
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:DNMT1-FDX1L (FusionGDB2 ID:23695) |
Fusion Gene Summary for DNMT1-FDX1L |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: DNMT1-FDX1L | Fusion gene ID: 23695 | Hgene | Tgene | Gene symbol | DNMT1 | FDX1L | Gene ID | 1786 | 112812 |
| Gene name | DNA methyltransferase 1 | ferredoxin 2 | |
| Synonyms | ADCADN|AIM|CXXC9|DNMT|HSN1E|MCMT|m.HsaI | FDX1L|MEOAL | |
| Cytomap | 19p13.2 | 19p13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | DNA (cytosine-5)-methyltransferase 1CXXC-type zinc finger protein 9DNA (cytosine-5-)-methyltransferase 1DNA MTase HsaIDNA methyltransferase HsaI | ferredoxin-2, mitochondrialadrenodoxin-like protein, mitochondrialferredoxin 1 like | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P26358 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000340748, ENST00000359526, ENST00000540357, ENST00000589538, | ENST00000492239, ENST00000494368, ENST00000393708, ENST00000541276, | |
| Fusion gene scores | * DoF score | 14 X 12 X 11=1848 | 6 X 3 X 5=90 |
| # samples | 17 | 7 | |
| ** MAII score | log2(17/1848*10)=-3.44235810527836 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/90*10)=-0.362570079384708 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: DNMT1 [Title/Abstract] AND FDX1L [Title/Abstract] AND fusion [Title/Abstract] | ||
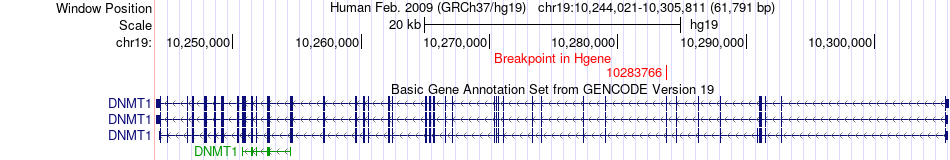
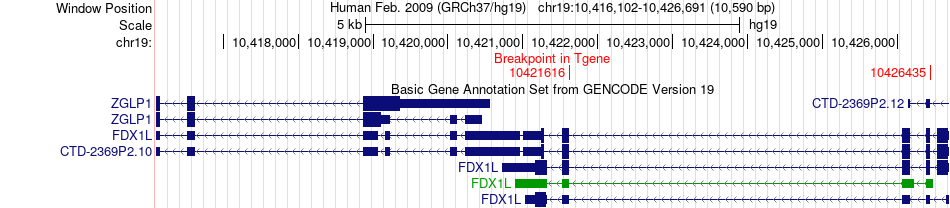
| Most frequent breakpoint | DNMT1(10283766)-FDX1L(10421616), # samples:1 DNMT1(10283766)-FDX1L(10426435), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | DNMT1-FDX1L seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. DNMT1-FDX1L seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. DNMT1-FDX1L seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. DNMT1-FDX1L seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. DNMT1-FDX1L seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. DNMT1-FDX1L seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | DNMT1 | GO:0010216 | maintenance of DNA methylation | 18754681|21745816 |
 Fusion gene breakpoints across DNMT1 (5'-gene) Fusion gene breakpoints across DNMT1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across FDX1L (3'-gene) Fusion gene breakpoints across FDX1L (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LIHC | TCGA-DD-A3A5-01A | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - |
| ChimerDB4 | UVM | TCGA-V4-A9EX-01A | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - |
Top |
Fusion Gene ORF analysis for DNMT1-FDX1L |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000340748 | ENST00000492239 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - |
| 5CDS-5UTR | ENST00000340748 | ENST00000492239 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - |
| 5CDS-5UTR | ENST00000340748 | ENST00000494368 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - |
| 5CDS-5UTR | ENST00000340748 | ENST00000494368 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - |
| 5CDS-5UTR | ENST00000359526 | ENST00000492239 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - |
| 5CDS-5UTR | ENST00000359526 | ENST00000492239 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - |
| 5CDS-5UTR | ENST00000359526 | ENST00000494368 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - |
| 5CDS-5UTR | ENST00000359526 | ENST00000494368 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - |
| 5CDS-5UTR | ENST00000540357 | ENST00000492239 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - |
| 5CDS-5UTR | ENST00000540357 | ENST00000492239 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - |
| 5CDS-5UTR | ENST00000540357 | ENST00000494368 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - |
| 5CDS-5UTR | ENST00000540357 | ENST00000494368 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - |
| Frame-shift | ENST00000340748 | ENST00000393708 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - |
| Frame-shift | ENST00000340748 | ENST00000393708 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - |
| Frame-shift | ENST00000359526 | ENST00000393708 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - |
| Frame-shift | ENST00000359526 | ENST00000393708 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - |
| Frame-shift | ENST00000540357 | ENST00000393708 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - |
| Frame-shift | ENST00000540357 | ENST00000393708 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - |
| In-frame | ENST00000340748 | ENST00000541276 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - |
| In-frame | ENST00000340748 | ENST00000541276 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - |
| In-frame | ENST00000359526 | ENST00000541276 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - |
| In-frame | ENST00000359526 | ENST00000541276 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - |
| In-frame | ENST00000540357 | ENST00000541276 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - |
