
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:DTX3L-FAM162A (FusionGDB2 ID:24413) |
Fusion Gene Summary for DTX3L-FAM162A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: DTX3L-FAM162A | Fusion gene ID: 24413 | Hgene | Tgene | Gene symbol | DTX3L | FAM162A | Gene ID | 151636 | 26355 |
| Gene name | deltex E3 ubiquitin ligase 3L | family with sequence similarity 162 member A | |
| Synonyms | BBAP|RNF143 | C3orf28|E2IG5|HGTD-P | |
| Cytomap | 3q21.1 | 3q21.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | E3 ubiquitin-protein ligase DTX3LB-lymphoma- and BAL-associated proteinRING-type E3 ubiquitin transferase DTX3Ldeltex 3 like, E3 ubiquitin ligasedeltex 3-likerhysin-2rhysin2 | protein FAM162AE2-induced gene 5 proteinHIF-1 alpha-responsive proapoptotic moleculegrowth and transformation-dependent protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8TDB6 | Q96A26 | |
| Ensembl transtripts involved in fusion gene | ENST00000296161, ENST00000383661, | ENST00000469967, ENST00000232125, ENST00000477892, | |
| Fusion gene scores | * DoF score | 4 X 5 X 2=40 | 5 X 5 X 2=50 |
| # samples | 5 | 5 | |
| ** MAII score | log2(5/40*10)=0.321928094887362 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(5/50*10)=0 | |
| Context | PubMed: DTX3L [Title/Abstract] AND FAM162A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | DTX3L(122289519)-FAM162A(122126128), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | DTX3L-FAM162A seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | DTX3L | GO:0010390 | histone monoubiquitination | 19818714|28525742 |
| Hgene | DTX3L | GO:0070936 | protein K48-linked ubiquitination | 26479788 |
| Hgene | DTX3L | GO:1901666 | positive regulation of NAD+ ADP-ribosyltransferase activity | 28525742 |
| Tgene | FAM162A | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 15082785 |
| Tgene | FAM162A | GO:0043065 | positive regulation of apoptotic process | 15082785 |
| Tgene | FAM162A | GO:0071456 | cellular response to hypoxia | 15082785 |
| Tgene | FAM162A | GO:0090200 | positive regulation of release of cytochrome c from mitochondria | 15082785 |
 Fusion gene breakpoints across DTX3L (5'-gene) Fusion gene breakpoints across DTX3L (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
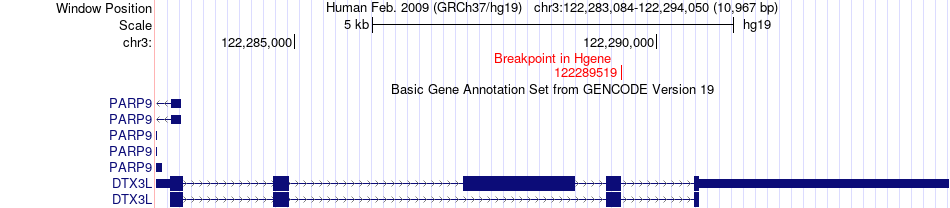 |
