
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:DNAH10-LIN7A (FusionGDB2 ID:23318) |
Fusion Gene Summary for DNAH10-LIN7A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: DNAH10-LIN7A | Fusion gene ID: 23318 | Hgene | Tgene | Gene symbol | DNAH10 | LIN7A | Gene ID | 196385 | 8825 |
| Gene name | dynein axonemal heavy chain 10 | lin-7 homolog A, crumbs cell polarity complex component | |
| Synonyms | - | LIN-7A|LIN7|MALS-1|MALS1|TIP-33|VELI1 | |
| Cytomap | 12q24.31 | 12q21.31 | |
| Type of gene | protein-coding | protein-coding | |
| Description | dynein heavy chain 10, axonemalaxonemal beta dynein heavy chain 10ciliary dynein heavy chain 10dynein, axonemal, heavy polypeptide 10 | protein lin-7 homolog Amammalian lin-seven protein 1tax interaction protein 33vertebrate LIN7 homolog 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8IVF4 | O14910 | |
| Ensembl transtripts involved in fusion gene | ENST00000409039, ENST00000538983, | ENST00000552864, | |
| Fusion gene scores | * DoF score | 4 X 4 X 2=32 | 12 X 5 X 4=240 |
| # samples | 4 | 7 | |
| ** MAII score | log2(4/32*10)=0.321928094887362 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(7/240*10)=-1.77760757866355 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: DNAH10 [Title/Abstract] AND LIN7A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | DNAH10(124323236)-LIN7A(81239718), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across DNAH10 (5'-gene) Fusion gene breakpoints across DNAH10 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
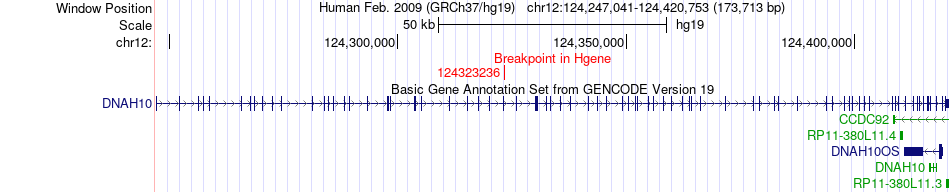 |
 Fusion gene breakpoints across LIN7A (3'-gene) Fusion gene breakpoints across LIN7A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
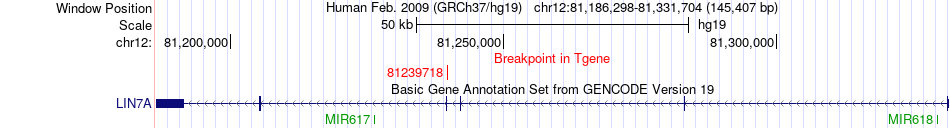 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-3B-A9HO-01A | DNAH10 | chr12 | 124323236 | - | LIN7A | chr12 | 81239718 | - |
| ChimerDB4 | SARC | TCGA-3B-A9HO-01A | DNAH10 | chr12 | 124323236 | + | LIN7A | chr12 | 81239718 | - |
Top |
Fusion Gene ORF analysis for DNAH10-LIN7A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000409039 | ENST00000552864 | DNAH10 | chr12 | 124323236 | + | LIN7A | chr12 | 81239718 | - |
| intron-3CDS | ENST00000538983 | ENST00000552864 | DNAH10 | chr12 | 124323236 | + | LIN7A | chr12 | 81239718 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000409039 | DNAH10 | chr12 | 124323236 | + | ENST00000552864 | LIN7A | chr12 | 81239718 | - | 10443 | 4807 | 25 | 5235 | 1736 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000409039 | ENST00000552864 | DNAH10 | chr12 | 124323236 | + | LIN7A | chr12 | 81239718 | - | 0.000138039 | 0.99986196 |