| In-frame | ENST00000540357 | ENST00000541276 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - |
| intron-3CDS | ENST00000589538 | ENST00000393708 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - |
| intron-3CDS | ENST00000589538 | ENST00000393708 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - |
| intron-3CDS | ENST00000589538 | ENST00000541276 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - |
| intron-3CDS | ENST00000589538 | ENST00000541276 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - |
| intron-5UTR | ENST00000589538 | ENST00000492239 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - |
| intron-5UTR | ENST00000589538 | ENST00000492239 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - |
| intron-5UTR | ENST00000589538 | ENST00000494368 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - |
| intron-5UTR | ENST00000589538 | ENST00000494368 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000340748 | DNMT1 | chr19 | 10283766 | - | ENST00000541276 | FDX1L | chr19 | 10421616 | - | 2592 | 956 | 44 | 1003 | 319 |
| ENST00000540357 | DNMT1 | chr19 | 10283766 | - | ENST00000541276 | FDX1L | chr19 | 10421616 | - | 2536 | 900 | 180 | 947 | 255 |
| ENST00000359526 | DNMT1 | chr19 | 10283766 | - | ENST00000541276 | FDX1L | chr19 | 10421616 | - | 2584 | 948 | 180 | 995 | 271 |
| ENST00000340748 | DNMT1 | chr19 | 10283766 | - | ENST00000541276 | FDX1L | chr19 | 10426435 | - | 2754 | 956 | 44 | 1015 | 323 |
| ENST00000540357 | DNMT1 | chr19 | 10283766 | - | ENST00000541276 | FDX1L | chr19 | 10426435 | - | 2698 | 900 | 180 | 959 | 259 |
| ENST00000359526 | DNMT1 | chr19 | 10283766 | - | ENST00000541276 | FDX1L | chr19 | 10426435 | - | 2746 | 948 | 180 | 1007 | 275 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000340748 | ENST00000541276 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - | 0.028503852 | 0.97149616 |
| ENST00000540357 | ENST00000541276 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - | 0.01209541 | 0.9879046 |
| ENST00000359526 | ENST00000541276 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10421616 | - | 0.011585866 | 0.98841405 |
| ENST00000340748 | ENST00000541276 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - | 0.034134082 | 0.9658659 |
| ENST00000540357 | ENST00000541276 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - | 0.012932624 | 0.9870674 |
| ENST00000359526 | ENST00000541276 | DNMT1 | chr19 | 10283766 | - | FDX1L | chr19 | 10426435 | - | 0.009857947 | 0.99014205 |
Top |
Fusion Genomic Features for DNMT1-FDX1L |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
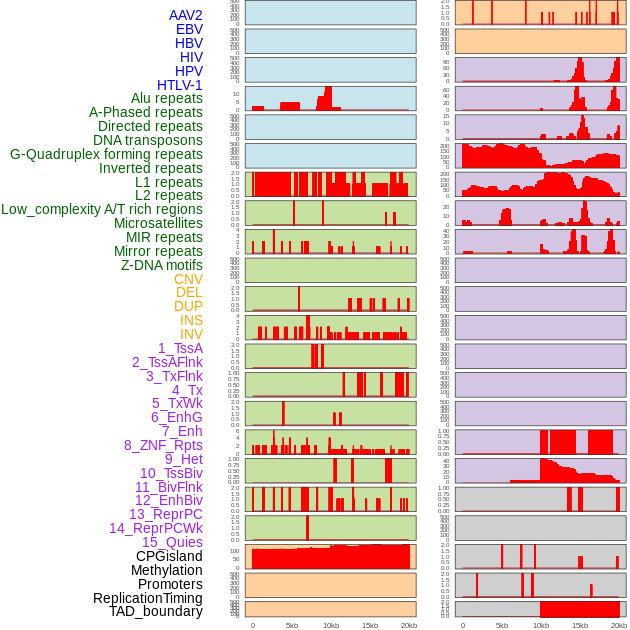
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for DNMT1-FDX1L |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:10283766/chr19:10421616) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| DNMT1 | . |
| FUNCTION: Methylates CpG residues. Preferentially methylates hemimethylated DNA. Associates with DNA replication sites in S phase maintaining the methylation pattern in the newly synthesized strand, that is essential for epigenetic inheritance. Associates with chromatin during G2 and M phases to maintain DNA methylation independently of replication. It is responsible for maintaining methylation patterns established in development. DNA methylation is coordinated with methylation of histones. Mediates transcriptional repression by direct binding to HDAC2. In association with DNMT3B and via the recruitment of CTCFL/BORIS, involved in activation of BAG1 gene expression by modulating dimethylation of promoter histone H3 at H3K4 and H3K9. Probably forms a corepressor complex required for activated KRAS-mediated promoter hypermethylation and transcriptional silencing of tumor suppressor genes (TSGs) or other tumor-related genes in colorectal cancer (CRC) cells (PubMed:24623306). Also required to maintain a transcriptionally repressive state of genes in undifferentiated embryonic stem cells (ESCs) (PubMed:24623306). Associates at promoter regions of tumor suppressor genes (TSGs) leading to their gene silencing (PubMed:24623306). Promotes tumor growth (PubMed:24623306). {ECO:0000269|PubMed:16357870, ECO:0000269|PubMed:18413740, ECO:0000269|PubMed:18754681, ECO:0000269|PubMed:24623306}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 16_109 | 240 | 1617.0 | Domain | DMAP1-binding |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 16_109 | 256 | 1633.0 | Domain | DMAP1-binding |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 16_109 | 240 | 1617.0 | Domain | DMAP1-binding |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 16_109 | 256 | 1633.0 | Domain | DMAP1-binding |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 177_205 | 240 | 1617.0 | Motif | Nuclear localization signal |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 177_205 | 256 | 1633.0 | Motif | Nuclear localization signal |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 177_205 | 240 | 1617.0 | Motif | Nuclear localization signal |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 177_205 | 256 | 1633.0 | Motif | Nuclear localization signal |