 Fusion gene breakpoints across FAM162A (3'-gene) Fusion gene breakpoints across FAM162A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
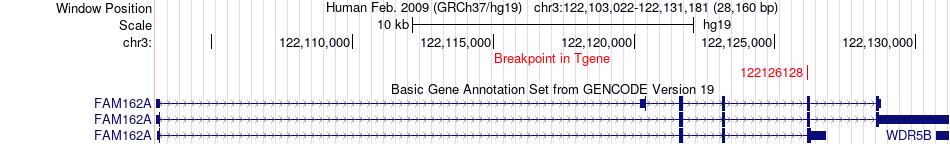 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | CESC | TCGA-VS-A9UV-01A | DTX3L | chr3 | 122289519 | + | FAM162A | chr3 | 122126128 | + |
| ChimerDB4 | CESC | TCGA-VS-A9UV | DTX3L | chr3 | 122289519 | + | FAM162A | chr3 | 122126128 | + |
Top |
Fusion Gene ORF analysis for DTX3L-FAM162A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000296161 | ENST00000469967 | DTX3L | chr3 | 122289519 | + | FAM162A | chr3 | 122126128 | + |
| Frame-shift | ENST00000383661 | ENST00000469967 | DTX3L | chr3 | 122289519 | + | FAM162A | chr3 | 122126128 | + |
| In-frame | ENST00000296161 | ENST00000232125 | DTX3L | chr3 | 122289519 | + | FAM162A | chr3 | 122126128 | + |
| In-frame | ENST00000296161 | ENST00000477892 | DTX3L | chr3 | 122289519 | + | FAM162A | chr3 | 122126128 | + |
| In-frame | ENST00000383661 | ENST00000232125 | DTX3L | chr3 | 122289519 | + | FAM162A | chr3 | 122126128 | + |
| In-frame | ENST00000383661 | ENST00000477892 | DTX3L | chr3 | 122289519 | + | FAM162A | chr3 | 122126128 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000296161 | DTX3L | chr3 | 122289519 | + | ENST00000232125 | FAM162A | chr3 | 122126128 | + | 2630 | 2342 | 189 | 2543 | 784 |
| ENST00000296161 | DTX3L | chr3 | 122289519 | + | ENST00000477892 | FAM162A | chr3 | 122126128 | + | 5047 | 2342 | 189 | 2543 | 784 |
| ENST00000383661 | DTX3L | chr3 | 122289519 | + | ENST00000232125 | FAM162A | chr3 | 122126128 | + | 905 | 617 | 0 | 818 | 272 |
| ENST00000383661 | DTX3L | chr3 | 122289519 | + | ENST00000477892 | FAM162A | chr3 | 122126128 | + | 3322 | 617 | 0 | 818 | 272 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000296161 | ENST00000232125 | DTX3L | chr3 | 122289519 | + | FAM162A | chr3 | 122126128 | + | 0.002529334 | 0.99747074 |
| ENST00000296161 | ENST00000477892 | DTX3L | chr3 | 122289519 | + | FAM162A | chr3 | 122126128 | + | 0.000628404 | 0.9993716 |
| ENST00000383661 | ENST00000232125 | DTX3L | chr3 | 122289519 | + | FAM162A | chr3 | 122126128 | + | 0.001030163 | 0.9989698 |
| ENST00000383661 | ENST00000477892 | DTX3L | chr3 | 122289519 | + | FAM162A | chr3 | 122126128 | + | 0.000174633 | 0.9998254 |