Top |
Fusion Genomic Features for DNAH10-LIN7A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
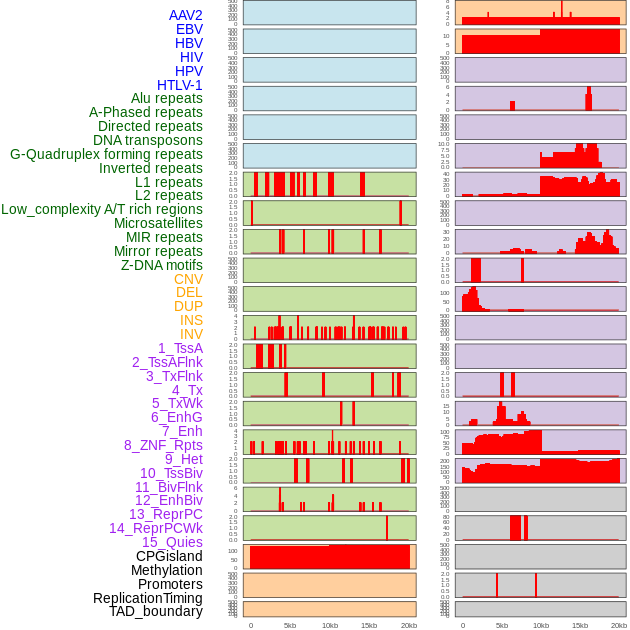 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for DNAH10-LIN7A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:124323236/chr12:81239718) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| DNAH10 | LIN7A |
| FUNCTION: Force generating protein of respiratory cilia. Produces force towards the minus ends of microtubules. Dynein has ATPase activity; the force-producing power stroke is thought to occur on release of ADP. Involved in sperm motility; implicated in sperm flagellar assembly (By similarity). Probable inner arm dynein heavy chain. {ECO:0000250}. | FUNCTION: Plays a role in establishing and maintaining the asymmetric distribution of channels and receptors at the plasma membrane of polarized cells. Forms membrane-associated multiprotein complexes that may regulate delivery and recycling of proteins to the correct membrane domains. The tripartite complex composed of LIN7 (LIN7A, LIN7B or LIN7C), CASK and APBA1 associates with the motor protein KIF17 to transport vesicles containing N-methyl-D-aspartate (NMDA) receptor subunit NR2B along microtubules (By similarity). This complex may have the potential to couple synaptic vesicle exocytosis to cell adhesion in brain. Ensures the proper localization of GRIN2B (subunit 2B of the NMDA receptor) to neuronal postsynaptic density and may function in localizing synaptic vesicles at synapses where it is recruited by beta-catenin and cadherin. Required to localize Kir2 channels, GABA transporter (SLC6A12) and EGFR/ERBB1, ERBB2, ERBB3 and ERBB4 to the basolateral membrane of epithelial cells. {ECO:0000250|UniProtKB:Q8JZS0, ECO:0000269|PubMed:12967566}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 1071_1106 | 1594 | 4472.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 1217_1245 | 1594 | 4472.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 203_223 | 1594 | 4472.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 602_622 | 1594 | 4472.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 18_25 | 1594 | 4472.0 | Compositional bias | Note=Poly-Glu |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 949_952 | 1594 | 4472.0 | Compositional bias | Note=Poly-Glu |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 1221_1254 | 1594 | 4472.0 | Repeat | Note=TPR 1 |
