCCCAGTGGCCCCGAGGAGCCTGGGGAGAAGCGCCCCAAGTTT CTGGACAAATTTGCCAGCATAAGAATTCCAGGGAGCAAGAAAGAGAGACCTCCACTTCCCAACCTGAAGACTGCATTTGCAAGCAGTGAT TGCTCAGCAGCACCTTTAGAGATGATGGAGAACTTTCCAAAGCCACTGTCAGAGAATGAACTCTTAGAACTTTTTGAAAAAATGATGGAA GATATGAATTTAAATGAAGATAAAAAGGCACCATTGCGGGAAAAGGACTTCAGTATCAAAAAAGAAATGGTGATGCAGTACATTAATACT GCTTCTAAGACAGGAAGTCTTAAGAGAAGCCGACAGATCTCACCTCAGGAATTCATTCATGAGCTGAAAATGGGGTCTGCAGATGAGAGA CTTGTCACATGCCTGGAGTCTCTCCGAGTGTCTTTGACCAGCAATCCTGTGAGTTGGGTGGAAAGCTTTGGACATGAAGGGCTTGGATTA TTATTAGACATTTTGGAAAAACTGATTAGTGGAAAAATCCAAGAAAAAGTTGTAAAGAAAAATCAACATAAAGTCATACAGTGTCTAAAA GCCCTGATGAATACGCAGTATGGCTTGGAAAGAATTATGAGTGAGGAGAGGAGCCTTTCCTTATTGGCCAAAGCCGTGGATCCCAGACAC CCCAATATGATGACAGATGTGGTTAAACTTCTCTCTGCGGTATGCATTGTAGGGGAAGAAAGCATCCTTGAAGAAGTTTTAGAAGCTTTA ACTTCAGCTGGTGAAGAAAAAAAAATTGACAGATTTTTTTGTATTGTGGAAGGCCTCCGGCACAATTCAGTTCAACTGCAAATGTTTCCA GCCCTCTGAAGCTTCATCGAACAGAGACTTTTCCAGCCTACAGATCTGAGCACTGACAGTAACTGCCAAGAATTGTTAACACACTTGTGA TGTGTCAGCCACAGATTCACCCAGGAGGTCACAGAATGACAGCAAGGGAAATGACGACCAAGAGAAGAAGCTTAAAGTCCTGGCTAATTG TGTGGTCATTGGAAAACTCTGCAATACAATAATTTTCTTTATTTTCTTTTTCTTTTTTAAATTCTTAGTGTAATTGAAACGTGCTCTATA GATATTGACTCTGTGTTCCCTCTTTTACAGCTGGACAGAAAGAAGTCAATGTCACGAAATGATTTTCTATTGTAGATACTTTGTCCCTTG CACTTCTCTGAATCTGTCCTTTTGTGGATTCTTGTGATTTTCCTTCCAAGTGTTTCAGTTGTATGACAGTCAGTATTGACAATAAAATGG >22769_22769_2_DIAPH3-DOK5_DIAPH3_chr13_60582677_ENST00000377908_DOK5_chr20_53266953_ENST00000395939_length(amino acids)=332AA_BP= MERHQPRLHHPAQGSAAGTPYPSSASLRGCRESKMPRRKGPQHPPPPSGPEEPGEKRPKFLDKFASIRIPGSKKERPPLPNLKTAFASSD CSAAPLEMMENFPKPLSENELLELFEKMMEDMNLNEDKKAPLREKDFSIKKEMVMQYINTASKTGSLKRSRQISPQEFIHELKMGSADER LVTCLESLRVSLTSNPVSWVESFGHEGLGLLLDILEKLISGKIQEKVVKKNQHKVIQCLKALMNTQYGLERIMSEERSLSLLAKAVDPRH -------------------------------------------------------------- >22769_22769_3_DIAPH3-DOK5_DIAPH3_chr13_60582677_ENST00000400319_DOK5_chr20_53266953_ENST00000395939_length(transcript)=1280nt_BP=804nt ATGGAACGGCACCAGCCGCGGCTGCACCACCCGGCCCAAGGCTCAGCCGCTGGGACTCCCTACCCTTCCTCAGCCTCTCTCCGCGGCTGC CGGGAAAGCAAGATGCCGCGCAGGAAGGGCCCCCAACACCCTCCGCCGCCCAGTGGCCCCGAGGAGCCTGGGGAGAAGCGCCCCAAGTTT