| Tgene | FDX1L | chr19:10283766 | chr19:10426435 | ENST00000393708 | 0 | 5 | 71_173 | 48 | 184.0 | Domain | 2Fe-2S ferredoxin-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 1139_1599 | 240 | 1617.0 | Domain | SAM-dependent MTase C5-type |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 755_880 | 240 | 1617.0 | Domain | BAH 1 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 972_1100 | 240 | 1617.0 | Domain | BAH 2 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 1139_1599 | 256 | 1633.0 | Domain | SAM-dependent MTase C5-type |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 755_880 | 256 | 1633.0 | Domain | BAH 1 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 972_1100 | 256 | 1633.0 | Domain | BAH 2 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 1139_1599 | 240 | 1617.0 | Domain | SAM-dependent MTase C5-type |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 755_880 | 240 | 1617.0 | Domain | BAH 1 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 972_1100 | 240 | 1617.0 | Domain | BAH 2 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 1139_1599 | 256 | 1633.0 | Domain | SAM-dependent MTase C5-type |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 755_880 | 256 | 1633.0 | Domain | BAH 1 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 972_1100 | 256 | 1633.0 | Domain | BAH 2 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 1109_1120 | 240 | 1617.0 | Region | Note=6 X 2 AA tandem repeats of K-G |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 1139_1616 | 240 | 1617.0 | Region | Note=Catalytic |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 1150_1151 | 240 | 1617.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 1168_1169 | 240 | 1617.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 1190_1191 | 240 | 1617.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 310_502 | 240 | 1617.0 | Region | Note=Homodimerization |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 331_550 | 240 | 1617.0 | Region | DNA replication foci-targeting sequence |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 651_697 | 240 | 1617.0 | Region | Note=Required for activity |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 693_754 | 240 | 1617.0 | Region | Note=Autoinhibitory linker |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 1109_1120 | 256 | 1633.0 | Region | Note=6 X 2 AA tandem repeats of K-G |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 1139_1616 | 256 | 1633.0 | Region | Note=Catalytic |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 1150_1151 | 256 | 1633.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 1168_1169 | 256 | 1633.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 1190_1191 | 256 | 1633.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 310_502 | 256 | 1633.0 | Region | Note=Homodimerization |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 331_550 | 256 | 1633.0 | Region | DNA replication foci-targeting sequence |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 651_697 | 256 | 1633.0 | Region | Note=Required for activity |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 693_754 | 256 | 1633.0 | Region | Note=Autoinhibitory linker |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 1109_1120 | 240 | 1617.0 | Region | Note=6 X 2 AA tandem repeats of K-G |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 1139_1616 | 240 | 1617.0 | Region | Note=Catalytic |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 1150_1151 | 240 | 1617.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 1168_1169 | 240 | 1617.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 1190_1191 | 240 | 1617.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 310_502 | 240 | 1617.0 | Region | Note=Homodimerization |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 331_550 | 240 | 1617.0 | Region | DNA replication foci-targeting sequence |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 651_697 | 240 | 1617.0 | Region | Note=Required for activity |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 693_754 | 240 | 1617.0 | Region | Note=Autoinhibitory linker |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 1109_1120 | 256 | 1633.0 | Region | Note=6 X 2 AA tandem repeats of K-G |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 1139_1616 | 256 | 1633.0 | Region | Note=Catalytic |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 1150_1151 | 256 | 1633.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 1168_1169 | 256 | 1633.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 1190_1191 | 256 | 1633.