| Tgene | LIN7A | chr12:124323236 | chr12:81239718 | ENST00000552864 | 2 | 6 | 210_213 | 91 | 1908.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | LIN7A | chr12:124323236 | chr12:81239718 | ENST00000552864 | 2 | 6 | 217_229 | 91 | 1908.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | LIN7A | chr12:124323236 | chr12:81239718 | ENST00000552864 | 2 | 6 | 108_190 | 91 | 1908.0 | Domain | PDZ |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 2747_2770 | 1594 | 4472.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 3045_3131 | 1594 | 4472.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 3257_3327 | 1594 | 4472.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 3567_3638 | 1594 | 4472.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 4235_4260 | 1594 | 4472.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 1832_1839 | 1594 | 4472.0 | Motif | Note=GPAGTGKT motif |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 1882_1888 | 1594 | 4472.0 | Motif | Note=CFDEFNR motif |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 2113_2120 | 1594 | 4472.0 | Nucleotide binding | ATP |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 2455_2462 | 1594 | 4472.0 | Nucleotide binding | ATP |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 2803_2810 | 1594 | 4472.0 | Nucleotide binding | ATP |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 1794_2015 | 1594 | 4472.0 | Region | AAA 1 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 1_1793 | 1594 | 4472.0 | Region | Stem |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 2075_2294 | 1594 | 4472.0 | Region | AAA 2 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 2417_2665 | 1594 | 4472.0 | Region | AAA 3 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 2765_3014 | 1594 | 4472.0 | Region | AAA 4 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 3029_3313 | 1594 | 4472.0 | Region | Stalk |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 3399_3629 | 1594 | 4472.0 | Region | AAA 5 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 3845_4062 | 1594 | 4472.0 | Region | AAA 6 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 2736_2769 | 1594 | 4472.0 | Repeat | Note=TPR 2 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 2771_2797 | 1594 | 4472.0 | Repeat | Note=TPR 3 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 3802_3837 | 1594 | 4472.0 | Repeat | Note=TPR 4 |
| Hgene | DNAH10 | chr12:124323236 | chr12:81239718 | ENST00000409039 | + | 28 | 78 | 4074_4108 | 1594 | 4472.0 | Repeat | Note=TPR 5 |
| Tgene | LIN7A | chr12:124323236 | chr12:81239718 | ENST00000552864 | 2 | 6 | 25_80 | 91 | 1908.0 | Domain | L27 | |