Top |
Fusion Genomic Features for DTX3L-FAM162A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| DTX3L | chr3 | 122289519 | + | FAM162A | chr3 | 122126127 | + | 0.02028826 | 0.9797117 |
| DTX3L | chr3 | 122289519 | + | FAM162A | chr3 | 122126127 | + | 0.02028826 | 0.9797117 |
| DTX3L | chr3 | 122289519 | + | FAM162A | chr3 | 122126127 | + | 0.02028826 | 0.9797117 |
| DTX3L | chr3 | 122289519 | + | FAM162A | chr3 | 122126127 | + | 0.02028826 | 0.9797117 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
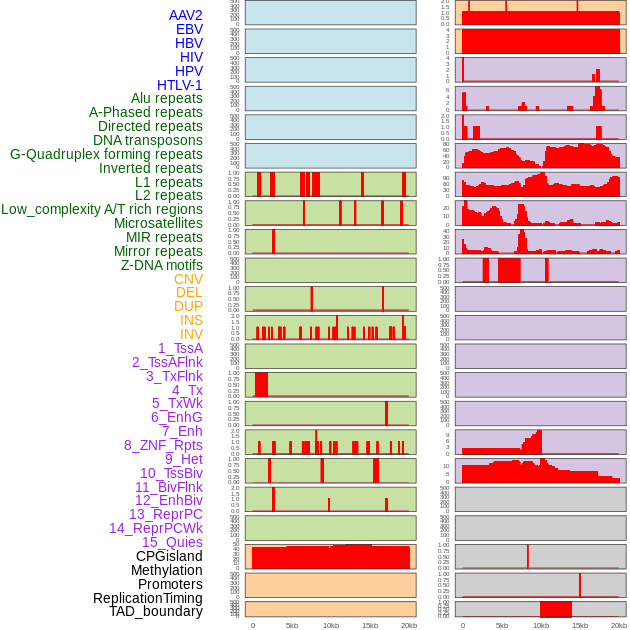 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
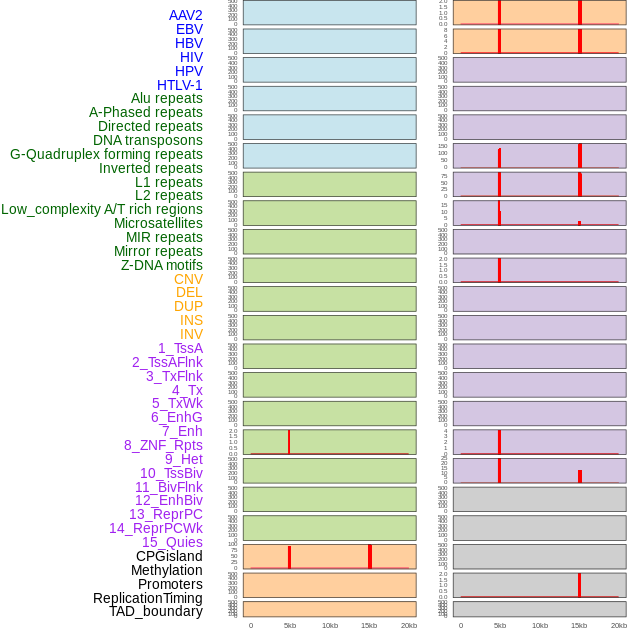 |
Top |
Fusion Protein Features for DTX3L-FAM162A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:122289519/chr3:122126128) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| DTX3L | FAM162A |
| FUNCTION: E3 ubiquitin-protein ligase which, in association with ADP-ribosyltransferase PARP9, plays a role in DNA damage repair and in interferon-mediated antiviral responses (PubMed:12670957, PubMed:19818714, PubMed:26479788, PubMed:23230272). Monoubiquitinates several histones, including histone H2A, H2B, H3 and H4 (PubMed:28525742). In response to DNA damage, mediates monoubiquitination of 'Lys-91' of histone H4 (H4K91ub1) (PubMed:19818714). The exact role of H4K91ub1 in DNA damage response is still unclear but it may function as a licensing signal for additional histone H4 post-translational modifications such as H4 'Lys-20' methylation (H4K20me) (PubMed:19818714). PARP1-dependent PARP9-DTX3L-mediated ubiquitination promotes the rapid and specific recruitment of 53BP1/TP53BP1, UIMC1/RAP80, and BRCA1 to DNA damage sites (PubMed:23230272). By monoubiquitinating histone H2B H2BC9/H2BJ and thereby promoting chromatin remodeling, positively regulates STAT1-dependent interferon-stimulated gene transcription and thus STAT1-mediated control of viral replication (PubMed:26479788). Independently of its catalytic activity, promotes the sorting of chemokine receptor CXCR4 from early endosome to lysosome following CXCL12 stimulation by reducing E3 ligase ITCH activity and thus ITCH-mediated ubiquitination of endosomal sorting complex required for transport ESCRT-0 components HGS and STAM (PubMed:24790097). In addition, required