
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:DOC2B-ABR (FusionGDB2 ID:23735) |
Fusion Gene Summary for DOC2B-ABR |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: DOC2B-ABR | Fusion gene ID: 23735 | Hgene | Tgene | Gene symbol | DOC2B | ABR | Gene ID | 8447 | 29 |
| Gene name | double C2 domain beta | ABR activator of RhoGEF and GTPase | |
| Synonyms | DOC2BL | MDB | |
| Cytomap | 17p13.3 | 17p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | double C2-like domain-containing protein betadoc2-betadouble C2-like domains, beta | active breakpoint cluster region-related proteinABR, RhoGEF and GTPase activating proteinactive BCR-related | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q14184 | Q15018 | |
| Ensembl transtripts involved in fusion gene | ENST00000343572, ENST00000609727, | ENST00000291107, ENST00000536794, ENST00000543210, ENST00000572441, ENST00000573895, ENST00000574437, ENST00000302538, ENST00000544583, | |
| Fusion gene scores | * DoF score | 4 X 3 X 3=36 | 17 X 14 X 9=2142 |
| # samples | 4 | 21 | |
| ** MAII score | log2(4/36*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(21/2142*10)=-3.35049724708413 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: DOC2B [Title/Abstract] AND ABR [Title/Abstract] AND fusion [Title/Abstract] | ||
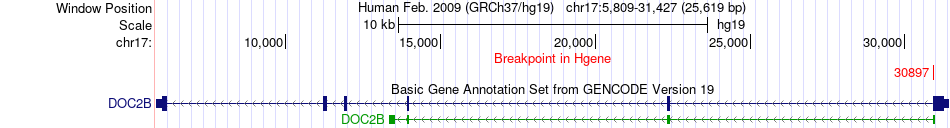
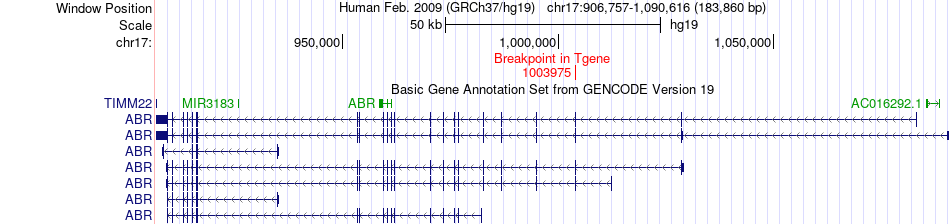
| Most frequent breakpoint | DOC2B(30897)-ABR(1003975), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | ABR | GO:0090630 | activation of GTPase activity | 7479768 |
 Fusion gene breakpoints across DOC2B (5'-gene) Fusion gene breakpoints across DOC2B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across ABR (3'-gene) Fusion gene breakpoints across ABR (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-IN-AB1X | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - |
Top |
Fusion Gene ORF analysis for DOC2B-ABR |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000343572 | ENST00000291107 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - |
| 5CDS-intron | ENST00000343572 | ENST00000536794 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - |
| 5CDS-intron | ENST00000343572 | ENST00000543210 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - |
| 5CDS-intron | ENST00000343572 | ENST00000572441 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - |
| 5CDS-intron | ENST00000343572 | ENST00000573895 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - |
| 5CDS-intron | ENST00000343572 | ENST00000574437 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - |
| 5UTR-3CDS | ENST00000609727 | ENST00000302538 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - |
| 5UTR-3CDS | ENST00000609727 | ENST00000544583 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - |
| 5UTR-intron | ENST00000609727 | ENST00000291107 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - |
| 5UTR-intron | ENST00000609727 | ENST00000536794 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - |
| 5UTR-intron | ENST00000609727 | ENST00000543210 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - |
| 5UTR-intron | ENST00000609727 | ENST00000572441 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - |
| 5UTR-intron | ENST00000609727 | ENST00000573895 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - |
| 5UTR-intron | ENST00000609727 | ENST00000574437 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - |
| In-frame | ENST00000343572 | ENST00000302538 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - |
| In-frame | ENST00000343572 | ENST00000544583 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000343572 | DOC2B | chr17 | 30897 | - | ENST00000302538 | ABR | chr17 | 1003975 | - | 5426 | 530 | 314 | 2863 | 849 |
| ENST00000343572 | DOC2B | chr17 | 30897 | - | ENST00000544583 | ABR | chr17 | 1003975 | - | 5417 | 530 | 314 | 2863 | 849 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000343572 | ENST00000302538 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - | 0.005844597 | 0.9941554 |
| ENST00000343572 | ENST00000544583 | DOC2B | chr17 | 30897 | - | ABR | chr17 | 1003975 | - | 0.005801449 | 0.99419856 |
Top |
Fusion Genomic Features for DOC2B-ABR |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
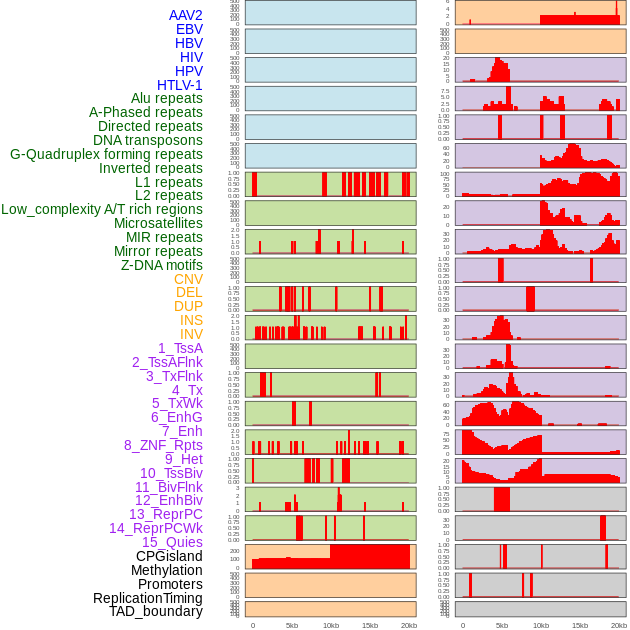 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for DOC2B-ABR |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:30897/chr17:1003975) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| DOC2B | ABR |
| FUNCTION: Calcium sensor which positively regulates SNARE-dependent fusion of vesicles with membranes. Binds phospholipids in a calcium-dependent manner and may act at the priming stage of fusion by modifying membrane curvature to stimulate fusion. Involved in calcium-triggered exocytosis in chromaffin cells and calcium-dependent spontaneous release of neurotransmitter in absence of action potentials in neuronal cells. Involved both in glucose-stimulated insulin secretion in pancreatic cells and insulin-dependent GLUT4 transport to the plasma membrane in adipocytes (By similarity). {ECO:0000250, ECO:0000269|PubMed:9804756}. | FUNCTION: Component of the BRISC complex, a multiprotein complex that specifically cleaves 'Lys-63'-linked polyubiquitin, leaving the last ubiquitin chain attached to its substrates (PubMed:19214193, PubMed:20032457, PubMed:20656690, PubMed:24075985). May act as a central scaffold protein that assembles the various components of the BRISC complex and retains them in the cytoplasm (PubMed:20656690). Plays a role in regulating the onset of apoptosis via its role in modulating 'Lys-63'-linked ubiquitination of target proteins (By similarity). Required for normal mitotic spindle assembly and microtubule attachment to kinetochores via its role in deubiquitinating NUMA1 (PubMed:26195665). Plays a role in interferon signaling via its role in the