| Tgene | LIN7A | chr12:124323236 | chr12:81239718 | ENST00000552864 | 2 | 6 | 14_28 | 91 | 1908.0 | Motif | Note=Kinase interacting site |
Top |
Fusion Gene Sequence for DNAH10-LIN7A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >23318_23318_1_DNAH10-LIN7A_DNAH10_chr12_124323236_ENST00000409039_LIN7A_chr12_81239718_ENST00000552864_length(transcript)=10443nt_BP=4807nt CTCGGCGCTCTTCATCTACCGCACTATGGTGCCGGAGGAGGTGGAGGTGGAGATTGATGAGATACCTGTCCTGTCTGAAGAGGGAGAAGA GGAAGAAGAGACTTATTCTCAAAAAGTGGAGTCCGTGGATAAAGTGCGAGCTAAGCGTGTGTCACTGAGAACCGAATCTCTAGGCCAACC TCTAAACAGAGAGGATGAAGAAATGGACAAAGAGATTTCAGAAAAACTCCCTTCCAAAAGAACTGCGAAGCACATCATGGAAAAGATGCA TCTCCACATGCTCTGTACCCCTCTTCCCGAGGAGTTCCTGGACCAAAACGTGGTGTTTTTCCTCAGAAATACCAAAGAGGCAATCTCTGA AGCTACCGACATGAAGGAAGCTATGGAAATTATGCCAGAAACACTGGAGTATGGAATTATAAACGCTAATGTGCTCCATTTTCTGAAGAA TATTATATGTCAGGTTTTTTTGCCAGCATTGTCCTTCAATCAGCACAGGACGAGTACAACCGTGGGAGTCACATCTGGAGAAGTCTCTAA TTCCTCTGAGCATGAATCAGACCTGCCGCCCATGCCTGGGGAGGCAGTAGAATATCACAGTATTCAATTAATACGGGATGAATTTTTAAT GAACGTGCAGAAATTTGCAAGTAATATTCAAAGAACCATGCAGCAACTTGAAGGTGAGATCAAGTTAGAAATGCCAATCATCAGTGTGGA GGGAGAGGTGTCTGACCTGGCAGCTGACCCGGAAACCGTTGACATCTTGGAGCAGTGTGTGATAAACTGGCTGAATCAGATATCCACAGC GGTTGAGGCCCAACTGAAGAAGACACCTCAGGGTAAAGGCCCTCTGGCTGAAATTGAATTCTGGAGGGAAAGAAATGCAACCTTAAGTGC GCTGCATGAACAAACAAAGCTTCCAATAGTCAGAAAAGTCTTGGATGTGATCAAGGAATCCGACTCCATGCTTGTGGCTAATCTGCAGCC AGTGTTCACCGAGTTATTCAAGTTCCACACGGAGGCCTCAGACAATGTGCGCTTTCTCTCCACCGTGGAGCGTTATTTCAAGAACATAAC GCACGGGTCTGGCTTCCACGTGGTCCTGGACACCATCCCCGCCATGATGAGTGCCCTGCGGATGGTGTGGATCATCTCCCGACACTACAA CAAAGACGAGAGGATGATTCCGCTCATGGAGCGCATCGCCTGGGAAATCGCTGAGAGAGTCTGCCGAGTGGTCAACCTGCGGACTTTGTT CAAAGAAAATCGAGCGAGTGCCCAAAGCAAAACCTTGGAAGCCAGGAACACCCTCAGGCTGTGGAAAAAGGCCTATTTTGACACCCGGGC CAAGATAGAGGCTTCGGGGAGGGAAGATCGGTGGGAGTTTGACCGGAAGCGGCTGTTCGAGAGGACGGATTATATGGCCACCATCTGCCA GGACCTCTCCGACGTTCTGCAGATTTTGGAGGAATTTTATAACATATTTGGTCCAGAACTAAAGGCAGTGACGGGGGACCCCAAGCGCAT TGATGATGTCCTATGCAGAGTGGACGGCCTAGTCACCCCCATGGAAAACCTGACCTTTGACCCCTTCAGCATCAAGTCCTCCCAGTTCTG GAAATATGTGATGGATGAATTCAAGATTGAAGTTCTGATTGACATCATTAATAAAATCTTTGTCCAGAACCTTGAAAATCCACCACTGTA TAAGAATCACCCTCCAGTAGCAGGTGCAATATACTGGGAACGATCTCTGTTCTTTCGGATTAAGCATACCATCCTCCGATTTCAAGAGGT ACAAGAGATACTGGACAGTGATCGAGGACAGGAGGTCAAACAAAAATATTTGGAAGTAGGTAGGACAATGAAGGAGTATGAAGACAGAAA GTATGAGCAGTGGATGGAGGTGACGGAGCAGGTGCTGCCAGCTCTCATGAAGAAGAGCCTTTTGACCAAGTCTTCCATCGCCACAGAGGA GCCTTCGACTTTAGAAAGGGGAGCTGTTTTTGCAATCAACTTTTCACCGGCTCTCAGAGAGATTATTAATGAAACAAAGTACTTAGAGCA GCTGGGGTTCACTGTCCCTGAATTAGCAAGAAATGTTGCTCTCCAGGAAGACAAATTCCTTAGGTACACAGCTGGGATACAGCGCATGTT GGATCATTATCACATGCTCATAGGAACGTTAAACGATGCGGAGTCTGTGCTTCTCAAAGATCATTCCCAGGAACTGCTCCGAGTGTTTAG