for the recruitment of HGS and STAM to early endosomes (PubMed:24790097). In association with PARP9, plays a role in antiviral responses by mediating 'Lys-48'-linked ubiquitination of encephalomyocarditis virus (EMCV) and human rhinovirus (HRV) C3 proteases and thus promoting their proteosomal-mediated degradation (PubMed:26479788). {ECO:0000269|PubMed:12670957, ECO:0000269|PubMed:19818714, ECO:0000269|PubMed:23230272, ECO:0000269|PubMed:24790097, ECO:0000269|PubMed:26479788, ECO:0000269|PubMed:28525742}. | FUNCTION: Proposed to be involved in regulation of apoptosis; the exact mechanism may differ between cell types/tissues (PubMed:15082785). May be involved in hypoxia-induced cell death of transformed cells implicating cytochrome C release and caspase activation (such as CASP9) and inducing mitochondrial permeability transition (PubMed:15082785). May be involved in hypoxia-induced cell death of neuronal cells probably by promoting release of AIFM1 from mitochondria to cytoplasm and its translocation to the nucleus; however, the involvement of caspases has been reported conflictingly (By similarity). {ECO:0000250|UniProtKB:Q9D6U8, ECO:0000269|PubMed:15082785}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DTX3L | chr3:122289519 | chr3:122126128 | ENST00000296161 | + | 4 | 5 | 561_600 | 717 | 741.0 | Zinc finger | RING-type |
| Tgene | FAM162A | chr3:122289519 | chr3:122126128 | ENST00000477892 | 2 | 5 | 103_120 | 87 | 155.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DTX3L | chr3:122289519 | chr3:122126128 | ENST00000383661 | + | 3 | 4 | 561_600 | 205 | 229.0 | Zinc finger | RING-type |
| Tgene | FAM162A | chr3:122289519 | chr3:122126128 | ENST00000477892 | 2 | 5 | 76_102 | 87 | 155.0 | Region | Required for proapoptotic activity |
Top |
Fusion Gene Sequence for DTX3L-FAM162A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >24413_24413_1_DTX3L-FAM162A_DTX3L_chr3_122289519_ENST00000296161_FAM162A_chr3_122126128_ENST00000232125_length(transcript)=2630nt_BP=2342nt TACTCACCCGGCAGGCCGCTCTCCTCGGTGCAGACAGCACAGGGAGGAGGGGGAAGCGGCTCTGCCGGGAACAGGGAGGGACCTCCAGGG AAGCGAAACTGAAACTTTGCGCCCAGTCCGCAGGGCGGGCCGCGCCTTTACCGCCCAGCTGCCTCCCGGAGCCCCCGCGCCCTCCCGACG CGCAGAGCCATGGCCTCCCACCTGCGCCCGCCGTCCCCGCTCCTCGTGCGGGTGTACAAGTCCGGCCCCCGAGTACGAAGGAAGCTGGAG