deubiquitination of the interferon receptor IFNAR1; deubiquitination increases IFNAR1 activities by enhancing its stability and cell surface expression (PubMed:24075985, PubMed:26344097). Down-regulates the response to bacterial lipopolysaccharide (LPS) via its role in IFNAR1 deubiquitination (PubMed:24075985). Required for normal induction of p53/TP53 in response to DNA damage (PubMed:25283148). Independent of the BRISC complex, promotes interaction between USP7 and p53/TP53, and thereby promotes deubiquitination of p53/TP53, preventing its degradation and resulting in increased p53/TP53-mediated transcription regulation and p53/TP53-dependent apoptosis in response to DNA damage (PubMed:25283148). {ECO:0000250|UniProtKB:Q3TCJ1, ECO:0000269|PubMed:19214193, ECO:0000269|PubMed:20032457, ECO:0000269|PubMed:20656690, ECO:0000269|PubMed:24075985, ECO:0000269|PubMed:25283148}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DOC2B | chr17:30897 | chr17:1003975 | ENST00000343572 | - | 1 | 6 | 1_36 | 124 | 309.0 | Region | Negatively regulates targeting to plasma membrane |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000291107 | 0 | 22 | 417_420 | 45 | 823.0 | Compositional bias | Note=Poly-Leu | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000302538 | 1 | 23 | 417_420 | 82 | 860.0 | Compositional bias | Note=Poly-Leu | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000543210 | 0 | 8 | 417_420 | 0 | 311.0 | Compositional bias | Note=Poly-Leu | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000544583 | 1 | 23 | 417_420 | 36 | 814.0 | Compositional bias | Note=Poly-Leu | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000574437 | 0 | 22 | 417_420 | 36 | 814.0 | Compositional bias | Note=Poly-Leu | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000291107 | 0 | 22 | 301_459 | 45 | 823.0 | Domain | PH | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000291107 | 0 | 22 | 484_613 | 45 | 823.0 | Domain | C2 | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000291107 | 0 | 22 | 647_845 | 45 | 823.0 | Domain | Rho-GAP | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000291107 | 0 | 22 | 91_284 | 45 | 823.0 | Domain | DH | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000302538 | 1 | 23 | 301_459 | 82 | 860.0 | Domain | PH | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000302538 | 1 | 23 | 484_613 | 82 | 860.0 | Domain | C2 | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000302538 | 1 | 23 | 647_845 | 82 | 860.0 | Domain | Rho-GAP | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000302538 | 1 | 23 | 91_284 | 82 | 860.0 | Domain | DH | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000543210 | 0 | 8 | 301_459 | 0 | 311.0 | Domain | PH | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000543210 | 0 | 8 | 484_613 | 0 | 311.0 | Domain | C2 | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000543210 | 0 | 8 | 647_845 | 0 | 311.0 | Domain | Rho-GAP | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000543210 | 0 | 8 | 91_284 | 0 | 311.0 | Domain | DH | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000544583 | 1 | 23 | 301_459 | 36 | 814.0 | Domain | PH | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000544583 | 1 | 23 | 484_613 | 36 | 814.0 | Domain | C2 | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000544583 | 1 | 23 | 647_845 | 36 | 814.0 | Domain | Rho-GAP | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000544583 | 1 | 23 | 91_284 | 36 | 814.0 | Domain | DH | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000574437 | 0 | 22 | 301_459 | 36 | 814.0 | Domain | PH | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000574437 | 0 | 22 | 484_613 | 36 | 814.0 | Domain | C2 | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000574437 | 0 | 22 | 647_845 | 36 | 814.0 | Domain | Rho-GAP | |
| Tgene | ABR | chr17:30897 | chr17:1003975 | ENST00000574437 | 0 | 22 | 91_284 | 36 | 814.0 | Domain | DH |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DOC2B | chr17:30897 | chr17:1003975 | ENST00000343572 | - | 1 | 6 | 126_250 | 124 | 309.0 | Domain | C2 1 |
| Hgene | DOC2B | chr17:30897 | chr17:1003975 | ENST00000343572 | - | 1 | 6 | 266_399 | 124 | 309.0 | Domain | C2 2 |
Top |
Fusion Gene Sequence for DOC2B-ABR |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >23735_23735_1_DOC2B-ABR_DOC2B_chr17_30897_ENST00000343572_ABR_chr17_1003975_ENST00000302538_length(transcript)=5426nt_BP=530nt TCCGCAGCGGAGCGGGCAGCGGCCAAGTCAGGGCCGTCCGGGGGCGCGGCCGGCGATGCCCGCAGCCCCCGCCGCGCCCCGCCGGGCCTG CTGAGCCGCCCCCGGGCCGGGGTCGCGCCGGGCCGGGCCGCGCCCGGGGCGGGGCGGCGCTGCCTGCATGACCCTCCGGCGGCGCGGGGA GAAGGCGACCATCAGCATCCAGGAGCATATGGCCATCGACGTGTGCCCCGGCCCCATCCGTCCCATCAAGCAGATCTCCGACTACTTCCC CCGCTTCCCGCGGGGCCTGCCCCCGGACGCCGGGCCCCGAGCCGCTGCACCCCCGGACGCCCCCGCGCGCCCGGCTGTGGCCGGTGCCGG CCGCCGCAGCCCCTCCGACGGCGCCCGCGAGGACGACGAGGATGTGGACCAGCTCTTCGGAGCCTACGGCTCCAGCCCGGGCCCCAGCCC GGGTCCCAGCCCCGCGCGGCCGCCAGCCAAGCCGCCGGAGGACGAGCCGGACGCCGACGGCTACGAGTCGGACGACTGCAGTGGAAGCAG GGAAAGGCCTGGAGATGAGGAAGCTGGTTCTCTCGGGGTTCTTGGCCAGCGAAGAGATCTACATTAACCAGCTGGAAGCCCTGTTGCTGC CCATGAAACCCCTGAAGGCCACCGCCACCACCTCCCAGCCCGTGCTCACCATCCAGCAGATCGAGACCATCTTCTACAAGATCCAGGACA TCTATGAGATCCACAAGGAGTTCTATGACAACCTGTGCCCCAAGGTGCAACAGTGGGACAGCCAGGTCACCATGGGCCACCTCTTCCAGA AGCTGGCCAGCCAGCTCGGTGTGTACAAAGCGTTTGTCGATAACTATAAAGTCGCTCTGGAGACAGCTGAGAAGTGCAGCCAGTCCAACA ACCAGTTCCAGAAGATCTCAGAGGAACTCAAAGTGAAAGGTCCCAAGGACTCCAAGGACAGCCACACGTCTGTCACCATGGAAGCTCTGC TCTACAAGCCCATTGACCGGGTCACTCGGAGCACCCTAGTCCTACACGACCTGCTGAAGCACACACCTGTGGACCACCCCGACTACCCGC TGCTGCAGGATGCCCTCCGCATCTCCCAGAACTTCCTGTCCAGCATCAACGAGGACATCGACCCCCGCCGGACTGCAGTGACAACGCCCA AGGGGGAGACGCGACAGCTGGTGAAGGACGGCTTCCTGGTGGAAGTGTCAGAGAGCTCCCGGAAGCTGCGGCACGTCTTCCTCTTTACAG ATGTCCTACTGTGTGCCAAGCTGAAGAAGACCTCTGCAGGGAAGCACCAGCAGTATGACTGTAAGTGGTACATCCCCCTGGCCGACCTGG TGTTTCCATCCCCCGAGGAGTCTGAGGCCAGCCCCCAGGTGCACCCCTTCCCAGACCATGAGCTGGAGGACATGAAGATGAAGATCTCTG