GTCGGGATATAAGAGGTTGAACTGGAACTCACTAGGTATCGGTGACTATATAACTGGTTGCAAACAGGCCATTGGGAAATTTGAGTCTCT CGTCCACCAGATTCATAAGAATGCAGATGACATTTCTTCCAGGCTGACATTAATAGAGGCCATAAATCTCTTTAAATATCCAGCCGCTAA AAGTGAGGAAGAACTCCCAGGCGTGAAGGAATTTTTTGAACACATTGAGCGAGAAAGGGCCAGCGACGTGGACCACATGGTCCGGTGGTA TCTTGCCATTGGACCACTGCTGACCAAAGTTGAGGGCCTGGTCGTCCACACCAACACAGGCAAGGCCCCCAAGCTGGCCTCCTACTACAA ATACTGGGAAAAGAAAATTTATGAGGTCCTGACAAAGCTCATCCTGAAGAACTTGCAGTCTTTTAATTCTTTGATCCTTGGAAATGTCCC TCTGTTCCACACTGAAACCATTCTGACGGCACCTGAGATCATCCTTCATCCCAACACAAATGAGATCGACAAGATGTGCTTCCATTGTGT CCGGAATTGCGTGGAGATCACCAAGCATTTTGTTCGTTGGATGAATGGCAGCTGCATAGAATGCCCACCTCAGAAGGGGGAGGAAGAGGA AGTTGTTATAATAAACTTTTACAATGATATCTCTCTGAACCCTCAGATAATTGAACAAGCTGTTATGATCCCCCAAAATGTCCACAGGAT TCTGATCAATCTTATGAAGTATCTACAAAAATGGAAGCGGTATCGACCTCTCTGGAAATTGGACAAAGCTATTGTGATGGAGAAATTTGC TGCCAAGAAACCTCCTTGTGTAGCATATGATGAAAAGTTGCAGTTCTATTCCAAGATAGCTTATGAGGTTATGCGCCACCCTCTAATTAA GGATGAGCATTGCATCAGACTTCAGCTCAGGCATCTGGCAAACACAGTGCAGGAAAATGCCAAGTCCTGGGTGATTTCGCTTGGAAAACT TCTCAATGAGTCAGCAAAAGAGGAGCTCTATAATCTCCATGAAGAGATGGAGCACCTGGCCAAAAACCTTAGGAAGATCCCCAATACCCT TGAAGATCTCAAGTTTGTCCTTGCAACAATTGCAGAAATTAGAAGTAAATCTCTAGTCATGGAACTCAGATATAGGGACGTCCAGGAGCG ATACCGTACCATGGCAATGTATAACCTCTTTCCTCCTGATGCAGAGAAAGAACTGGTTGATAAGATTGAGAGCATATGGTCCAATCTGTT TAATGATTCAGTGAATGTGGAGCATGCTCTTGGGGACATAAAGAGAACTTTCACAGAGCTTACTCGAGGCGAAATAATGAACTACAGAGT TCAGATAGAGGAGTTTGCAAAGCGTTTTTACAGTGAAGGCCCTGGTTCTGTTGGTGATGATCTTGATAAAGGAGTAGAGCTTTTAGGTGT TTATGAAAGAGAGCTGGCAAGACATGAAAAGAGCCGTCAGGAACTGGCTAACGCTGAGAAACTTTTCGATCTTCCTATTACAATGTACCC AGAGCTGCTGAAAGTGCAGAAGGAAATGAGTGGGCTGAGGATGATTTACGAGCTCTATGAAGGACTAAAGGTTGCAAAAGAAGAATGGTC TCAGACCCTTTGGATCAACCTGAATGTGCAGATTCTCCAGGAAGGAATTGAAGGTTTTCTCAGGGCTCTCAGAAAGCTACCTCGGCCAGT CCGTGGCTTATCAGTGACCTACTACTTGGAAGCAAAAATGAAGGCATTCAAAGACTCGATTCCTTTACTTCTTGACTTGAAAAACGAGGC ACTAAGAGACAGGCATTGGAAAGAACTTATGGAAAAAACGTCTGTCTTTTTTGAAATGACCGAAACGTTCACCTTGGAAAATATGTTTGC TATGGAACTGCACAAACACACAGATGTTCTCAATGAGATTGTCACAGCAGCAATCAAGGAGGTTGCCATTGAGAAGGCTGTGAAGGAAAT CCTAGACACGTGGGAAAATATGAAATTCACTGTAGTCAAGTATTGCAAAGGCACACAGGAGCGAGGCTACATCCTGGGTTCTGTTGACGA AATTATTCAGTCTCTTGATGACAACACTTTCAACCTGCAGAGCATCTCAGGAAGCAGATTTGTGGGGCCTTTTCTGCAAACTGTTCACAA ATGGGAAAAAACGCTTTCTCTAATAGGGGAAGTCATTGAGATTTGGATGTTGGTTCAGAGAAAATGGATGTATCTTGAAAGTATTTTTAT TGGTGGAGATATAAGATCACAACTTCCGGAAGAGGCAAAAAAGTTTGACAACATCGATAAAGTATTTAAAAGGATCATGGGTGAGACCTT AAAAGACCCCGTGATCAAGAGGTGCTGTGAAGCCCCAAACCGCCTCAGTGACCTACAGAACGTCAGCGAGGGCCTGGAGAAATGCCAGAA AAGCCTCAACGACTACTTAGATTCGAAGAGAAATGCTTTCCCAAGGTTCTTCTTCATTTCTGACGATGAGTTGCTTAGCATTCTGGGGAG CAGCGACCCACTCTGCGTCCAGGAGCACATGATCAAGGCAACAGTTGCAGCTTTTGCAGCTAGTGAAGGCCACTCCCACCCTCGAGTAGT