AGCTACTTCCAGAGCTCTAAGTCCTCGGGCGGCGGGGAGTGCACGGTCAGCACCCAGGAACACGAAGCCCCGGGCACCTTCCGGGTGGAG TTCAGTGAAAGGGCAGCTAAGGAGAGAGTGTTGAAAAAAGGAGAGCACCAAATACTTGTTGACGAAAAACCTGTGCCCATTTTCCTGGTA CCCACTGAAAATTCAATAAAGAAGAACACGAGACCTCAAATTTCTTCACTGACACAATCACAAGCAGAAACACCGTCTGGTGATATGCAT CAACATGAAGGACATATTCCTAATGCTGTGGATTCCTGTCTCCAAAAGATCTTTCTTACTGTAACAGCTGACCTGAACTGTAACCTGTTC TCCAAAGAGCAGAGGGCATACATAACCACACTGTGCCCTAGTATCAGAAAAATGGAAGGTCACGATGGAATTGAGAAGGTGTGTGGTGAC TTCCAAGACATTGAAAGAATACATCAATTTTTGAGTGAGCAGTTCCTGGAAAGTGAGCAGAAACAACAATTTTCCCCTTCAATGACAGAG AGGAAGCCACTCAGTCAGCAGGAGAGGGACAGCTGCATTTCTCCTTCTGAACCAGAAACCAAGGCAGAACAAAAAAGCAACTATTTTGAA GTTCCCTTGCCTTACTTTGAATACTTTAAATATATCTGTCCTGATAAAATCAACTCAATAGAGAAAAGATTTGGTGTAAACATTGAAATC CAGGAGAGTTCTCCAAATATGGTCTGTTTAGATTTCACCTCAAGTCGATCAGGTGACCTGGAAGCAGCTCGTGAGTCTTTTGCTAGTGAA TTTCAGAAGAACACAGAACCTCTGAAGCAAGAATGTGTCTCTTTAGCAGACAGTAAGCAGGCAAATAAATTCAAACAGGAATTGAATCAC CAGTTTACAAAGCTCCTTATAAAGGAGAAAGGAGGCGAATTAACTCTCCTTGGGACCCAAGATGACATTTCAGCTGCCAAACAAAAAATC TCTGAAGCTTTTGTCAAGATACCTGTGAAACTATTTGCTGCCAATTACATGATGAATGTAATTGAGGTTGATAGTGCCCACTATAAACTT TTAGAAACTGAATTACTACAGGAGATATCAGAGATCGAAAAAAGGTATGACATTTGCAGCAAGGTTTCTGAGAAAGGTCAGAAAACCTGC ATTCTGTTTGAATCCAAGGACAGGCAGGTAGATCTATCTGTGCATGCTTATGCAAGTTTCATCGATGCCTTTCAACATGCCTCATGTCAG TTGATGAGAGAAGTTCTTTTACTGAAGTCTTTGGGCAAGGAGAGAAAGCACTTACATCAGACCAAGTTTGCTGATGACTTTAGAAAAAGA CATCCAAATGTACACTTTGTGCTAAATCAAGAGTCAATGACTTTGACTGGTTTGCCAAATCACCTTGCAAAGGCGAAGCAGTATGTTCTA AAAGGAGGAGGAATGTCTTCATTGGCTGGAAAGAAATTGAAAGAGGGTCATGAAACACCGATGGACATTGATAGCGATGATTCCAAAGCA GCTTCTCCGCCACTCAAGGGCTCTGTGAGTTCTGAGGCCTCAGAACTGGACAAGAAGGAAAAGGGCATCTGTGTCATCTGTATGGACACC ATTAGTAACAAAAAAGTGCTACCAAAGTGCAAGCATGAATTCTGCGCCCCTTGTATCAACAAAGCCATGTCATATAAGCCAATCTGTCCC ACATGCCAGACTTCCTATGGTATTCAGAAAGGAAATCAGCCAGAGGGAAGCATGGTTTTCACTGTTTCAAGAGACTCACTTCCAGGTTAT GAGTCCTTTGGCACCATTGTGATTACTTATTCTATGAAAGCAGGCATACAAACAGAAGAACACCCAAACCCAGGAAAGAGATACCCTGGA ATACAGCGAACTGCATACTTGCCTGATAATAAGGAAGGAAGGAAGGTTTTGAAACTGCTTTATAGGGCCTTTGACCAAAAGCTGATTTTT ACAGTGGGGTACTCTCGCGTATTAGGAGTCTCAGATGTCATCACTTGGAATGATATTCACCACAAAACATCCCGGTTTGGAGGACCAGAA ATGTTGGAGATGCTTGATGCTGCAAAGAACAAGATGCGAGTGAAGATCAGCTATCTAATGATTGCCCTGACGGTGGTAGGATGCATCTTC ATGGTTATTGAGGGCAAGAAGGCTGCCCAAAGACACGAGACTTTAACAAGCTTGAACTTAGAAAAGAAAGCTCGTCTGAAAGAGGAAGCA GCTATGAAGGCCAAAACAGAGTAGCAGAGGTATCCGTGTTGGCTGGATTTTGAAAATCCAGGAATTATGTTATAACGTGCCTGTATTAAA >24413_24413_1_DTX3L-FAM162A_DTX3L_chr3_122289519_ENST00000296161_FAM162A_chr3_122126128_ENST00000232125_length(amino acids)=784AA_BP=718 MASHLRPPSPLLVRVYKSGPRVRRKLESYFQSSKSSGGGECTVSTQEHEAPGTFRVEFSERAAKERVLKKGEHQILVDEKPVPIFLVPTE NSIKKNTRPQISSLTQSQAETPSGDMHQHEGHIPNAVDSCLQKIFLTVTADLNCNLFSKEQRAYITTLCPSIRKMEGHDGIEKVCGDFQD IERIHQFLSEQFLESEQKQQFSPSMTERKPLSQQERDSCISPSEPETKAEQKSNYFEVPLPYFEYFKYICPDKINSIEKRFGVNIEIQES SPNMVCLDFTSSRSGDLEAARESFASEFQKNTEPLKQECVSLADSKQANKFKQELNHQFTKLLIKEKGGELTLLGTQDDISAAKQKISEA FVKIPVKLFAANYMMNVIEVDSAHYKLLETELLQEISEIEKRYDICSKVSEKGQKTCILFESKDRQVDLSVHAYASFIDAFQHASCQLMR EVLLLKSLGKERKHLHQTKFADDFRKRHPNVHFVLNQESMTLTGLPNHLAKAKQYVLKGGGMSSLAGKKLKEGHETPMDIDSDDSKAASP PLKGSVSSEASELDKKEKGICVICMDTISNKKVLPKCKHEFCAPCINKAMSYKPICPTCQTSYGIQKGNQPEGSMVFTVSRDSLPGYESF GTIVITYSMKAGIQTEEHPNPGKRYPGIQRTAYLPDNKEGRKVLKLLYRAFDQKLIFTVGYSRVLGVSDVITWNDIHHKTSRFGGPEMLE -------------------------------------------------------------- >24413_24413_2_DTX3L-FAM162A_DTX3L_chr3_122289519_ENST00000296161_FAM162A_chr3_122126128_ENST00000477892_length(transcript)=5047nt_BP=2342nt TACTCACCCGGCAGGCCGCTCTCCTCGGTGCAGACAGCACAGGGAGGAGGGGGAAGCGGCTCTGCCGGGAACAGGGAGGGACCTCCAGGG AAGCGAAACTGAAACTTTGCGCCCAGTCCGCAGGGCGGGCCGCGCCTTTACCGCCCAGCTGCCTCCCGGAGCCCCCGCGCCCTCCCGACG CGCAGAGCCATGGCCTCCCACCTGCGCCCGCCGTCCCCGCTCCTCGTGCGGGTGTACAAGTCCGGCCCCCGAGTACGAAGGAAGCTGGAG AGCTACTTCCAGAGCTCTAAGTCCTCGGGCGGCGGGGAGTGCACGGTCAGCACCCAGGAACACGAAGCCCCGGGCACCTTCCGGGTGGAG TTCAGTGAAAGGGCAGCTAAGGAGAGAGTGTTGAAAAAAGGAGAGCACCAAATACTTGTTGACGAAAAACCTGTGCCCATTTTCCTGGTA CCCACTGAAAATTCAATAAAGAAGAACACGAGACCTCAAATTTCTTCACTGACACAATCACAAGCAGAAACACCGTCTGGTGATATGCAT CAACATGAAGGACATATTCCTAATGCTGTGGATTCCTGTCTCCAAAAGATCTTTCTTACTGTAACAGCTGACCTGAACTGTAACCTGTTC TCCAAAGAGCAGAGGGCATACATAACCACACTGTGCCCTAGTATCAGAAAAATGGAAGGTCACGATGGAATTGAGAAGGTGTGTGGTGAC TTCCAAGACATTGAAAGAATACATCAATTTTTGAGTGAGCAGTTCCTGGAAAGTGAGCAGAAACAACAATTTTCCCCTTCAATGACAGAG AGGAAGCCACTCAGTCAGCAGGAGAGGGACAGCTGCATTTCTCCTTCTGAACCAGAAACCAAGGCAGAACAAAAAAGCAACTATTTTGAA GTTCCCTTGCCTTACTTTGAATACTTTAAATATATCTGTCCTGATAAAATCAACTCAATAGAGAAAAGATTTGGTGTAAACATTGAAATC CAGGAGAGTTCTCCAAATATGGTCTGTTTAGATTTCACCTCAAGTCGATCAGGTGACCTGGAAGCAGCTCGTGAGTCTTTTGCTAGTGAA TTTCAGAAGAACACAGAACCTCTGAAGCAAGAATGTGTCTCTTTAGCAGACAGTAAGCAGGCAAATAAATTCAAACAGGAATTGAATCAC