CCCTCAAGAGTGAAATCCAGAAGGAGAAAGCCAACAAAGGCCAGAGCCGGGCCATCGAGCGCCTGAAGAAGAAGATGTTTGAGAATGAGT TCCTGCTGCTGCTCAACTCCCCCACAATCCCGTTCAGGATCCACAATCGGAATGGAAAGAGTTACCTGTTCCTACTGTCCTCGGACTACG AGAGGTCAGAGTGGAGAGAAGCAATTCAGAAACTACAGAAGAAGGATCTCCAGGCCTTTGTCCTGAGCTCAGTGGAGCTCCAGGTGCTCA CAGGATCCTGTTTCAAGCTTAGGACTGTACACAACATTCCTGTCACCAGCAATAAAGACGACGATGAGTCTCCAGGACTCTATGGCTTCC TTCATGTCATCGTCCACTCTGCCAAGGGATTTAAGCAATCAGCCAACCTGTACTGTACCCTGGAGGTGGATTCCTTCGGCTATTTTGTCA GCAAAGCCAAAACCAGGGTGTTCCGGGACACAGCGGAGCCCAAGTGGGATGAGGAGTTTGAGATCGAGCTGGAGGGCTCCCAGTCCCTGA GGATCCTGTGCTATGAGAAGTGCTATGACAAGACCAAGGTCAACAAGGACAACAATGAGATCGTGGACAAGATCATGGGCAAAGGACAGA TCCAGCTGGACCCACAAACCGTGGAGACCAAGAACTGGCACACGGACGTGATTGAGATGAACGGGATCAAAGTGGAATTTTCCATGAAAT TCACCAGCCGAGATATGAGCCTGAAGAGGACCCCGTCCAAAAAGCAGACCGGCGTCTTCGGTGTGAAGATCAGCGTGGTGACGAAGCGGG AGCGCTCCAAGGTGCCCTACATCGTCCGGCAGTGTGTGGAGGAGGTGGAGAAGAGGGGTATCGAGGAGGTTGGCATCTACAGGATATCGG GCGTGGCCACGGACATCCAGGCGCTCAAGGCCGTCTTCGATGCCAATAACAAGGACATCCTGCTGATGCTGAGTGACATGGACATCAACG CCATCGCCGGGACGCTCAAGCTGTACTTCCGGGAACTGCCCGAGCCGCTCCTCACGGACCGACTCTACCCAGCCTTCATGGAGGGCATCG CCCTGTCAGACCCTGCTGCCAAGGAAAACTGCATGATGCACCTGCTCCGCTCCCTGCCCGACCCCAACCTCATCACCTTCCTCTTCCTGC TGGAACACTTGAAAAGGGTTGCCGAGAAGGAGCCCATCAACAAAATGTCACTTCACAACCTGGCTACCGTGTTTGGACCCACGTTACTGA GACCCTCAGAAGTGGAGAGCAAAGCACACCTCACCTCGGCTGCGGACATCTGGTCCCATGACGTCATGGCGCAGGTCCAGGTCCTCCTCT ACTACCTGCAGCACCCCCCCATTTCCTTCGCAGAACTCAAGCGGAACACACTGTACTTCTCCACCGACGTGTAGCCCGAGGCAGGGTGGC TGCGGGCGGGTGGTGGAACCAGCCCCTCCAGCCTGGGGTCCAACTCAGACTTGAAAGACTGCAATAGAAAACTCCCAAACCCAGCACTCC AGACTCGAGGGAAGCCAGCTTCCAAGAACTGGAATGCGTACGTCTTTTGTGCCACCTTGTACAAAGCCGGCTGCCCAGCCCCAGCCTCAC CACCGCATCCCACCTCCTGCCCTCCATACCTCTAGTTGTGTCTGATGCTCCGTGCTGTTCGGGAATTGTTTTATGTACACTTGTCAGGCA GAAAAGGTAGTGACCGGCCCGGCGTGGGCACACAGACAGCCCGCTTTGTTCTTTCATTTCCTCCAGCACTTTCTTTCCGCCTGAGTCCAG CCCAAGGCCTTTTATTTTGCGCTGTGTAACTGCTGCCAGCTTCTCTCTTGGCCCTGCTCCCAGATGGCGGTCTCCTGGCAGCCTCCCCTC AGTCTTCCTCCACCCGCTCTTCCTTCCCAGCCTGCCTGCATGCATGTGCACCCTTGGTCTTCGCTCCATCGCCTTGAAAGCTCTGAAGAG GCCCTGGGTTGCCGCGGCAGCAGTGGTCTGTTTGATGCTGCCGTTTGCCGCTGCCGGCCCCTCCTCAGACTCCGCCTTTGGGAGCACACC TGCTTTGCCTTGCTGCCTGTGCAAATGTTGGACAAGCAGACACACTCACACTCGTCCCCAGCTTAGCACAGAGCTGGAGCGCCCATTTCT GGAATTTTCCGTTTGGGAATCTCCACTTCTGGGGTTTACCTGTTCGGCCTCCTGTCTATCAGTGAGGCATCTCTGACTGTTTCTTCTACT GCTTTTCAGTTCCCTTCCCTGCTGTTCTATTTCCTTTGAGTGTAAAGACTCACAGGTGACCTGCTATCGAGATAGCCAGAGGGTCAGGAG AGAATGGGGGAGGAGGCGGTCAGGCTGCTGAGGAAACACCACAGGCTGAACGGGGGAGGAATGCACATGCCACGCTGGGTGTCCCGGGTC GCGGGGAGGCAGCTCAGCTCTTAGGAGCAAGTTGTGGGGGCTTTTCAAGAGGGGCCAGGCTTCCTGGAGGGTGACTGATGTGGCCGAAGC AGGTGTCCAGGCAGGTAGGCTGCAGCCAGGAGCTCCCTGGCACCGCAGGACCTCGTGGTACTCTTGCCTTAGATTTTACACACACTCCAC AGCCAAGCACTGCCACGGTCCTCCAGGACCTGGGAAGCAAAGGCACAGGCCCACGGTGGCCAGCCATTGTGGTGCCGCCCCAGCTTCTGG ATACAGCCTTTTGGGTAAACACTGGGAACTCCAGAAGTTGTGGGGAGAGTGGGGAATCAGACAGCCGCCTCTAGGGGCTGGGTTCTGCTG GGGCCTCCTTGTTGGTGCTGTAGGCACCCGCCAGGGAGCAGGGACCCGACTTGCAGACGCATTGCCCGGTACTAGGAAGGAGTGAGGTGT