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 310_502 | 256 | 1633.0 | Region | Note=Homodimerization |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 331_550 | 256 | 1633.0 | Region | DNA replication foci-targeting sequence |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 651_697 | 256 | 1633.0 | Region | Note=Required for activity |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 693_754 | 256 | 1633.0 | Region | Note=Autoinhibitory linker |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 1109_1110 | 240 | 1617.0 | Repeat | Note=1 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 1111_1112 | 240 | 1617.0 | Repeat | Note=2 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 1113_1114 | 240 | 1617.0 | Repeat | Note=3 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 1115_1116 | 240 | 1617.0 | Repeat | Note=4 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 1117_1118 | 240 | 1617.0 | Repeat | Note=5 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 1119_1120 | 240 | 1617.0 | Repeat | Note=6%3B approximate |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 1109_1110 | 256 | 1633.0 | Repeat | Note=1 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 1111_1112 | 256 | 1633.0 | Repeat | Note=2 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 1113_1114 | 256 | 1633.0 | Repeat | Note=3 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 1115_1116 | 256 | 1633.0 | Repeat | Note=4 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 1117_1118 | 256 | 1633.0 | Repeat | Note=5 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 1119_1120 | 256 | 1633.0 | Repeat | Note=6%3B approximate |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 1109_1110 | 240 | 1617.0 | Repeat | Note=1 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 1111_1112 | 240 | 1617.0 | Repeat | Note=2 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 1113_1114 | 240 | 1617.0 | Repeat | Note=3 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 1115_1116 | 240 | 1617.0 | Repeat | Note=4 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 1117_1118 | 240 | 1617.0 | Repeat | Note=5 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 1119_1120 | 240 | 1617.0 | Repeat | Note=6%3B approximate |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 1109_1110 | 256 | 1633.0 | Repeat | Note=1 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 1111_1112 | 256 | 1633.0 | Repeat | Note=2 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 1113_1114 | 256 | 1633.0 | Repeat | Note=3 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 1115_1116 | 256 | 1633.0 | Repeat | Note=4 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 1117_1118 | 256 | 1633.0 | Repeat | Note=5 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 1119_1120 | 256 | 1633.0 | Repeat | Note=6%3B approximate |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 646_692 | 240 | 1617.0 | Zinc finger | CXXC-type |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 646_692 | 256 | 1633.0 | Zinc finger | CXXC-type |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 646_692 | 240 | 1617.0 | Zinc finger | CXXC-type |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 646_692 | 256 | 1633.0 | Zinc finger | CXXC-type |
| Tgene | FDX1L | chr19:10283766 | chr19:10421616 | ENST00000393708 | 2 | 5 | 71_173 | 102 | 184.0 | Domain | 2Fe-2S ferredoxin-type |
Top |
Fusion Gene Sequence for DNMT1-FDX1L |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >23695_23695_1_DNMT1-FDX1L_DNMT1_chr19_10283766_ENST00000340748_FDX1L_chr19_10421616_ENST00000541276_length(transcript)=2592nt_BP=956nt TCCGCGTGGGGGGGGTGTGTGCCCGCCTTGCGCATGCGTGTTCCCTGGGCATGGCCGGCTCCGTTCCATCCTTCTGCACAGGGTATCGCC TCTCTCCGTTTGGTACATCCCCTCCTCCCCCACGCCCGGACTGGGGTGGTAGACGCCGCCTCCGCTCATCGCCCCTCCCCATCGGTTTCC GCGCGAAAAGCCGGGGCGCCTGCGCTGCCGCCGCCGCGTCTGCTGAAGCCTCCGAGATGCCGGCGCGTACCGCCCCAGCCCGGGTGCCCA CACTGGCCGTCCCGGCCATCTCGCTGCCCGACGATGTCCGCAGGCGGCTCAAAGATTTGGAAAGAGACAGCTTAACAGAAAAGGAATGTG TGAAGGAGAAATTGAATCTCTTGCACGAATTTCTGCAAACAGAAATAAAGAATCAGTTATGTGACTTGGAAACCAAATTACGTAAAGAAG AATTATCCGAGGAGGGCTACCTGGCTAAAGTCAAATCCCTTTTAAATAAAGATTTGTCCTTGGAGAACGGTGCTCATGCTTACAACCGGG AAGTGAATGGACGTCTAGAAAACGGGAACCAAGCAAGAAGTGAAGCCCGTAGAGTGGGAATGGCAGATGCCAACAGCCCCCCCAAACCCC TTTCCAAACCTCGCACGCCCAGGAGGAGCAAGTCCGATGGAGAGGCTAAGCCTGAACCTTCACCTAGCCCCAGGATTACAAGGAAAAGCA CCAGGCAAACCACCATCACATCTCATTTTGCAAAGGGCCCTGCCAAACGGAAACCTCAGGAAGAGTCTGAAAGAGCCAAATCGGATGAGT CCATCAAGGAAGAAGACAAAGACCAGGATGAGAAGAGACGTAGAGTTACATCCAGAGAACGAGTTGCTAGACCGCTTCCTGCAGAAGAAC CTGAAAGAGCAAAATCAGGAACGCGCACTGAAAAGGAAGAAGAAAGAGATGAAAAAGGGCCTGTGAAGCCTCCCTGGCCTGCTCCACCTG CCATGTGTATGTGAGTGAAGACCACCTGGATCTCCTGCCTCCTCCCGAGGAGAGGAGAACTCGCGGCTGGGCTGCCAGATTGTGCTGACA CCGGAGCTGGAAGGAGCGGAATTCACCCTGCCCAAGATCACCAGGAACTTCTACGTGGATGGCCATGTCCCCAAGCCCCACTGACATGAA CACCTGGACCATTCCACATTGCCATGGCCCCAGGGCCCAGATTGAGGGAATAGCCAGGTGCCAGCCCTGCCCAGAGTGCGGACAGGCCCG GGAGAGACGTGGAAGCCCCTGTGAAGGACAACACCCCTGCTTGGGAGAGAGTCCCATGTCCAGGCCCCTGAGAATCAGGGTTTGAGTAGG AGTGGACTCATATTGGAGCTGCAATAAATCGATAACACAGGCCCCAGCATGTTGAGTGTCCTTGGGGGACAGATCTGGGGTCTACAGGTG GCTCACACCTGTAATCCTAGCACTTTGGGAAGCCAAGATGGGAGGATCACTTGAGGCCAGGAGCTTAAGACCATCCTGGGCAACATGGCG AGACCTCGTCTCTATAAAAACAGTAAAAATTAGCGTGGAGTGGTGGTAAGTGCCTGTAGTCCCAGCTACTCGGGAGACTGAGGTGGGAGG