CAGTTTACAAAGCTCCTTATAAAGGAGAAAGGAGGCGAATTAACTCTCCTTGGGACCCAAGATGACATTTCAGCTGCCAAACAAAAAATC TCTGAAGCTTTTGTCAAGATACCTGTGAAACTATTTGCTGCCAATTACATGATGAATGTAATTGAGGTTGATAGTGCCCACTATAAACTT TTAGAAACTGAATTACTACAGGAGATATCAGAGATCGAAAAAAGGTATGACATTTGCAGCAAGGTTTCTGAGAAAGGTCAGAAAACCTGC ATTCTGTTTGAATCCAAGGACAGGCAGGTAGATCTATCTGTGCATGCTTATGCAAGTTTCATCGATGCCTTTCAACATGCCTCATGTCAG TTGATGAGAGAAGTTCTTTTACTGAAGTCTTTGGGCAAGGAGAGAAAGCACTTACATCAGACCAAGTTTGCTGATGACTTTAGAAAAAGA CATCCAAATGTACACTTTGTGCTAAATCAAGAGTCAATGACTTTGACTGGTTTGCCAAATCACCTTGCAAAGGCGAAGCAGTATGTTCTA AAAGGAGGAGGAATGTCTTCATTGGCTGGAAAGAAATTGAAAGAGGGTCATGAAACACCGATGGACATTGATAGCGATGATTCCAAAGCA GCTTCTCCGCCACTCAAGGGCTCTGTGAGTTCTGAGGCCTCAGAACTGGACAAGAAGGAAAAGGGCATCTGTGTCATCTGTATGGACACC ATTAGTAACAAAAAAGTGCTACCAAAGTGCAAGCATGAATTCTGCGCCCCTTGTATCAACAAAGCCATGTCATATAAGCCAATCTGTCCC ACATGCCAGACTTCCTATGGTATTCAGAAAGGAAATCAGCCAGAGGGAAGCATGGTTTTCACTGTTTCAAGAGACTCACTTCCAGGTTAT GAGTCCTTTGGCACCATTGTGATTACTTATTCTATGAAAGCAGGCATACAAACAGAAGAACACCCAAACCCAGGAAAGAGATACCCTGGA ATACAGCGAACTGCATACTTGCCTGATAATAAGGAAGGAAGGAAGGTTTTGAAACTGCTTTATAGGGCCTTTGACCAAAAGCTGATTTTT ACAGTGGGGTACTCTCGCGTATTAGGAGTCTCAGATGTCATCACTTGGAATGATATTCACCACAAAACATCCCGGTTTGGAGGACCAGAA ATGTTGGAGATGCTTGATGCTGCAAAGAACAAGATGCGAGTGAAGATCAGCTATCTAATGATTGCCCTGACGGTGGTAGGATGCATCTTC ATGGTTATTGAGGGCAAGAAGGCTGCCCAAAGACACGAGACTTTAACAAGCTTGAACTTAGAAAAGAAAGCTCGTCTGAAAGAGGAAGCA GCTATGAAGGCCAAAACAGAGTAGCAGAGGTATCCGTGTTGGCTGGATTTTGAAAATCCAGGAATTATGTTATAACGTGCCTGTATTAAA AAGGATGTGGTATGAGGATCCATTTCATAAAGTATGATTTGCCCAAACCTGTACCATTTCCGTATTTCTGCTGTAGAAGTAGAAATAAAT TTTCTTAAATAATGACTGTGTTTTATTGTTTTGATCCAAGTCAAGTGTAGCCTCTCAGCCTTTTGGAGAAAAATGAAACAATTTATTTCA ACTTGAATACCAACTCTTCAGAGAAGCAGGTACCATATCTTACTTCTCTTCCCATTATCAAAGTAACCTCTCCACAGGCGAACTCATTTT GAGGAGATAAGGAAGGCCAGTTGAGTATATGGCCCAGGTGCAGCTTGGAGGGCCAGCCTTCTCAAATGTCCTGGTGTACTGGTGGGGAGG CTGCCCCCTAATAGAGCGAGCGCTGAGAAGCAGCCAGACCACATTCTATAATCTTTGAACATGCTGGCAGATGGAGGCTTTTAGGAGAAT AGTTTTTTAAACATTCTGACAAGGTCCAGCCAGGAATTTCTAAGGACTTAGAAATCCCAGCGTCATGGAGTTGCTTGGGGTCAACACAAG TCACCTCTTAGCTTCATGCCTTAGAATTCTCAACTGGCTGTTTTCTTACCACCTGAGAAACTAAATTTCTCAATTATAAAAATCTCATGT ATTATTACTGTTAGAAGTATTTGCACCTCATATATATCTCATTTACAGTTTGACCTTGGGATATCGAGTTGTCTCATATTTAGGTATGAA TACAGGTTTCTATTTTAAACATTGATTCTCACATGCTTCTTAATTATTCATGGATGACAAGCCATTATAGTAGTCCCTTCTTATCCACGG CTTTGTTTTCTGTGGTTTCAGTTACCTGAGGTCTGAAAATATTAAATGGAAAATTCCTATAACAATGCATAAGTTATAGATTACATACCA