TGAACTGCCAAAGACTGATGAAGGCCTTGGTTTTAATGTGATGGGAGGAAAGGAGCAAAATTCCCCCATTTATATCTCTCGCATAATTCC TGGAGGGGTGGCTGAAAGACACGGAGGCCTCAAAAGAGGAGACCAGCTGCTATCAGTGAACGGAGTGAGTGTGGAAGGAGAACACCATGA GAAAGCTGTGGAACTACTCAAGGCTGCTAAAGACAGCGTCAAGCTGGTGGTGCGATACACCCCAAAAGTTCTGGAAGAAATGGAGGCTCG CTTTGAAAAGCTACGAACAGCCAGGCGTCGGCAGCAGCAGCAATTGCTAATTCAGCAGCAGCAACAGCAGCAGCAGCAACAAACACAACA AAACCACATGTCATAGGCCCTTGAGGGAAAGCTACTTGATCAAACATCCGATAGTCACAAATTTGAAACCGTGCTTCAGAATCCCAGCAC ATAGTAAAAGACAACACTGATAATTATACCTGTCAAGAAGCTGTGAACACATGGTGTATAAATTCTTTACCAAGGCAACTCAACACCTTC TTTCTCTGGGCTTGAACCGCCACTGCTCACGTGGGCTTTACATACATTGACCTTCCATTCACTGCAGTGGGAATTCTCAGTGTGCAGAGG GAGAGGTTTTCTAGTCTGCAAACTGAAACAGTGTAAGAAGAATAAAGTCTATGACTTTTAAATAAATCACCAGGGCCAAGATTGAGTGAA TCTTGGGTCCATAGCTTTTCCAGATATTAGCAAACCCCTCATTAGGCTATAATTCAAACTGCATCAGTTCAAAGTGAAACCTACACATTG CTCCTTACCCTCACCTGCTCCACAATCCATAGTGCTGAGTTGTCTGAATTGGAATTCCAGTTAATTGCAAGTTCAACCTGTTTTCCTCTT GAAAATCGCAAGGTATCATACTATATTTATATTTTCTTCAGGTTCTAGAAGCAATCGGCTTTTTCTTTTGTTTCCATGAATTTTATTATG CATTAATAGAATGGACCAATTTTAGCTCAACTTTTAGTTTGTTAGAAGCAAGTGTAGGAACTCTAGCACTGTAGTTTTTAATTATTGCTT GTATCTATTATTATTAATTCCAACAGAGTATAATGTATATTTATTCTATAAAATATATATTATCAGAGTGCATTTGTTACAACTTAGGTT CTTTTCTTACCAAGTATTAAGAATCTAGTAAGAGAATACTAGCAAAGGACCTAGCCCTGTGAACAGATTCTCGTATGTTATATACATAAA CCCACTCTCAATATTTTTTGAGTTTTATACTTTTTGATTGATAAACACTGAAGAAATCTAAACTCTACTCACACTGATGTGCTGTCATAG AAGCCTATTTCATATTTTAAAGCTTAGCAAAACTAAATTATATGTGCATATATATTCAAATATTTGTGTCACACAAAGTTTAGGTCAATT TCCCCAAATATAAACCAGATATTTTTGGCACTCTCAGATAAACAAGTATTATTTCTCTTATAATTGGGCAGGTGGAAACCAGTGATATCA ATATTGTGAATGGCAGATGCTGCTTCCTCTAGAAAAATAAAATAAAATCACCAACTTTAATATATTCCCATAGGTAAATAGACAATGAGA TATCATTTTTCAGAAAATATCATTCACTGGAAAATAATCTGTTATAACTACATTTACAGGTCATAATTTTTGGTTTATGGTAAATCTAGG CACCTAAGTCTGATTTTACTTTATGAAATAAATAGGAAGCAATAATTGTTTGGGCATTTGAATACCTTAAGCACATAAATGAACCTAAGA AAAGAATAATTGCTTAGTTGAAGTATTCAAAATAGATCAGTAAACCACAGGCAAATATTTGAATGTTCCCTTTTCTCAAATACAGAAATT GATTCCCCAAGATACAATACAATTTGACACTTCAGATAATCCCCCAAAATAGAAATGTATGAAGCTCAGTTTAGACTGTCTTCTTGAAAA TAAAACAAAACCAATCAAAACTACCCAGCACATTTTATTATGCCCTGTTGGGTGATAGTATTGTCAATACCAGGTTTTCCCTGCTCTTCA CAATGCATCCCTCACCAGAGGAAAAACAGCAAGAAAACTAGGACATTTGCTAATTTCAACAATACACCCCTGTGATAACAGATTCCCATC CTTGAATACATATCATTTTTTACTAGAGAGAACGGGGGTGGTGAGGTGGGGGCAGTTAGGACCCTGTATGTTATGTAAAATAGATTTACA AAAAGGACAAGCATGATAAAAGGGAATGAATGGAGACGATGTATTATTCAGTTTGAAAGAGATAGTTCACATTTTTATAAGAAAAATTTA ATAAAACATATATTTTAGTTATCATTTAACAAAGATATATTAAAGCTATTCAGGCAATGCCACAGGATTCTCATGAAGGGAAATCACTGA ACCCTTGTCATGAGAAAATGGTCATTTCAACACATTTCAATACATTGTATAAATTTGGAGCTTAAGGTGAGTGAGTATGTTTTCACTTAG