GAAGATATGAATTTAAATGAAGATAAAAAGGCACCATTGCGGGAAAAGGACTTCAGTATCAAAAAAGAAATGGTGATGCAGTACATTAAT ACTGCTTCTAAGACAGGAAGTCTTAAGAGAAGCCGACAGATCTCACCTCAGGAATTCATTCATGAGCTGAAAATGGGGTCTGCAGATGAG AGACTTGTCACATGCCTGGAGTCTCTCCGAGTGTCTTTGACCAGCAATCCTGTGAGTTGGGTGGAAAGCTTTGGACATGAAGGGCTTGGA TTATTATTAGACATTTTGGAAAAACTGATTAGTGGAAAAATCCAAGAAAAAGTTGTAAAGAAAAATCAACATAAAGTCATACAGTGTCTA AAAGCCCTGATGAATACGCAGTATGGCTTGGAAAGAATTATGAGTGAGGAGAGGAGCCTTTCCTTATTGGCCAAAGCCGTGGATCCCAGA CACCCCAATATGATGACAGATGTGGTTAAACTTCTCTCTGCGGTATGCATTGTAGGGGAAGAAAGCATCCTTGAAGAAGTTTTAGAAGCT TTAACTTCAGCTGGTGAAGAAAAAAAAATTGACAGATTTTTTTGTATTGTGGAAGGCCTCCGGCACAATTCAGTTCAACTGCAAATGTTT CCAGCCCTCTGAAGCTTCATCGAACAGAGACTTTTCCAGCCTACAGATCTGAGCACTGACAGTAACTGCCAAGAATTGTTAACACACTTG TGATGTGTCAGCCACAGATTCACCCAGGAGGTCACAGAATGACAGCAAGGGAAATGACGACCAAGAGAAGAAGCTTAAAGTCCTGGCTAA TTGTGTGGTCATTGGAAAACTCTGCAATACAATAATTTTCTTTATTTTCTTTTTCTTTTTTAAATTCTTAGTGTAATTGAAACGTGCTCT ATAGATATTGACTCTGTGTTCCCTCTTTTACAGCTGGACAGAAAGAAGTCAATGTCACGAAATGATTTTCTATTGTAGATACTTTGTCCC TTGCACTTCTCTGAATCTGTCCTTTTGTGGATTCTTGTGATTTTCCTTCCAAGTGTTTCAGTTGTATGACAGTCAGTATTGACAATAAAA >22769_22769_3_DIAPH3-DOK5_DIAPH3_chr13_60582677_ENST00000400319_DOK5_chr20_53266953_ENST00000395939_length(amino acids)=273AA_BP= MERHQPRLHHPAQGSAAGTPYPSSASLRGCRESKMPRRKGPQHPPPPSGPEEPGEKRPKFEDMNLNEDKKAPLREKDFSIKKEMVMQYIN TASKTGSLKRSRQISPQEFIHELKMGSADERLVTCLESLRVSLTSNPVSWVESFGHEGLGLLLDILEKLISGKIQEKVVKKNQHKVIQCL KALMNTQYGLERIMSEERSLSLLAKAVDPRHPNMMTDVVKLLSAVCIVGEESILEEVLEALTSAGEEKKIDRFFCIVEGLRHNSVQLQMF -------------------------------------------------------------- >22769_22769_4_DIAPH3-DOK5_DIAPH3_chr13_60582677_ENST00000400320_DOK5_chr20_53266953_ENST00000395939_length(transcript)=1352nt_BP=876nt ATGGAACGGCACCAGCCGCGGCTGCACCACCCGGCCCAAGGCTCAGCCGCTGGGACTCCCTACCCTTCCTCAGCCTCTCTCCGCGGCTGC CGGGAAAGCAAGATGCCGCGCAGGAAGGGCCCCCAACACCCTCCGCCGCCCAGTGGCCCCGAGGAGCCTGGGGAGAAGCGCCCCAAGTTT CTGGACAAATTTGCCAGCATAAGAATTCCAGGGAGCAAGAAAGAGAGACCTCCACTTCCCAACCTGAAGACTGCATTTGCAAGCAGTGAT TGCTCAGCAGCACCTTTAGAGATGATGGAGAACTTTCCAAAGCCACTGTCAGAGAATGAACTCTTAGAACTTTTTGAAAAAATGATGGGA AGTCTTAAGAGAAGCCGACAGATCTCACCTCAGGAATTCATTCATGAGCTGAAAATGGGGTCTGCAGATGAGAGACTTGTCACATGCCTG