ATCACTTGAGCCCAGGGGATTTAGGCTGCAGTGGAGCGGTGATCAGGCCATTCCACTCCAGCCTGGGTGACAGAGTGAGGCCCTGTCTCA AAAAAAAAAAAAACAAAAACAAAAACAAACCACACCTGCATCTGCTGTGCTGGGCCCCCGGTTCTCTTTTCCCTCTGGGATCTAAGATGC CGCTGGGGATCCAAGGCAGAGGCCTGGGAGCCCCCATTTCCGAGGGGGAGGGCTAGCGTGGAAGGCCCCGCCCCGGCAGCTGCGGAGGGA TTTGGTGGGGTCCAGGCCGCCCTTGCCACGCAGGGGTGCGCGGATGACCTGAAAAAAGCCAGAAAGAGGAAAGGTGCCGCCGAGGCTGGT CGCTCTCAGCTGGGGCTCATCGCATCCTCTACCAAGAGCGCTCCCACCACACAGGTCCCGCCCCTCCCAGCAAAGCTTCTGGCGCCGCCC TGTCTGAACCTGGAGCGGCTGGGAGGATCCCCCTCCCAGGAGAAGCCGAGGACCCCAGGATGCTGCAGAATCAGTGAGAGGTCCCTCTGG CCTGCATGCCAGGAGTCGGTCACCGCCCTGTGCTTCCTCCAAGAGACAGTGGAGAGGCTGGGGATTCCAAGGACACACAGACCAGGATCA GCCAAAAGGGCCGCCGTCTGCAGCCCCCGGGGACTCCCTCGGCCCCACCCCAGAGAAGGCCCCGGAAACAGCTGAACCCCTGCCGGGGCA CCGAGAGAGTGGACCCTGGGTTCGAGGGGGTGACTCTGAAGTTTCAGATAAAGCCGGACTCCAGCCTGCAGATCATCCCCACGTACAGCC TGCCCTGCAGTAGCCGTTCTCAGGAATCCCCTGCAGATGCTGTTGGGGGCCCTGCAGCCCACCCAGGAGGCACCGAGGCCCACTCAGCAG >23695_23695_1_DNMT1-FDX1L_DNMT1_chr19_10283766_ENST00000340748_FDX1L_chr19_10421616_ENST00000541276_length(amino acids)=319AA_BP=304 MGMAGSVPSFCTGYRLSPFGTSPPPPRPDWGGRRRLRSSPLPIGFRAKSRGACAAAAASAEASEMPARTAPARVPTLAVPAISLPDDVRR RLKDLERDSLTEKECVKEKLNLLHEFLQTEIKNQLCDLETKLRKEELSEEGYLAKVKSLLNKDLSLENGAHAYNREVNGRLENGNQARSE ARRVGMADANSPPKPLSKPRTPRRSKSDGEAKPEPSPSPRITRKSTRQTTITSHFAKGPAKRKPQEESERAKSDESIKEEDKDQDEKRRR -------------------------------------------------------------- >23695_23695_2_DNMT1-FDX1L_DNMT1_chr19_10283766_ENST00000340748_FDX1L_chr19_10426435_ENST00000541276_length(transcript)=2754nt_BP=956nt TCCGCGTGGGGGGGGTGTGTGCCCGCCTTGCGCATGCGTGTTCCCTGGGCATGGCCGGCTCCGTTCCATCCTTCTGCACAGGGTATCGCC TCTCTCCGTTTGGTACATCCCCTCCTCCCCCACGCCCGGACTGGGGTGGTAGACGCCGCCTCCGCTCATCGCCCCTCCCCATCGGTTTCC GCGCGAAAAGCCGGGGCGCCTGCGCTGCCGCCGCCGCGTCTGCTGAAGCCTCCGAGATGCCGGCGCGTACCGCCCCAGCCCGGGTGCCCA CACTGGCCGTCCCGGCCATCTCGCTGCCCGACGATGTCCGCAGGCGGCTCAAAGATTTGGAAAGAGACAGCTTAACAGAAAAGGAATGTG TGAAGGAGAAATTGAATCTCTTGCACGAATTTCTGCAAACAGAAATAAAGAATCAGTTATGTGACTTGGAAACCAAATTACGTAAAGAAG AATTATCCGAGGAGGGCTACCTGGCTAAAGTCAAATCCCTTTTAAATAAAGATTTGTCCTTGGAGAACGGTGCTCATGCTTACAACCGGG AAGTGAATGGACGTCTAGAAAACGGGAACCAAGCAAGAAGTGAAGCCCGTAGAGTGGGAATGGCAGATGCCAACAGCCCCCCCAAACCCC TTTCCAAACCTCGCACGCCCAGGAGGAGCAAGTCCGATGGAGAGGCTAAGCCTGAACCTTCACCTAGCCCCAGGATTACAAGGAAAAGCA CCAGGCAAACCACCATCACATCTCATTTTGCAAAGGGCCCTGCCAAACGGAAACCTCAGGAAGAGTCTGAAAGAGCCAAATCGGATGAGT CCATCAAGGAAGAAGACAAAGACCAGGATGAGAAGAGACGTAGAGTTACATCCAGAGAACGAGTTGCTAGACCGCTTCCTGCAGAAGAAC CTGAAAGAGCAAAATCAGGAACGCGCACTGAAAAGGAAGAAGAAAGAGATGAAAAAGCTCGCGCCCGGCTGGAGAGGAGGACGCGGGCGG CCCGGAGCGGCCCGGGGACGTGGTGAACGTGGTGTTCGTAGACCGCTCAGGCCAGCGGATCCCAGTGAGTGGCAGAGTCGGGGACAATGT TCTTCACCTGGCCCAGCGCCACGGGGTGGACCTGGAAGGGGCCTGTGAAGCCTCCCTGGCCTGCTCCACCTGCCATGTGTATGTGAGTGA AGACCACCTGGATCTCCTGCCTCCTCCCGAGGAGAGGAGAACTCGCGGCTGGGCTGCCAGATTGTGCTGACACCGGAGCTGGAAGGAGCG GAATTCACCCTGCCCAAGATCACCAGGAACTTCTACGTGGATGGCCATGTCCCCAAGCCCCACTGACATGAACACCTGGACCATTCCACA TTGCCATGGCCCCAGGGCCCAGATTGAGGGAATAGCCAGGTGCCAGCCCTGCCCAGAGTGCGGACAGGCCCGGGAGAGACGTGGAAGCCC CTGTGAAGGACAACACCCCTGCTTGGGAGAGAGTCCCATGTCCAGGCCCCTGAGAATCAGGGTTTGAGTAGGAGTGGACTCATATTGGAG CTGCAATAAATCGATAACACAGGCCCCAGCATGTTGAGTGTCCTTGGGGGACAGATCTGGGGTCTACAGGTGGCTCACACCTGTAATCCT AGCACTTTGGGAAGCCAAGATGGGAGGATCACTTGAGGCCAGGAGCTTAAGACCATCCTGGGCAACATGGCGAGACCTCGTCTCTATAAA AACAGTAAAAATTAGCGTGGAGTGGTGGTAAGTGCCTGTAGTCCCAGCTACTCGGGAGACTGAGGTGGGAGGATCACTTGAGCCCAGGGG ATTTAGGCTGCAGTGGAGCGGTGATCAGGCCATTCCACTCCAGCCTGGGTGACAGAGTGAGGCCCTGTCTCAAAAAAAAAAAAAACAAAA ACAAAAACAAACCACACCTGCATCTGCTGTGCTGGGCCCCCGGTTCTCTTTTCCCTCTGGGATCTAAGATGCCGCTGGGGATCCAAGGCA GAGGCCTGGGAGCCCCCATTTCCGAGGGGGAGGGCTAGCGTGGAAGGCCCCGCCCCGGCAGCTGCGGAGGGATTTGGTGGGGTCCAGGCC GCCCTTGCCACGCAGGGGTGCGCGGATGACCTGAAAAAAGCCAGAAAGAGGAAAGGTGCCGCCGAGGCTGGTCGCTCTCAGCTGGGGCTC ATCGCATCCTCTACCAAGAGCGCTCCCACCACACAGGTCCCGCCCCTCCCAGCAAAGCTTCTGGCGCCGCCCTGTCTGAACCTGGAGCGG CTGGGAGGATCCCCCTCCCAGGAGAAGCCGAGGACCCCAGGATGCTGCAGAATCAGTGAGAGGTCCCTCTGGCCTGCATGCCAGGAGTCG GTCACCGCCCTGTGCTTCCTCCAAGAGACAGTGGAGAGGCTGGGGATTCCAAGGACACACAGACCAGGATCAGCCAAAAGGGCCGCCGTC TGCAGCCCCCGGGGACTCCCTCGGCCCCACCCCAGAGAAGGCCCCGGAAACAGCTGAACCCCTGCCGGGGCACCGAGAGAGTGGACCCTG GGTTCGAGGGGGTGACTCTGAAGTTTCAGATAAAGCCGGACTCCAGCCTGCAGATCATCCCCACGTACAGCCTGCCCTGCAGTAGCCGTT CTCAGGAATCCCCTGCAGATGCTGTTGGGGGCCCTGCAGCCCACCCAGGAGGCACCGAGGCCCACTCAGCAGGCAGCGAGGCCCTGGAGC >23695_23695_2_DNMT1-FDX1L_DNMT1_chr19_10283766_ENST00000340748_FDX1L_chr19_10426435_ENST00000541276_length(amino acids)=323AA_BP=304 MGMAGSVPSFCTGYRLSPFGTSPPPPRPDWGGRRRLRSSPLPIGFRAKSRGACAAAAASAEASEMPARTAPARVPTLAVPAISLPDDVRR RLKDLERDSLTEKECVKEKLNLLHEFLQTEIKNQLCDLETKLRKEELSEEGYLAKVKSLLNKDLSLENGAHAYNREVNGRLENGNQARSE ARRVGMADANSPPKPLSKPRTPRRSKSDGEAKPEPSPSPRITRKSTRQTTITSHFAKGPAKRKPQEESERAKSDESIKEEDKDQDEKRRR -------------------------------------------------------------- >23695_23695_3_DNMT1-FDX1L_DNMT1_chr19_10283766_ENST00000359526_FDX1L_chr19_10421616_ENST00000541276_length(transcript)=2584nt_BP=948nt GGCTCCGTTCCATCCTTCTGCACAGGGTATCGCCTCTCTCCGTTTGGTACATCCCCTCCTCCCCCACGCCCGGACTGGGGTGGTAGACGC