GTTCCCACCGTACACTTCCCACACGAGCTGCGGCTGCCAGCCTCGGGCCATCAGCCTAGGAGAGCAGATGCAGCTCCAGGGGCTCGACTT ATAGCCAGTTACAGCTCCCCGGCTCTTCTGTGTGGCAGAGCGTCGTTTCCGGGCCCTCAGGGCTGGGGAGCTCAGTTCCCATTGCTTGTG CTCAGGGCTGAGTCTTAAAGAAGGGTTTGCCGGCCCTAACGCTGCAGCGCGTGCGCGGTGAGAGGCCCTTTTTGAGCCTGTTTACTCCTG TGGCCTTGGGCAGAACAGTAAATACTCTGTGCACGGAGGAAAGACATGCCCAAGAGGAAGGAAGTACTGACCATCGGCTGCCTGTGAGCA GCTTAGCAAGGAGCCCTTGCTCCCTGGGAAAGGCGGTGAACTTGAGTCTAAAGATGCAGTGCCTGGCCCTTCCTAAGGTCCCTGCCTGGC ATCCGAGTGTCGGTGTGTGGCACAGAAGGCTCCTGCTTGCTTCCAAAGTGATGGACAGGAAGGGGCAGAGTGAGTCACGGCCCAGACTGG GCACCTTCGCGTCTCAGCCTCAGGGAGCCCCACAGCCCCAAGCTCGCTGAGGCAACGTGAGAACAGGCTATGGGAAGGCTGCAAAGGCTG AGAAATGCAAAGGCTCATATTTATAAATCCCACCCCCAGAGTGGGGAGGGTCAGGTGCCAGACCTGGACTAAACTGCACCAAGGAAACAC CCAGCAGGGTCTCCTGTGAGCCGGGGACCATGCAGCCCGAAACCTCCAGTCACTGCGCCCGGCAGGAGTCAGGAGCCAGGGACTGTGCAG CCTGGAACCTCCAGTCACTGTGCCCAGCAGGGTGGGCTGTGCCCAGCAGGAGTCAGGCTAAGAAACGCCAGGTCTGCCTGTTCTTGCTGG GCAATGGCTGATGGCTGCCAGTTTCTGCTGATACACAGGTAGGATGGGACCCTTCATGAATATCTGACTTTAATAAGTTGGTAAGGATAT ATTTTTTTGTCTATGTTCTGTTTCAACTTATGTAGATTATTATAAATTGATGTAAACCACGTGAGAGGAAAATGTTAATAAAAAATGCAA >23735_23735_1_DOC2B-ABR_DOC2B_chr17_30897_ENST00000343572_ABR_chr17_1003975_ENST00000302538_length(amino acids)=849AA_BP=4 MHPRTPPRARLWPVPAAAAPPTAPARTTRMWTSSSEPTAPARAPARVPAPRGRQPSRRRTSRTPTATSRTTAVEAGKGLEMRKLVLSGFL ASEEIYINQLEALLLPMKPLKATATTSQPVLTIQQIETIFYKIQDIYEIHKEFYDNLCPKVQQWDSQVTMGHLFQKLASQLGVYKAFVDN YKVALETAEKCSQSNNQFQKISEELKVKGPKDSKDSHTSVTMEALLYKPIDRVTRSTLVLHDLLKHTPVDHPDYPLLQDALRISQNFLSS INEDIDPRRTAVTTPKGETRQLVKDGFLVEVSESSRKLRHVFLFTDVLLCAKLKKTSAGKHQQYDCKWYIPLADLVFPSPEESEASPQVH PFPDHELEDMKMKISALKSEIQKEKANKGQSRAIERLKKKMFENEFLLLLNSPTIPFRIHNRNGKSYLFLLSSDYERSEWREAIQKLQKK DLQAFVLSSVELQVLTGSCFKLRTVHNIPVTSNKDDDESPGLYGFLHVIVHSAKGFKQSANLYCTLEVDSFGYFVSKAKTRVFRDTAEPK WDEEFEIELEGSQSLRILCYEKCYDKTKVNKDNNEIVDKIMGKGQIQLDPQTVETKNWHTDVIEMNGIKVEFSMKFTSRDMSLKRTPSKK QTGVFGVKISVVTKRERSKVPYIVRQCVEEVEKRGIEEVGIYRISGVATDIQALKAVFDANNKDILLMLSDMDINAIAGTLKLYFRELPE PLLTDRLYPAFMEGIALSDPAAKENCMMHLLRSLPDPNLITFLFLLEHLKRVAEKEPINKMSLHNLATVFGPTLLRPSEVESKAHLTSAA -------------------------------------------------------------- >23735_23735_2_DOC2B-ABR_DOC2B_chr17_30897_ENST00000343572_ABR_chr17_1003975_ENST00000544583_length(transcript)=5417nt_BP=530nt TCCGCAGCGGAGCGGGCAGCGGCCAAGTCAGGGCCGTCCGGGGGCGCGGCCGGCGATGCCCGCAGCCCCCGCCGCGCCCCGCCGGGCCTG CTGAGCCGCCCCCGGGCCGGGGTCGCGCCGGGCCGGGCCGCGCCCGGGGCGGGGCGGCGCTGCCTGCATGACCCTCCGGCGGCGCGGGGA GAAGGCGACCATCAGCATCCAGGAGCATATGGCCATCGACGTGTGCCCCGGCCCCATCCGTCCCATCAAGCAGATCTCCGACTACTTCCC CCGCTTCCCGCGGGGCCTGCCCCCGGACGCCGGGCCCCGAGCCGCTGCACCCCCGGACGCCCCCGCGCGCCCGGCTGTGGCCGGTGCCGG CCGCCGCAGCCCCTCCGACGGCGCCCGCGAGGACGACGAGGATGTGGACCAGCTCTTCGGAGCCTACGGCTCCAGCCCGGGCCCCAGCCC GGGTCCCAGCCCCGCGCGGCCGCCAGCCAAGCCGCCGGAGGACGAGCCGGACGCCGACGGCTACGAGTCGGACGACTGCAGTGGAAGCAG GGAAAGGCCTGGAGATGAGGAAGCTGGTTCTCTCGGGGTTCTTGGCCAGCGAAGAGATCTACATTAACCAGCTGGAAGCCCTGTTGCTGC CCATGAAACCCCTGAAGGCCACCGCCACCACCTCCCAGCCCGTGCTCACCATCCAGCAGATCGAGACCATCTTCTACAAGATCCAGGACA