GAGTCTCTCCGAGTGTCTTTGACCAGCAATCCTGTGAGTTGGGTGGAAAGCTTTGGACATGAAGGGCTTGGATTATTATTAGACATTTTG GAAAAACTGATTAGTGGAAAAATCCAAGAAAAAGTTGTAAAGAAAAATCAACATAAAGTCATACAGTGTCTAAAAGCCCTGATGAATACG CAGTATGGCTTGGAAAGAATTATGAGTGAGGAGAGGAGCCTTTCCTTATTGGCCAAAGCCGTGGATCCCAGACACCCCAATATGATGACA GATGTGGTTAAACTTCTCTCTGCGGTATGCATTGTAGGGGAAGAAAGCATCCTTGAAGAAGTTTTAGAAGCTTTAACTTCAGCTGGTGAA GAAAAAAAAATTGACAGATTTTTTTGTATTGTGGAAGGCCTCCGGCACAATTCAGTTCAACTGCAAATGTTTCCAGCCCTCTGAAGCTTC ATCGAACAGAGACTTTTCCAGCCTACAGATCTGAGCACTGACAGTAACTGCCAAGAATTGTTAACACACTTGTGATGTGTCAGCCACAGA TTCACCCAGGAGGTCACAGAATGACAGCAAGGGAAATGACGACCAAGAGAAGAAGCTTAAAGTCCTGGCTAATTGTGTGGTCATTGGAAA ACTCTGCAATACAATAATTTTCTTTATTTTCTTTTTCTTTTTTAAATTCTTAGTGTAATTGAAACGTGCTCTATAGATATTGACTCTGTG TTCCCTCTTTTACAGCTGGACAGAAAGAAGTCAATGTCACGAAATGATTTTCTATTGTAGATACTTTGTCCCTTGCACTTCTCTGAATCT GTCCTTTTGTGGATTCTTGTGATTTTCCTTCCAAGTGTTTCAGTTGTATGACAGTCAGTATTGACAATAAAATGGCTTTTAATTATTTGT >22769_22769_4_DIAPH3-DOK5_DIAPH3_chr13_60582677_ENST00000400320_DOK5_chr20_53266953_ENST00000395939_length(amino acids)=297AA_BP= MERHQPRLHHPAQGSAAGTPYPSSASLRGCRESKMPRRKGPQHPPPPSGPEEPGEKRPKFLDKFASIRIPGSKKERPPLPNLKTAFASSD CSAAPLEMMENFPKPLSENELLELFEKMMGSLKRSRQISPQEFIHELKMGSADERLVTCLESLRVSLTSNPVSWVESFGHEGLGLLLDIL EKLISGKIQEKVVKKNQHKVIQCLKALMNTQYGLERIMSEERSLSLLAKAVDPRHPNMMTDVVKLLSAVCIVGEESILEEVLEALTSAGE -------------------------------------------------------------- >22769_22769_5_DIAPH3-DOK5_DIAPH3_chr13_60582677_ENST00000400324_DOK5_chr20_53266953_ENST00000395939_length(transcript)=1711nt_BP=1235nt ATTGATCCGGCGGCGCGGGATTGGCCAGTCCCGGGCCAACACGGGTGGCGGGAGTTTTCAGATGTGTTGGCTGCGGCCCCGACTTCAGCG GTTGGGCTAGGCTGTGCTGACTGTTTGGGTGGCGGACCCCGGGAGTAAAACCTGTTGTCGATCCCTCAGCTTCCAGCTTGCGGCTTGCTG AGTGGCCACCTTCTTTCCGGTCCCCGGAGCTGCGGGGAAAGATGGAACGGCACCAGCCGCGGCTGCACCACCCGGCCCAAGGCTCAGCCG CTGGGACTCCCTACCCTTCCTCAGCCTCTCTCCGCGGCTGCCGGGAAAGCAAGATGCCGCGCAGGAAGGGCCCCCAACACCCTCCGCCGC CCAGTGGCCCCGAGGAGCCTGGGGAGAAGCGCCCCAAGTTTCATTTAAATATTAGGACACTGACGGATGATATGCTGGACAAATTTGCCA GCATAAGAATTCCAGGGAGCAAGAAAGAGAGACCTCCACTTCCCAACCTGAAGACTGCATTTGCAAGCAGTGATTGCTCAGCAGCACCTT