CGCCTCCGCTCATCGCCCCTCCCCATCGGTTTCCGCGCGAAAAGCCGGGGCGCCTGCGCTGCCGCCGCCGCGTCTGCTGAAGCCTCCGAG ATGCCGGCGCGTACCGCCCCAGCCCGGGTGCCCACACTGGCCGTCCCGGCCATCTCGCTGCCCGACGATGTCCGCAGGCGGCTCAAAGAT TTGGAAAGAGACAGCTTAACAGAAAAGGAATGTGTGAAGGAGAAATTGAATCTCTTGCACGAATTTCTGCAAACAGAAATAAAGAATCAG TTATGTGACTTGGAAACCAAATTACGTAAAGAAGAATTATCCGAGGAGGGCTACCTGGCTAAAGTCAAATCCCTTTTAAATAAAGATTTG TCCTTGGAGAACGGTGCTCATGCTTACAACCGGGAAGTGAATGGACGTCTAGAAAACGGGAACCAAGCAAGAAGTGAAGCCCGTAGAGTG GGAATGGCAGATGCCAACAGCCCCCCCAAACCCCTTTCCAAACCTCGCACGCCCAGGAGGAGCAAGTCCGATGGAGAGGCTAAGCGTTCA AGAGACCCTCCTGCCTCAGCCTCCCAAGTAACTGGGATTAGAGCTGAACCTTCACCTAGCCCCAGGATTACAAGGAAAAGCACCAGGCAA ACCACCATCACATCTCATTTTGCAAAGGGCCCTGCCAAACGGAAACCTCAGGAAGAGTCTGAAAGAGCCAAATCGGATGAGTCCATCAAG GAAGAAGACAAAGACCAGGATGAGAAGAGACGTAGAGTTACATCCAGAGAACGAGTTGCTAGACCGCTTCCTGCAGAAGAACCTGAAAGA GCAAAATCAGGAACGCGCACTGAAAAGGAAGAAGAAAGAGATGAAAAAGGGCCTGTGAAGCCTCCCTGGCCTGCTCCACCTGCCATGTGT ATGTGAGTGAAGACCACCTGGATCTCCTGCCTCCTCCCGAGGAGAGGAGAACTCGCGGCTGGGCTGCCAGATTGTGCTGACACCGGAGCT GGAAGGAGCGGAATTCACCCTGCCCAAGATCACCAGGAACTTCTACGTGGATGGCCATGTCCCCAAGCCCCACTGACATGAACACCTGGA CCATTCCACATTGCCATGGCCCCAGGGCCCAGATTGAGGGAATAGCCAGGTGCCAGCCCTGCCCAGAGTGCGGACAGGCCCGGGAGAGAC GTGGAAGCCCCTGTGAAGGACAACACCCCTGCTTGGGAGAGAGTCCCATGTCCAGGCCCCTGAGAATCAGGGTTTGAGTAGGAGTGGACT CATATTGGAGCTGCAATAAATCGATAACACAGGCCCCAGCATGTTGAGTGTCCTTGGGGGACAGATCTGGGGTCTACAGGTGGCTCACAC CTGTAATCCTAGCACTTTGGGAAGCCAAGATGGGAGGATCACTTGAGGCCAGGAGCTTAAGACCATCCTGGGCAACATGGCGAGACCTCG TCTCTATAAAAACAGTAAAAATTAGCGTGGAGTGGTGGTAAGTGCCTGTAGTCCCAGCTACTCGGGAGACTGAGGTGGGAGGATCACTTG AGCCCAGGGGATTTAGGCTGCAGTGGAGCGGTGATCAGGCCATTCCACTCCAGCCTGGGTGACAGAGTGAGGCCCTGTCTCAAAAAAAAA AAAAACAAAAACAAAAACAAACCACACCTGCATCTGCTGTGCTGGGCCCCCGGTTCTCTTTTCCCTCTGGGATCTAAGATGCCGCTGGGG ATCCAAGGCAGAGGCCTGGGAGCCCCCATTTCCGAGGGGGAGGGCTAGCGTGGAAGGCCCCGCCCCGGCAGCTGCGGAGGGATTTGGTGG GGTCCAGGCCGCCCTTGCCACGCAGGGGTGCGCGGATGACCTGAAAAAAGCCAGAAAGAGGAAAGGTGCCGCCGAGGCTGGTCGCTCTCA GCTGGGGCTCATCGCATCCTCTACCAAGAGCGCTCCCACCACACAGGTCCCGCCCCTCCCAGCAAAGCTTCTGGCGCCGCCCTGTCTGAA CCTGGAGCGGCTGGGAGGATCCCCCTCCCAGGAGAAGCCGAGGACCCCAGGATGCTGCAGAATCAGTGAGAGGTCCCTCTGGCCTGCATG CCAGGAGTCGGTCACCGCCCTGTGCTTCCTCCAAGAGACAGTGGAGAGGCTGGGGATTCCAAGGACACACAGACCAGGATCAGCCAAAAG GGCCGCCGTCTGCAGCCCCCGGGGACTCCCTCGGCCCCACCCCAGAGAAGGCCCCGGAAACAGCTGAACCCCTGCCGGGGCACCGAGAGA GTGGACCCTGGGTTCGAGGGGGTGACTCTGAAGTTTCAGATAAAGCCGGACTCCAGCCTGCAGATCATCCCCACGTACAGCCTGCCCTGC AGTAGCCGTTCTCAGGAATCCCCTGCAGATGCTGTTGGGGGCCCTGCAGCCCACCCAGGAGGCACCGAGGCCCACTCAGCAGGCAGCGAG >23695_23695_3_DNMT1-FDX1L_DNMT1_chr19_10283766_ENST00000359526_FDX1L_chr19_10421616_ENST00000541276_length(amino acids)=271AA_BP=256 MPARTAPARVPTLAVPAISLPDDVRRRLKDLERDSLTEKECVKEKLNLLHEFLQTEIKNQLCDLETKLRKEELSEEGYLAKVKSLLNKDL SLENGAHAYNREVNGRLENGNQARSEARRVGMADANSPPKPLSKPRTPRRSKSDGEAKRSRDPPASASQVTGIRAEPSPSPRITRKSTRQ TTITSHFAKGPAKRKPQEESERAKSDESIKEEDKDQDEKRRRVTSRERVARPLPAEEPERAKSGTRTEKEEERDEKGPVKPPWPAPPAMC -------------------------------------------------------------- >23695_23695_4_DNMT1-FDX1L_DNMT1_chr19_10283766_ENST00000359526_FDX1L_chr19_10426435_ENST00000541276_length(transcript)=2746nt_BP=948nt GGCTCCGTTCCATCCTTCTGCACAGGGTATCGCCTCTCTCCGTTTGGTACATCCCCTCCTCCCCCACGCCCGGACTGGGGTGGTAGACGC CGCCTCCGCTCATCGCCCCTCCCCATCGGTTTCCGCGCGAAAAGCCGGGGCGCCTGCGCTGCCGCCGCCGCGTCTGCTGAAGCCTCCGAG ATGCCGGCGCGTACCGCCCCAGCCCGGGTGCCCACACTGGCCGTCCCGGCCATCTCGCTGCCCGACGATGTCCGCAGGCGGCTCAAAGAT TTGGAAAGAGACAGCTTAACAGAAAAGGAATGTGTGAAGGAGAAATTGAATCTCTTGCACGAATTTCTGCAAACAGAAATAAAGAATCAG TTATGTGACTTGGAAACCAAATTACGTAAAGAAGAATTATCCGAGGAGGGCTACCTGGCTAAAGTCAAATCCCTTTTAAATAAAGATTTG TCCTTGGAGAACGGTGCTCATGCTTACAACCGGGAAGTGAATGGACGTCTAGAAAACGGGAACCAAGCAAGAAGTGAAGCCCGTAGAGTG GGAATGGCAGATGCCAACAGCCCCCCCAAACCCCTTTCCAAACCTCGCACGCCCAGGAGGAGCAAGTCCGATGGAGAGGCTAAGCGTTCA AGAGACCCTCCTGCCTCAGCCTCCCAAGTAACTGGGATTAGAGCTGAACCTTCACCTAGCCCCAGGATTACAAGGAAAAGCACCAGGCAA ACCACCATCACATCTCATTTTGCAAAGGGCCCTGCCAAACGGAAACCTCAGGAAGAGTCTGAAAGAGCCAAATCGGATGAGTCCATCAAG GAAGAAGACAAAGACCAGGATGAGAAGAGACGTAGAGTTACATCCAGAGAACGAGTTGCTAGACCGCTTCCTGCAGAAGAACCTGAAAGA GCAAAATCAGGAACGCGCACTGAAAAGGAAGAAGAAAGAGATGAAAAAGCTCGCGCCCGGCTGGAGAGGAGGACGCGGGCGGCCCGGAGC GGCCCGGGGACGTGGTGAACGTGGTGTTCGTAGACCGCTCAGGCCAGCGGATCCCAGTGAGTGGCAGAGTCGGGGACAATGTTCTTCACC TGGCCCAGCGCCACGGGGTGGACCTGGAAGGGGCCTGTGAAGCCTCCCTGGCCTGCTCCACCTGCCATGTGTATGTGAGTGAAGACCACC TGGATCTCCTGCCTCCTCCCGAGGAGAGGAGAACTCGCGGCTGGGCTGCCAGATTGTGCTGACACCGGAGCTGGAAGGAGCGGAATTCAC CCTGCCCAAGATCACCAGGAACTTCTACGTGGATGGCCATGTCCCCAAGCCCCACTGACATGAACACCTGGACCATTCCACATTGCCATG GCCCCAGGGCCCAGATTGAGGGAATAGCCAGGTGCCAGCCCTGCCCAGAGTGCGGACAGGCCCGGGAGAGACGTGGAAGCCCCTGTGAAG GACAACACCCCTGCTTGGGAGAGAGTCCCATGTCCAGGCCCCTGAGAATCAGGGTTTGAGTAGGAGTGGACTCATATTGGAGCTGCAATA AATCGATAACACAGGCCCCAGCATGTTGAGTGTCCTTGGGGGACAGATCTGGGGTCTACAGGTGGCTCACACCTGTAATCCTAGCACTTT GGGAAGCCAAGATGGGAGGATCACTTGAGGCCAGGAGCTTAAGACCATCCTGGGCAACATGGCGAGACCTCGTCTCTATAAAAACAGTAA AAATTAGCGTGGAGTGGTGGTAAGTGCCTGTAGTCCCAGCTACTCGGGAGACTGAGGTGGGAGGATCACTTGAGCCCAGGGGATTTAGGC TGCAGTGGAGCGGTGATCAGGCCATTCCACTCCAGCCTGGGTGACAGAGTGAGGCCCTGTCTCAAAAAAAAAAAAAACAAAAACAAAAAC AAACCACACCTGCATCTGCTGTGCTGGGCCCCCGGTTCTCTTTTCCCTCTGGGATCTAAGATGCCGCTGGGGATCCAAGGCAGAGGCCTG