TCATAAATGTTTGGTCAAAATTAAAGTTATCATATGGTTTCACATCAATACAAATATTGTCACAGTGTTACAGAAACAACTATCAAATGG TAAGATAGTTTACATTTGTGTTTTCTAGAAACCTTTGTATTTATGCTTCATTTGGTTAATGTTAAGTGCTGATAAAGGCATACCAGATTG TTTGTCAAAGTAAAGAAGCAATCCATCACACCTAGGACATCTAAGGATGGATTATACGTCAGAGTGGTCTCATATACAAATGACCATTTT CCTTCAGTAAAATCTTGTTGGTTGAAATGTAAGTCTTTTCCTAAAGTTTTAATCAGAGGTAGCCATCACTAAGACTTAAGCCACCTGTGG TTCTCTTAAGTTTCACTGAAGCCAGAAGAAGGAAATTACCACAACTTGTATTATACTAATTATCTTCATTATTAACGATCATATTAGAGC CACTGACATGTCCCAAATTATATTAAAATAAAAACCTGCATTGCTCTGACATGAAGCTCAATTCAATGTAATAAACAAATTAGGTATTAA ACGTTATAATTTAAAAAACTTCTACGATATCACCAGAAATCCTGGTGAAATTTAATTTTTTCCCTTTTTTAGGTCAGTGGCCGGTGGGAT AGATTTTTCTAAACTTTTTCCTGCTATGATTTTGAAGGCAAATACATAGAGCACAGTTCAGTAAAAGAGTTTCCAAGCCTTTTCACTGGA GCATTTACATTTTGATTCTGTTGTCATTTGAAATAGAACATCTGCAGGGAGATAATGAGCTTAAGTTACAATTGACTTTGGAAAGAATAA CAGACTTTTGTGTTTCCTCAATCATGTAGTCAATAGGGAATTCTCCAGTTATGTGAACAATCACAGTCTTTTAAGACAATACTTAGATCT AAATGCAAAATCTAGCATGCAGAGGCTTTTTAAGGATTTTAGACTGTCTCCAAAGAACTTGGCTCGAGATGGATTACACAGATTTACAAA TGCAGTTTTCAACTGTGACATCAAACTGATCTTATCTCATTTCATGTTACTCAACTGTGCCCGTAGGACCCCAAGAGGTGGAATATTTTG TCTACATTATTTTAAACATCTATAGATATGTGTGGATGTGTGAATAATGCCTAAATTTCTCATCTTTTAACTAGTTTGTTTTTATCATAC AGACACAGCCTGTTCATAAGACCAGCTTTAGACAACTTAGAACTCAATGCTCTGAAGATTCAGGGAGGCCAACATAGAAAAAGATCTTCC AAAATATGGGGTGGTCCAACCCATCTCATCCCTCTCTAGCTGGAAAACTATAGAGAAAGGGCTGTGACTGCCTTGACCACTGCCTGGCAC AATGTAGGCACTCGGTATTTGTTGGATGAATAAGTGGCTGAATCAGTGAATAGACAAAAAACATGGATTTTGCCTTCTGAAAATCTCACA AATTTTACCAGCTTATATTATACTGATTGTCACTATGATTTCTAAGTTTCATGGTTAGAACCATATTTTGAGTCTATATAAAAACTTTTT AGATGTATGAATTTGAAATTTCATTAATTACTTTAAAGGCTTCAAAGATTTGAGGACATTTGAACAGTGGATAAGGAACAATGTTTGGGG ATACAGAGTTTTTATGCTGAGTTGAACTCAACCTTTCTAGTTCTTTATACAAGCTTCCTGAATTATCTGGCAGGAATGATTCAAACATAA AACCCCATTTTTAAATTTTATAAAAGATATTTTCATTTTTATATAAAAAGATCATCAGGCATATTATCCCCTTTGATATTTATTAACAAC CATTCTCTAAGAAATTCATAATAATTGGAGACACCATCCCTAAATCTCTTCCTATTTTCAAAAGATCATTTATCCTTTCATATTTGATTC CTGGAATACACTAAGCTGAAGTTTAATCACATACTTACTCAGATGTTTCCATGTGTTTATTCATTTAACAAATACTTATTGTGTACCTAG TGCATGCTAGGTTCTGGAAATGCAACAGCAAACAAGACAGACACAATCCCTCTTGGAGGTTACTGTGAGAAACTCTCCAACATTTACTCT AGCAATAACTAGAGAGTTTACTGTCCATTGGGTCCTCTTCACACCTGATAGAAAAAATAAAATTCCTCACCCCATGGAAGTCATGAAATA CTCCCTGCACAGATTTGTGTGAGCTTGCTTTTCTAAAACCACTGCTCAAAAATGCTGCTTAGTTTATCTATAAACCACGGTTATACCCCA GTGAGGTAGGTAAGTAATTTATTCATAACTAACATTTATGCAGCTCTATTTTCTAGCCACTGTGCTGAGTGCTTTTATGGACTATCTCAC CAAATCCCTTCAACAACTCTATTCAGTAGGTTCCATTTTATAGATACAAAAACTGAAGCACAGATATATTATGTGACTTGTCCAAGGTCA CACAGCTAGCAGGTGGAACTGGGATTTGAACCCATGCAATCTGACACAAGAGTTCATGTGTCTAATCATTATCTGTGGTTTGGGGGACAC CATTACATTAAAATGAATGAGTCACTAAGTGAGTCACTGTATATAAAATACTTGGAAATTGTAAAGTAATATACAAAAGATATTAACAAA