TAGAGATGATGGAGAACTTTCCAAAGCCACTGTCAGAGAATGAACTCTTAGAACTTTTTGAAAAAATGATGGAAGATATGAATTTAAATG AAGATAAAAAGGCACCATTGCGGGAAAAGGACTTCAGTATCAAAAAAGAAATGGTGATGCAGTACATTAATACTGCTTCTAAGACAGGAA GTCTTAAGAGAAGCCGACAGATCTCACCTCAGGAATTCATTCATGAGCTGAAAATGGGGTCTGCAGATGAGAGACTTGTCACATGCCTGG AGTCTCTCCGAGTGTCTTTGACCAGCAATCCTGTGAGTTGGGTGGAAAGCTTTGGACATGAAGGGCTTGGATTATTATTAGACATTTTGG AAAAACTGATTAGTGGAAAAATCCAAGAAAAAGTTGTAAAGAAAAATCAACATAAAGTCATACAGTGTCTAAAAGCCCTGATGAATACGC AGTATGGCTTGGAAAGAATTATGAGTGAGGAGAGGAGCCTTTCCTTATTGGCCAAAGCCGTGGATCCCAGACACCCCAATATGATGACAG ATGTGGTTAAACTTCTCTCTGCGGTATGCATTGTAGGGGAAGAAAGCATCCTTGAAGAAGTTTTAGAAGCTTTAACTTCAGCTGGTGAAG AAAAAAAAATTGACAGATTTTTTTGTATTGTGGAAGGCCTCCGGCACAATTCAGTTCAACTGCAAATGTTTCCAGCCCTCTGAAGCTTCA TCGAACAGAGACTTTTCCAGCCTACAGATCTGAGCACTGACAGTAACTGCCAAGAATTGTTAACACACTTGTGATGTGTCAGCCACAGAT TCACCCAGGAGGTCACAGAATGACAGCAAGGGAAATGACGACCAAGAGAAGAAGCTTAAAGTCCTGGCTAATTGTGTGGTCATTGGAAAA CTCTGCAATACAATAATTTTCTTTATTTTCTTTTTCTTTTTTAAATTCTTAGTGTAATTGAAACGTGCTCTATAGATATTGACTCTGTGT TCCCTCTTTTACAGCTGGACAGAAAGAAGTCAATGTCACGAAATGATTTTCTATTGTAGATACTTTGTCCCTTGCACTTCTCTGAATCTG TCCTTTTGTGGATTCTTGTGATTTTCCTTCCAAGTGTTTCAGTTGTATGACAGTCAGTATTGACAATAAAATGGCTTTTAATTATTTGTT >22769_22769_5_DIAPH3-DOK5_DIAPH3_chr13_60582677_ENST00000400324_DOK5_chr20_53266953_ENST00000395939_length(amino acids)=380AA_BP= MFGWRTPGVKPVVDPSASSLRLAEWPPSFRSPELRGKMERHQPRLHHPAQGSAAGTPYPSSASLRGCRESKMPRRKGPQHPPPPSGPEEP GEKRPKFHLNIRTLTDDMLDKFASIRIPGSKKERPPLPNLKTAFASSDCSAAPLEMMENFPKPLSENELLELFEKMMEDMNLNEDKKAPL REKDFSIKKEMVMQYINTASKTGSLKRSRQISPQEFIHELKMGSADERLVTCLESLRVSLTSNPVSWVESFGHEGLGLLLDILEKLISGK IQEKVVKKNQHKVIQCLKALMNTQYGLERIMSEERSLSLLAKAVDPRHPNMMTDVVKLLSAVCIVGEESILEEVLEALTSAGEEKKIDRF -------------------------------------------------------------- >22769_22769_6_DIAPH3-DOK5_DIAPH3_chr13_60582677_ENST00000400330_DOK5_chr20_53266953_ENST00000395939_length(transcript)=1709nt_BP=1233nt TGATCCGGCGGCGCGGGATTGGCCAGTCCCGGGCCAACACGGGTGGCGGGAGTTTTCAGATGTGTTGGCTGCGGCCCCGACTTCAGCGGT TGGGCTAGGCTGTGCTGACTGTTTGGGTGGCGGACCCCGGGAGTAAAACCTGTTGTCGATCCCTCAGCTTCCAGCTTGCGGCTTGCTGAG TGGCCACCTTCTTTCCGGTCCCCGGAGCTGCGGGGAAAGATGGAACGGCACCAGCCGCGGCTGCACCACCCGGCCCAAGGCTCAGCCGCT