GGAGCCCCCATTTCCGAGGGGGAGGGCTAGCGTGGAAGGCCCCGCCCCGGCAGCTGCGGAGGGATTTGGTGGGGTCCAGGCCGCCCTTGC CACGCAGGGGTGCGCGGATGACCTGAAAAAAGCCAGAAAGAGGAAAGGTGCCGCCGAGGCTGGTCGCTCTCAGCTGGGGCTCATCGCATC CTCTACCAAGAGCGCTCCCACCACACAGGTCCCGCCCCTCCCAGCAAAGCTTCTGGCGCCGCCCTGTCTGAACCTGGAGCGGCTGGGAGG ATCCCCCTCCCAGGAGAAGCCGAGGACCCCAGGATGCTGCAGAATCAGTGAGAGGTCCCTCTGGCCTGCATGCCAGGAGTCGGTCACCGC CCTGTGCTTCCTCCAAGAGACAGTGGAGAGGCTGGGGATTCCAAGGACACACAGACCAGGATCAGCCAAAAGGGCCGCCGTCTGCAGCCC CCGGGGACTCCCTCGGCCCCACCCCAGAGAAGGCCCCGGAAACAGCTGAACCCCTGCCGGGGCACCGAGAGAGTGGACCCTGGGTTCGAG GGGGTGACTCTGAAGTTTCAGATAAAGCCGGACTCCAGCCTGCAGATCATCCCCACGTACAGCCTGCCCTGCAGTAGCCGTTCTCAGGAA TCCCCTGCAGATGCTGTTGGGGGCCCTGCAGCCCACCCAGGAGGCACCGAGGCCCACTCAGCAGGCAGCGAGGCCCTGGAGCCCCGGCGC >23695_23695_4_DNMT1-FDX1L_DNMT1_chr19_10283766_ENST00000359526_FDX1L_chr19_10426435_ENST00000541276_length(amino acids)=275AA_BP=256 MPARTAPARVPTLAVPAISLPDDVRRRLKDLERDSLTEKECVKEKLNLLHEFLQTEIKNQLCDLETKLRKEELSEEGYLAKVKSLLNKDL SLENGAHAYNREVNGRLENGNQARSEARRVGMADANSPPKPLSKPRTPRRSKSDGEAKRSRDPPASASQVTGIRAEPSPSPRITRKSTRQ TTITSHFAKGPAKRKPQEESERAKSDESIKEEDKDQDEKRRRVTSRERVARPLPAEEPERAKSGTRTEKEEERDEKARARLERRTRAARS -------------------------------------------------------------- >23695_23695_5_DNMT1-FDX1L_DNMT1_chr19_10283766_ENST00000540357_FDX1L_chr19_10421616_ENST00000541276_length(transcript)=2536nt_BP=900nt GGCTCCGTTCCATCCTTCTGCACAGGGTATCGCCTCTCTCCGTTTGGTACATCCCCTCCTCCCCCACGCCCGGACTGGGGTGGTAGACGC CGCCTCCGCTCATCGCCCCTCCCCATCGGTTTCCGCGCGAAAAGCCGGGGCGCCTGCGCTGCCGCCGCCGCGTCTGCTGAAGCCTCCGAG ATGCCGGCGCGTACCGCCCCAGCCCGGGTGCCCACACTGGCCGTCCCGGCCATCTCGCTGCCCGACGATGTCCGCAGGCGGCTCAAAGAT TTGGAAAGAGACAGCTTAACAGAAAAGGAATGTGTGAAGGAGAAATTGAATCTCTTGCACGAATTTCTGCAAACAGAAATAAAGAATCAG TTATGTGACTTGGAAACCAAATTACGTAAAGAAGAATTATCCGAGGAGGGCTACCTGGCTAAAGTCAAATCCCTTTTAAATAAAGATTTG TCCTTGGAGAACGGTGCTCATGCTTACAACCGGGAAGTGAATGGACGTCTAGAAAACGGGAACCAAGCAAGAAGTGAAGCCCGTAGAGTG GGAATGGCAGATGCCAACAGCCCCCCCAAACCCCTTTCCAAACCTCGCACGCCCAGGAGGAGCAAGTCCGATGGAGAGGCTAAGCCTGAA CCTTCACCTAGCCCCAGGATTACAAGGAAAAGCACCAGGCAAACCACCATCACATCTCATTTTGCAAAGGGCCCTGCCAAACGGAAACCT CAGGAAGAGTCTGAAAGAGCCAAATCGGATGAGTCCATCAAGGAAGAAGACAAAGACCAGGATGAGAAGAGACGTAGAGTTACATCCAGA GAACGAGTTGCTAGACCGCTTCCTGCAGAAGAACCTGAAAGAGCAAAATCAGGAACGCGCACTGAAAAGGAAGAAGAAAGAGATGAAAAA GGGCCTGTGAAGCCTCCCTGGCCTGCTCCACCTGCCATGTGTATGTGAGTGAAGACCACCTGGATCTCCTGCCTCCTCCCGAGGAGAGGA GAACTCGCGGCTGGGCTGCCAGATTGTGCTGACACCGGAGCTGGAAGGAGCGGAATTCACCCTGCCCAAGATCACCAGGAACTTCTACGT GGATGGCCATGTCCCCAAGCCCCACTGACATGAACACCTGGACCATTCCACATTGCCATGGCCCCAGGGCCCAGATTGAGGGAATAGCCA GGTGCCAGCCCTGCCCAGAGTGCGGACAGGCCCGGGAGAGACGTGGAAGCCCCTGTGAAGGACAACACCCCTGCTTGGGAGAGAGTCCCA TGTCCAGGCCCCTGAGAATCAGGGTTTGAGTAGGAGTGGACTCATATTGGAGCTGCAATAAATCGATAACACAGGCCCCAGCATGTTGAG TGTCCTTGGGGGACAGATCTGGGGTCTACAGGTGGCTCACACCTGTAATCCTAGCACTTTGGGAAGCCAAGATGGGAGGATCACTTGAGG CCAGGAGCTTAAGACCATCCTGGGCAACATGGCGAGACCTCGTCTCTATAAAAACAGTAAAAATTAGCGTGGAGTGGTGGTAAGTGCCTG TAGTCCCAGCTACTCGGGAGACTGAGGTGGGAGGATCACTTGAGCCCAGGGGATTTAGGCTGCAGTGGAGCGGTGATCAGGCCATTCCAC TCCAGCCTGGGTGACAGAGTGAGGCCCTGTCTCAAAAAAAAAAAAAACAAAAACAAAAACAAACCACACCTGCATCTGCTGTGCTGGGCC CCCGGTTCTCTTTTCCCTCTGGGATCTAAGATGCCGCTGGGGATCCAAGGCAGAGGCCTGGGAGCCCCCATTTCCGAGGGGGAGGGCTAG CGTGGAAGGCCCCGCCCCGGCAGCTGCGGAGGGATTTGGTGGGGTCCAGGCCGCCCTTGCCACGCAGGGGTGCGCGGATGACCTGAAAAA AGCCAGAAAGAGGAAAGGTGCCGCCGAGGCTGGTCGCTCTCAGCTGGGGCTCATCGCATCCTCTACCAAGAGCGCTCCCACCACACAGGT CCCGCCCCTCCCAGCAAAGCTTCTGGCGCCGCCCTGTCTGAACCTGGAGCGGCTGGGAGGATCCCCCTCCCAGGAGAAGCCGAGGACCCC AGGATGCTGCAGAATCAGTGAGAGGTCCCTCTGGCCTGCATGCCAGGAGTCGGTCACCGCCCTGTGCTTCCTCCAAGAGACAGTGGAGAG GCTGGGGATTCCAAGGACACACAGACCAGGATCAGCCAAAAGGGCCGCCGTCTGCAGCCCCCGGGGACTCCCTCGGCCCCACCCCAGAGA AGGCCCCGGAAACAGCTGAACCCCTGCCGGGGCACCGAGAGAGTGGACCCTGGGTTCGAGGGGGTGACTCTGAAGTTTCAGATAAAGCCG GACTCCAGCCTGCAGATCATCCCCACGTACAGCCTGCCCTGCAGTAGCCGTTCTCAGGAATCCCCTGCAGATGCTGTTGGGGGCCCTGCA GCCCACCCAGGAGGCACCGAGGCCCACTCAGCAGGCAGCGAGGCCCTGGAGCCCCGGCGCTGTGCTTCCTGTCGGACCCAGAGGACCCCG >23695_23695_5_DNMT1-FDX1L_DNMT1_chr19_10283766_ENST00000540357_FDX1L_chr19_10421616_ENST00000541276_length(amino acids)=255AA_BP=240 MPARTAPARVPTLAVPAISLPDDVRRRLKDLERDSLTEKECVKEKLNLLHEFLQTEIKNQLCDLETKLRKEELSEEGYLAKVKSLLNKDL SLENGAHAYNREVNGRLENGNQARSEARRVGMADANSPPKPLSKPRTPRRSKSDGEAKPEPSPSPRITRKSTRQTTITSHFAKGPAKRKP -------------------------------------------------------------- >23695_23695_6_DNMT1-FDX1L_DNMT1_chr19_10283766_ENST00000540357_FDX1L_chr19_10426435_ENST00000541276_length(transcript)=2698nt_BP=900nt GGCTCCGTTCCATCCTTCTGCACAGGGTATCGCCTCTCTCCGTTTGGTACATCCCCTCCTCCCCCACGCCCGGACTGGGGTGGTAGACGC CGCCTCCGCTCATCGCCCCTCCCCATCGGTTTCCGCGCGAAAAGCCGGGGCGCCTGCGCTGCCGCCGCCGCGTCTGCTGAAGCCTCCGAG ATGCCGGCGCGTACCGCCCCAGCCCGGGTGCCCACACTGGCCGTCCCGGCCATCTCGCTGCCCGACGATGTCCGCAGGCGGCTCAAAGAT TTGGAAAGAGACAGCTTAACAGAAAAGGAATGTGTGAAGGAGAAATTGAATCTCTTGCACGAATTTCTGCAAACAGAAATAAAGAATCAG TTATGTGACTTGGAAACCAAATTACGTAAAGAAGAATTATCCGAGGAGGGCTACCTGGCTAAAGTCAAATCCCTTTTAAATAAAGATTTG TCCTTGGAGAACGGTGCTCATGCTTACAACCGGGAAGTGAATGGACGTCTAGAAAACGGGAACCAAGCAAGAAGTGAAGCCCGTAGAGTG