GTATTATTCATGGTAAACAAATCACATCAGTAGCAAACACAGGGTGGGAACTCAATCTTATGTTATAGCTTACTCTTAAGAAGAATATGT GAAGCCAGGAACATGGTTAAAGGTACAGACCAGAAAATTTCTGACATGTGATAAATATTTCAGTGACTTTTCAGATTTATTTCTTGTTAG CCGCTGTGTCTATTTGGTGCTACAAAAACTGAAAGAAACAAAATCCCTGATGTAAGGGCTTATAATAAATACACAGTTTGGAGATGAACT AAGGATATTTTACATTTACGGGAAAGGATTGGACAATAGTATCATTTTCTCACCATTTGCATGTACATCATGTCTTTCCAATTGTTTTCA TTATTTTTCTGAAAGAGCTGCAAAATTGTGCAGTGGTTCAACCTGACCAAAGTGGTATTATGCTGCTGGGAAGTAAAAGTATTAGACACC >23318_23318_1_DNAH10-LIN7A_DNAH10_chr12_124323236_ENST00000409039_LIN7A_chr12_81239718_ENST00000552864_length(amino acids)=1736AA_BP=1594 MVPEEVEVEIDEIPVLSEEGEEEEETYSQKVESVDKVRAKRVSLRTESLGQPLNREDEEMDKEISEKLPSKRTAKHIMEKMHLHMLCTPL PEEFLDQNVVFFLRNTKEAISEATDMKEAMEIMPETLEYGIINANVLHFLKNIICQVFLPALSFNQHRTSTTVGVTSGEVSNSSEHESDL PPMPGEAVEYHSIQLIRDEFLMNVQKFASNIQRTMQQLEGEIKLEMPIISVEGEVSDLAADPETVDILEQCVINWLNQISTAVEAQLKKT PQGKGPLAEIEFWRERNATLSALHEQTKLPIVRKVLDVIKESDSMLVANLQPVFTELFKFHTEASDNVRFLSTVERYFKNITHGSGFHVV LDTIPAMMSALRMVWIISRHYNKDERMIPLMERIAWEIAERVCRVVNLRTLFKENRASAQSKTLEARNTLRLWKKAYFDTRAKIEASGRE DRWEFDRKRLFERTDYMATICQDLSDVLQILEEFYNIFGPELKAVTGDPKRIDDVLCRVDGLVTPMENLTFDPFSIKSSQFWKYVMDEFK IEVLIDIINKIFVQNLENPPLYKNHPPVAGAIYWERSLFFRIKHTILRFQEVQEILDSDRGQEVKQKYLEVGRTMKEYEDRKYEQWMEVT EQVLPALMKKSLLTKSSIATEEPSTLERGAVFAINFSPALREIINETKYLEQLGFTVPELARNVALQEDKFLRYTAGIQRMLDHYHMLIG TLNDAESVLLKDHSQELLRVFRSGYKRLNWNSLGIGDYITGCKQAIGKFESLVHQIHKNADDISSRLTLIEAINLFKYPAAKSEEELPGV KEFFEHIERERASDVDHMVRWYLAIGPLLTKVEGLVVHTNTGKAPKLASYYKYWEKKIYEVLTKLILKNLQSFNSLILGNVPLFHTETIL TAPEIILHPNTNEIDKMCFHCVRNCVEITKHFVRWMNGSCIECPPQKGEEEEVVIINFYNDISLNPQIIEQAVMIPQNVHRILINLMKYL QKWKRYRPLWKLDKAIVMEKFAAKKPPCVAYDEKLQFYSKIAYEVMRHPLIKDEHCIRLQLRHLANTVQENAKSWVISLGKLLNESAKEE LYNLHEEMEHLAKNLRKIPNTLEDLKFVLATIAEIRSKSLVMELRYRDVQERYRTMAMYNLFPPDAEKELVDKIESIWSNLFNDSVNVEH ALGDIKRTFTELTRGEIMNYRVQIEEFAKRFYSEGPGSVGDDLDKGVELLGVYERELARHEKSRQELANAEKLFDLPITMYPELLKVQKE MSGLRMIYELYEGLKVAKEEWSQTLWINLNVQILQEGIEGFLRALRKLPRPVRGLSVTYYLEAKMKAFKDSIPLLLDLKNEALRDRHWKE LMEKTSVFFEMTETFTLENMFAMELHKHTDVLNEIVTAAIKEVAIEKAVKEILDTWENMKFTVVKYCKGTQERGYILGSVDEIIQSLDDN TFNLQSISGSRFVGPFLQTVHKWEKTLSLIGEVIEIWMLVQRKWMYLESIFIGGDIRSQLPEEAKKFDNIDKVFKRIMGETLKDPVIKRC CEAPNRLSDLQNVSEGLEKCQKSLNDYLDSKRNAFPRFFFISDDELLSILGSSDPLCVQEHMIKATVAAFAASEGHSHPRVVELPKTDEG LGFNVMGGKEQNSPIYISRIIPGGVAERHGGLKRGDQLLSVNGVSVEGEHHEKAVELLKAAKDSVKLVVRYTPKVLEEMEARFEKLRTAR -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for DNAH10-LIN7A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for DNAH10-LIN7A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for DNAH10-LIN7A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