TCTATGAGATCCACAAGGAGTTCTATGACAACCTGTGCCCCAAGGTGCAACAGTGGGACAGCCAGGTCACCATGGGCCACCTCTTCCAGA AGCTGGCCAGCCAGCTCGGTGTGTACAAAGCGTTTGTCGATAACTATAAAGTCGCTCTGGAGACAGCTGAGAAGTGCAGCCAGTCCAACA ACCAGTTCCAGAAGATCTCAGAGGAACTCAAAGTGAAAGGTCCCAAGGACTCCAAGGACAGCCACACGTCTGTCACCATGGAAGCTCTGC TCTACAAGCCCATTGACCGGGTCACTCGGAGCACCCTAGTCCTACACGACCTGCTGAAGCACACACCTGTGGACCACCCCGACTACCCGC TGCTGCAGGATGCCCTCCGCATCTCCCAGAACTTCCTGTCCAGCATCAACGAGGACATCGACCCCCGCCGGACTGCAGTGACAACGCCCA AGGGGGAGACGCGACAGCTGGTGAAGGACGGCTTCCTGGTGGAAGTGTCAGAGAGCTCCCGGAAGCTGCGGCACGTCTTCCTCTTTACAG ATGTCCTACTGTGTGCCAAGCTGAAGAAGACCTCTGCAGGGAAGCACCAGCAGTATGACTGTAAGTGGTACATCCCCCTGGCCGACCTGG TGTTTCCATCCCCCGAGGAGTCTGAGGCCAGCCCCCAGGTGCACCCCTTCCCAGACCATGAGCTGGAGGACATGAAGATGAAGATCTCTG CCCTCAAGAGTGAAATCCAGAAGGAGAAAGCCAACAAAGGCCAGAGCCGGGCCATCGAGCGCCTGAAGAAGAAGATGTTTGAGAATGAGT TCCTGCTGCTGCTCAACTCCCCCACAATCCCGTTCAGGATCCACAATCGGAATGGAAAGAGTTACCTGTTCCTACTGTCCTCGGACTACG AGAGGTCAGAGTGGAGAGAAGCAATTCAGAAACTACAGAAGAAGGATCTCCAGGCCTTTGTCCTGAGCTCAGTGGAGCTCCAGGTGCTCA CAGGATCCTGTTTCAAGCTTAGGACTGTACACAACATTCCTGTCACCAGCAATAAAGACGACGATGAGTCTCCAGGACTCTATGGCTTCC TTCATGTCATCGTCCACTCTGCCAAGGGATTTAAGCAATCAGCCAACCTGTACTGTACCCTGGAGGTGGATTCCTTCGGCTATTTTGTCA GCAAAGCCAAAACCAGGGTGTTCCGGGACACAGCGGAGCCCAAGTGGGATGAGGAGTTTGAGATCGAGCTGGAGGGCTCCCAGTCCCTGA GGATCCTGTGCTATGAGAAGTGCTATGACAAGACCAAGGTCAACAAGGACAACAATGAGATCGTGGACAAGATCATGGGCAAAGGACAGA TCCAGCTGGACCCACAAACCGTGGAGACCAAGAACTGGCACACGGACGTGATTGAGATGAACGGGATCAAAGTGGAATTTTCCATGAAAT TCACCAGCCGAGATATGAGCCTGAAGAGGACCCCGTCCAAAAAGCAGACCGGCGTCTTCGGTGTGAAGATCAGCGTGGTGACGAAGCGGG AGCGCTCCAAGGTGCCCTACATCGTCCGGCAGTGTGTGGAGGAGGTGGAGAAGAGGGGTATCGAGGAGGTTGGCATCTACAGGATATCGG GCGTGGCCACGGACATCCAGGCGCTCAAGGCCGTCTTCGATGCCAATAACAAGGACATCCTGCTGATGCTGAGTGACATGGACATCAACG CCATCGCCGGGACGCTCAAGCTGTACTTCCGGGAACTGCCCGAGCCGCTCCTCACGGACCGACTCTACCCAGCCTTCATGGAGGGCATCG CCCTGTCAGACCCTGCTGCCAAGGAAAACTGCATGATGCACCTGCTCCGCTCCCTGCCCGACCCCAACCTCATCACCTTCCTCTTCCTGC TGGAACACTTGAAAAGGGTTGCCGAGAAGGAGCCCATCAACAAAATGTCACTTCACAACCTGGCTACCGTGTTTGGACCCACGTTACTGA GACCCTCAGAAGTGGAGAGCAAAGCACACCTCACCTCGGCTGCGGACATCTGGTCCCATGACGTCATGGCGCAGGTCCAGGTCCTCCTCT ACTACCTGCAGCACCCCCCCATTTCCTTCGCAGAACTCAAGCGGAACACACTGTACTTCTCCACCGACGTGTAGCCCGAGGCAGGGTGGC TGCGGGCGGGTGGTGGAACCAGCCCCTCCAGCCTGGGGTCCAACTCAGACTTGAAAGACTGCAATAGAAAACTCCCAAACCCAGCACTCC AGACTCGAGGGAAGCCAGCTTCCAAGAACTGGAATGCGTACGTCTTTTGTGCCACCTTGTACAAAGCCGGCTGCCCAGCCCCAGCCTCAC CACCGCATCCCACCTCCTGCCCTCCATACCTCTAGTTGTGTCTGATGCTCCGTGCTGTTCGGGAATTGTTTTATGTACACTTGTCAGGCA GAAAAGGTAGTGACCGGCCCGGCGTGGGCACACAGACAGCCCGCTTTGTTCTTTCATTTCCTCCAGCACTTTCTTTCCGCCTGAGTCCAG CCCAAGGCCTTTTATTTTGCGCTGTGTAACTGCTGCCAGCTTCTCTCTTGGCCCTGCTCCCAGATGGCGGTCTCCTGGCAGCCTCCCCTC AGTCTTCCTCCACCCGCTCTTCCTTCCCAGCCTGCCTGCATGCATGTGCACCCTTGGTCTTCGCTCCATCGCCTTGAAAGCTCTGAAGAG GCCCTGGGTTGCCGCGGCAGCAGTGGTCTGTTTGATGCTGCCGTTTGCCGCTGCCGGCCCCTCCTCAGACTCCGCCTTTGGGAGCACACC TGCTTTGCCTTGCTGCCTGTGCAAATGTTGGACAAGCAGACACACTCACACTCGTCCCCAGCTTAGCACAGAGCTGGAGCGCCCATTTCT GGAATTTTCCGTTTGGGAATCTCCACTTCTGGGGTTTACCTGTTCGGCCTCCTGTCTATCAGTGAGGCATCTCTGACTGTTTCTTCTACT