GGGACTCCCTACCCTTCCTCAGCCTCTCTCCGCGGCTGCCGGGAAAGCAAGATGCCGCGCAGGAAGGGCCCCCAACACCCTCCGCCGCCC AGTGGCCCCGAGGAGCCTGGGGAGAAGCGCCCCAAGTTTCATTTAAATATTAGGACACTGACGGATGATATGCTGGACAAATTTGCCAGC ATAAGAATTCCAGGGAGCAAGAAAGAGAGACCTCCACTTCCCAACCTGAAGACTGCATTTGCAAGCAGTGATTGCTCAGCAGCACCTTTA GAGATGATGGAGAACTTTCCAAAGCCACTGTCAGAGAATGAACTCTTAGAACTTTTTGAAAAAATGATGGAAGATATGAATTTAAATGAA GATAAAAAGGCACCATTGCGGGAAAAGGACTTCAGTATCAAAAAAGAAATGGTGATGCAGTACATTAATACTGCTTCTAAGACAGGAAGT CTTAAGAGAAGCCGACAGATCTCACCTCAGGAATTCATTCATGAGCTGAAAATGGGGTCTGCAGATGAGAGACTTGTCACATGCCTGGAG TCTCTCCGAGTGTCTTTGACCAGCAATCCTGTGAGTTGGGTGGAAAGCTTTGGACATGAAGGGCTTGGATTATTATTAGACATTTTGGAA AAACTGATTAGTGGAAAAATCCAAGAAAAAGTTGTAAAGAAAAATCAACATAAAGTCATACAGTGTCTAAAAGCCCTGATGAATACGCAG TATGGCTTGGAAAGAATTATGAGTGAGGAGAGGAGCCTTTCCTTATTGGCCAAAGCCGTGGATCCCAGACACCCCAATATGATGACAGAT GTGGTTAAACTTCTCTCTGCGGTATGCATTGTAGGGGAAGAAAGCATCCTTGAAGAAGTTTTAGAAGCTTTAACTTCAGCTGGTGAAGAA AAAAAAATTGACAGATTTTTTTGTATTGTGGAAGGCCTCCGGCACAATTCAGTTCAACTGCAAATGTTTCCAGCCCTCTGAAGCTTCATC GAACAGAGACTTTTCCAGCCTACAGATCTGAGCACTGACAGTAACTGCCAAGAATTGTTAACACACTTGTGATGTGTCAGCCACAGATTC ACCCAGGAGGTCACAGAATGACAGCAAGGGAAATGACGACCAAGAGAAGAAGCTTAAAGTCCTGGCTAATTGTGTGGTCATTGGAAAACT CTGCAATACAATAATTTTCTTTATTTTCTTTTTCTTTTTTAAATTCTTAGTGTAATTGAAACGTGCTCTATAGATATTGACTCTGTGTTC CCTCTTTTACAGCTGGACAGAAAGAAGTCAATGTCACGAAATGATTTTCTATTGTAGATACTTTGTCCCTTGCACTTCTCTGAATCTGTC >22769_22769_6_DIAPH3-DOK5_DIAPH3_chr13_60582677_ENST00000400330_DOK5_chr20_53266953_ENST00000395939_length(amino acids)=380AA_BP= MFGWRTPGVKPVVDPSASSLRLAEWPPSFRSPELRGKMERHQPRLHHPAQGSAAGTPYPSSASLRGCRESKMPRRKGPQHPPPPSGPEEP GEKRPKFHLNIRTLTDDMLDKFASIRIPGSKKERPPLPNLKTAFASSDCSAAPLEMMENFPKPLSENELLELFEKMMEDMNLNEDKKAPL REKDFSIKKEMVMQYINTASKTGSLKRSRQISPQEFIHELKMGSADERLVTCLESLRVSLTSNPVSWVESFGHEGLGLLLDILEKLISGK IQEKVVKKNQHKVIQCLKALMNTQYGLERIMSEERSLSLLAKAVDPRHPNMMTDVVKLLSAVCIVGEESILEEVLEALTSAGEEKKIDRF -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for DIAPH3-DOK5 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for DIAPH3-DOK5 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for DIAPH3-DOK5 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