GGAATGGCAGATGCCAACAGCCCCCCCAAACCCCTTTCCAAACCTCGCACGCCCAGGAGGAGCAAGTCCGATGGAGAGGCTAAGCCTGAA CCTTCACCTAGCCCCAGGATTACAAGGAAAAGCACCAGGCAAACCACCATCACATCTCATTTTGCAAAGGGCCCTGCCAAACGGAAACCT CAGGAAGAGTCTGAAAGAGCCAAATCGGATGAGTCCATCAAGGAAGAAGACAAAGACCAGGATGAGAAGAGACGTAGAGTTACATCCAGA GAACGAGTTGCTAGACCGCTTCCTGCAGAAGAACCTGAAAGAGCAAAATCAGGAACGCGCACTGAAAAGGAAGAAGAAAGAGATGAAAAA GCTCGCGCCCGGCTGGAGAGGAGGACGCGGGCGGCCCGGAGCGGCCCGGGGACGTGGTGAACGTGGTGTTCGTAGACCGCTCAGGCCAGC GGATCCCAGTGAGTGGCAGAGTCGGGGACAATGTTCTTCACCTGGCCCAGCGCCACGGGGTGGACCTGGAAGGGGCCTGTGAAGCCTCCC TGGCCTGCTCCACCTGCCATGTGTATGTGAGTGAAGACCACCTGGATCTCCTGCCTCCTCCCGAGGAGAGGAGAACTCGCGGCTGGGCTG CCAGATTGTGCTGACACCGGAGCTGGAAGGAGCGGAATTCACCCTGCCCAAGATCACCAGGAACTTCTACGTGGATGGCCATGTCCCCAA GCCCCACTGACATGAACACCTGGACCATTCCACATTGCCATGGCCCCAGGGCCCAGATTGAGGGAATAGCCAGGTGCCAGCCCTGCCCAG AGTGCGGACAGGCCCGGGAGAGACGTGGAAGCCCCTGTGAAGGACAACACCCCTGCTTGGGAGAGAGTCCCATGTCCAGGCCCCTGAGAA TCAGGGTTTGAGTAGGAGTGGACTCATATTGGAGCTGCAATAAATCGATAACACAGGCCCCAGCATGTTGAGTGTCCTTGGGGGACAGAT CTGGGGTCTACAGGTGGCTCACACCTGTAATCCTAGCACTTTGGGAAGCCAAGATGGGAGGATCACTTGAGGCCAGGAGCTTAAGACCAT CCTGGGCAACATGGCGAGACCTCGTCTCTATAAAAACAGTAAAAATTAGCGTGGAGTGGTGGTAAGTGCCTGTAGTCCCAGCTACTCGGG AGACTGAGGTGGGAGGATCACTTGAGCCCAGGGGATTTAGGCTGCAGTGGAGCGGTGATCAGGCCATTCCACTCCAGCCTGGGTGACAGA GTGAGGCCCTGTCTCAAAAAAAAAAAAAACAAAAACAAAAACAAACCACACCTGCATCTGCTGTGCTGGGCCCCCGGTTCTCTTTTCCCT CTGGGATCTAAGATGCCGCTGGGGATCCAAGGCAGAGGCCTGGGAGCCCCCATTTCCGAGGGGGAGGGCTAGCGTGGAAGGCCCCGCCCC GGCAGCTGCGGAGGGATTTGGTGGGGTCCAGGCCGCCCTTGCCACGCAGGGGTGCGCGGATGACCTGAAAAAAGCCAGAAAGAGGAAAGG TGCCGCCGAGGCTGGTCGCTCTCAGCTGGGGCTCATCGCATCCTCTACCAAGAGCGCTCCCACCACACAGGTCCCGCCCCTCCCAGCAAA GCTTCTGGCGCCGCCCTGTCTGAACCTGGAGCGGCTGGGAGGATCCCCCTCCCAGGAGAAGCCGAGGACCCCAGGATGCTGCAGAATCAG TGAGAGGTCCCTCTGGCCTGCATGCCAGGAGTCGGTCACCGCCCTGTGCTTCCTCCAAGAGACAGTGGAGAGGCTGGGGATTCCAAGGAC ACACAGACCAGGATCAGCCAAAAGGGCCGCCGTCTGCAGCCCCCGGGGACTCCCTCGGCCCCACCCCAGAGAAGGCCCCGGAAACAGCTG AACCCCTGCCGGGGCACCGAGAGAGTGGACCCTGGGTTCGAGGGGGTGACTCTGAAGTTTCAGATAAAGCCGGACTCCAGCCTGCAGATC ATCCCCACGTACAGCCTGCCCTGCAGTAGCCGTTCTCAGGAATCCCCTGCAGATGCTGTTGGGGGCCCTGCAGCCCACCCAGGAGGCACC >23695_23695_6_DNMT1-FDX1L_DNMT1_chr19_10283766_ENST00000540357_FDX1L_chr19_10426435_ENST00000541276_length(amino acids)=259AA_BP=240 MPARTAPARVPTLAVPAISLPDDVRRRLKDLERDSLTEKECVKEKLNLLHEFLQTEIKNQLCDLETKLRKEELSEEGYLAKVKSLLNKDL SLENGAHAYNREVNGRLENGNQARSEARRVGMADANSPPKPLSKPRTPRRSKSDGEAKPEPSPSPRITRKSTRQTTITSHFAKGPAKRKP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for DNMT1-FDX1L |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 1_120 | 240.0 | 1617.0 | DMAP1 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 1_120 | 256.0 | 1633.0 | DMAP1 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 1_120 | 240.0 | 1617.0 | DMAP1 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 1_120 | 256.0 | 1633.0 | DMAP1 |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 1_148 | 240.0 | 1617.0 | DNMT3A |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 1_148 | 256.0 | 1633.0 | DNMT3A |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 1_148 | 240.0 | 1617.0 | DNMT3A |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 1_148 | 256.0 | 1633.0 | DNMT3A |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 149_217 | 240.0 | 1617.0 | DNMT3B |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 149_217 | 256.0 | 1633.0 | DNMT3B |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 149_217 | 240.0 | 1617.0 | DNMT3B |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 149_217 | 256.0 | 1633.0 | DNMT3B |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 163_174 | 240.0 | 1617.0 | PCNA |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 163_174 | 256.0 | 1633.0 | PCNA |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 163_174 | 240.0 | 1617.0 | PCNA |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 163_174 | 256.0 | 1633.0 | PCNA |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 1121_1616 | 240.0 | 1617.0 | the PRC2/EED-EZH2 complex |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000340748 | - | 8 | 40 | 308_606 | 240.0 | 1617.0 | the PRC2/EED-EZH2 complex |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 1121_1616 | 256.0 | 1633.0 | the PRC2/EED-EZH2 complex |
| Hgene | DNMT1 | chr19:10283766 | chr19:10421616 | ENST00000359526 | - | 9 | 41 | 308_606 | 256.0 | 1633.0 | the PRC2/EED-EZH2 complex |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 1121_1616 | 240.0 | 1617.0 | the PRC2/EED-EZH2 complex |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000340748 | - | 8 | 40 | 308_606 | 240.0 | 1617.0 | the PRC2/EED-EZH2 complex |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 1121_1616 | 256.0 | 1633.0 | the PRC2/EED-EZH2 complex |
| Hgene | DNMT1 | chr19:10283766 | chr19:10426435 | ENST00000359526 | - | 9 | 41 | 308_606 | 256.0 | 1633.0 | the PRC2/EED-EZH2 complex |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for DNMT1-FDX1L |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for DNMT1-FDX1L |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