GCTTTTCAGTTCCCTTCCCTGCTGTTCTATTTCCTTTGAGTGTAAAGACTCACAGGTGACCTGCTATCGAGATAGCCAGAGGGTCAGGAG AGAATGGGGGAGGAGGCGGTCAGGCTGCTGAGGAAACACCACAGGCTGAACGGGGGAGGAATGCACATGCCACGCTGGGTGTCCCGGGTC GCGGGGAGGCAGCTCAGCTCTTAGGAGCAAGTTGTGGGGGCTTTTCAAGAGGGGCCAGGCTTCCTGGAGGGTGACTGATGTGGCCGAAGC AGGTGTCCAGGCAGGTAGGCTGCAGCCAGGAGCTCCCTGGCACCGCAGGACCTCGTGGTACTCTTGCCTTAGATTTTACACACACTCCAC AGCCAAGCACTGCCACGGTCCTCCAGGACCTGGGAAGCAAAGGCACAGGCCCACGGTGGCCAGCCATTGTGGTGCCGCCCCAGCTTCTGG ATACAGCCTTTTGGGTAAACACTGGGAACTCCAGAAGTTGTGGGGAGAGTGGGGAATCAGACAGCCGCCTCTAGGGGCTGGGTTCTGCTG GGGCCTCCTTGTTGGTGCTGTAGGCACCCGCCAGGGAGCAGGGACCCGACTTGCAGACGCATTGCCCGGTACTAGGAAGGAGTGAGGTGT GTTCCCACCGTACACTTCCCACACGAGCTGCGGCTGCCAGCCTCGGGCCATCAGCCTAGGAGAGCAGATGCAGCTCCAGGGGCTCGACTT ATAGCCAGTTACAGCTCCCCGGCTCTTCTGTGTGGCAGAGCGTCGTTTCCGGGCCCTCAGGGCTGGGGAGCTCAGTTCCCATTGCTTGTG CTCAGGGCTGAGTCTTAAAGAAGGGTTTGCCGGCCCTAACGCTGCAGCGCGTGCGCGGTGAGAGGCCCTTTTTGAGCCTGTTTACTCCTG TGGCCTTGGGCAGAACAGTAAATACTCTGTGCACGGAGGAAAGACATGCCCAAGAGGAAGGAAGTACTGACCATCGGCTGCCTGTGAGCA GCTTAGCAAGGAGCCCTTGCTCCCTGGGAAAGGCGGTGAACTTGAGTCTAAAGATGCAGTGCCTGGCCCTTCCTAAGGTCCCTGCCTGGC ATCCGAGTGTCGGTGTGTGGCACAGAAGGCTCCTGCTTGCTTCCAAAGTGATGGACAGGAAGGGGCAGAGTGAGTCACGGCCCAGACTGG GCACCTTCGCGTCTCAGCCTCAGGGAGCCCCACAGCCCCAAGCTCGCTGAGGCAACGTGAGAACAGGCTATGGGAAGGCTGCAAAGGCTG AGAAATGCAAAGGCTCATATTTATAAATCCCACCCCCAGAGTGGGGAGGGTCAGGTGCCAGACCTGGACTAAACTGCACCAAGGAAACAC CCAGCAGGGTCTCCTGTGAGCCGGGGACCATGCAGCCCGAAACCTCCAGTCACTGCGCCCGGCAGGAGTCAGGAGCCAGGGACTGTGCAG CCTGGAACCTCCAGTCACTGTGCCCAGCAGGGTGGGCTGTGCCCAGCAGGAGTCAGGCTAAGAAACGCCAGGTCTGCCTGTTCTTGCTGG GCAATGGCTGATGGCTGCCAGTTTCTGCTGATACACAGGTAGGATGGGACCCTTCATGAATATCTGACTTTAATAAGTTGGTAAGGATAT ATTTTTTTGTCTATGTTCTGTTTCAACTTATGTAGATTATTATAAATTGATGTAAACCACGTGAGAGGAAAATGTTAATAAAAAATGCAA >23735_23735_2_DOC2B-ABR_DOC2B_chr17_30897_ENST00000343572_ABR_chr17_1003975_ENST00000544583_length(amino acids)=849AA_BP=4 MHPRTPPRARLWPVPAAAAPPTAPARTTRMWTSSSEPTAPARAPARVPAPRGRQPSRRRTSRTPTATSRTTAVEAGKGLEMRKLVLSGFL ASEEIYINQLEALLLPMKPLKATATTSQPVLTIQQIETIFYKIQDIYEIHKEFYDNLCPKVQQWDSQVTMGHLFQKLASQLGVYKAFVDN YKVALETAEKCSQSNNQFQKISEELKVKGPKDSKDSHTSVTMEALLYKPIDRVTRSTLVLHDLLKHTPVDHPDYPLLQDALRISQNFLSS INEDIDPRRTAVTTPKGETRQLVKDGFLVEVSESSRKLRHVFLFTDVLLCAKLKKTSAGKHQQYDCKWYIPLADLVFPSPEESEASPQVH PFPDHELEDMKMKISALKSEIQKEKANKGQSRAIERLKKKMFENEFLLLLNSPTIPFRIHNRNGKSYLFLLSSDYERSEWREAIQKLQKK DLQAFVLSSVELQVLTGSCFKLRTVHNIPVTSNKDDDESPGLYGFLHVIVHSAKGFKQSANLYCTLEVDSFGYFVSKAKTRVFRDTAEPK WDEEFEIELEGSQSLRILCYEKCYDKTKVNKDNNEIVDKIMGKGQIQLDPQTVETKNWHTDVIEMNGIKVEFSMKFTSRDMSLKRTPSKK QTGVFGVKISVVTKRERSKVPYIVRQCVEEVEKRGIEEVGIYRISGVATDIQALKAVFDANNKDILLMLSDMDINAIAGTLKLYFRELPE PLLTDRLYPAFMEGIALSDPAAKENCMMHLLRSLPDPNLITFLFLLEHLKRVAEKEPINKMSLHNLATVFGPTLLRPSEVESKAHLTSAA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for DOC2B-ABR |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for DOC2B-ABR |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for DOC2B-ABR |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