TTCAGTGTAGCATGATGAAATCTCTTGCCAATCTGTTCCATCCACCTGGGATGTGAATCATCCCTCTCTCTGGTGTATTCACACTGTATG TGCTAGCCGTCTCTGTTACGAGATCAGCTGTCGTGGAATCAGTGCTAGTGTTCAGGTAACTCTTATTTAATAATTGCCTCAAAGTGCATT GTGCTGTGCCTAATTTATAAAATAAACGGTATCATAGGTATGTATGTATACGAAAAACATAGTATATGCAGGGTTGGGTACTATCCACAG TTTCAGGCATCTACTGGGGGGTCTTGGAATATATCCCCAGAGGGTAAGGGGTGACTACTGTATACAGCCATGCCAACATGCTGGTTCTCA GAGAGTTATAAGAGAGATTAAAAATACATGCAGCTGTTTGGGCACTTTCTTCCCAAATAAAATACCCAGTAAAATGTAACATACTACCTA TAGCTTGATAATCTATCTGAGGATGTTTATGATGTCTGTCTATATACTTTAATTTGGCCTTAGCTTCTTTGTAGGTATCAAGTTATCACT TGCAAATTTATTTTCAAAAAGCATAGTTTCTTGTACATTTTAGAAGATTAGTTTTTCTTATTTTGGGACACCTACACCAATGTTTTAAAT GTGGATAAATTTTAATGTGGAATTTCTTTTAAAAATAATCCTCTTCATAGAGCAATACAACAAAAATGTGTTGTATCTTGGAGATTTACA TTATAAATGTAGTAACATTTATGAGCTTAAATTTAATGAAACCTTTTTTTTTTTTTTGAGACAGTCTTGCACTGTCGCCCAGGCTAGAGT GCAGGGCATGATCTCAGCTCACTGCAACCTCCGTCTCCCGGGTTCAAGCAATTCTCGTGCCTCAGCCTCCTCAGTAACTGGCATTATAGG TGTGCACCACCACACCTGGCTAATTTTTGTATTTTTCATAAAGACGGGGGTTTCACCATGTTGGCCAGGCTGGTCTCAAACTCCTGACCT CAGGTGATCAGCCTCCCTCGGCCTCCCAAAGTGCTGGGATTACAGGCATGAGCCATGGTGCCTGGCCAAACGTTTTTATTACAAATATTA CATTAAACTAAAGCTCACAACTGTTATTATGCATCAGGAATTAACAGTCATTTCTCATTTGTGTTATTCTCCTATTTGATAACATATAGT GACTATCTCTTTATGGGTTAAATATCAGTTTCTTGGCCAGAATTCAAGATCTGCCCCTAACTAATCCTACTTCTCCCAGTACCCTTACAG CACTCCTACCCTACTAGTAACTCTGATATCTCATAGTCCTCAGATTTATCAAGCAAACGAAACTGAATGAGCACTACTATAAGGTACCAC AAACTACAAAAGGAGACAGTTTTGCCCTTGTTGCTTTCAGTTAAATTATTGTGCTATTTTTTGCTTAATTTCTAGGTTTTTTTTGCGTGT TTATCATGTCTCCGTAGGTAAAGTGGGCAGAAATCTTTTTCTACTTTACATTTCCTGTAAGTGCTGTTTGTTTGAATAAAGTTAATGTGT >24413_24413_2_DTX3L-FAM162A_DTX3L_chr3_122289519_ENST00000296161_FAM162A_chr3_122126128_ENST00000477892_length(amino acids)=784AA_BP=718 MASHLRPPSPLLVRVYKSGPRVRRKLESYFQSSKSSGGGECTVSTQEHEAPGTFRVEFSERAAKERVLKKGEHQILVDEKPVPIFLVPTE NSIKKNTRPQISSLTQSQAETPSGDMHQHEGHIPNAVDSCLQKIFLTVTADLNCNLFSKEQRAYITTLCPSIRKMEGHDGIEKVCGDFQD IERIHQFLSEQFLESEQKQQFSPSMTERKPLSQQERDSCISPSEPETKAEQKSNYFEVPLPYFEYFKYICPDKINSIEKRFGVNIEIQES SPNMVCLDFTSSRSGDLEAARESFASEFQKNTEPLKQECVSLADSKQANKFKQELNHQFTKLLIKEKGGELTLLGTQDDISAAKQKISEA FVKIPVKLFAANYMMNVIEVDSAHYKLLETELLQEISEIEKRYDICSKVSEKGQKTCILFESKDRQVDLSVHAYASFIDAFQHASCQLMR EVLLLKSLGKERKHLHQTKFADDFRKRHPNVHFVLNQESMTLTGLPNHLAKAKQYVLKGGGMSSLAGKKLKEGHETPMDIDSDDSKAASP PLKGSVSSEASELDKKEKGICVICMDTISNKKVLPKCKHEFCAPCINKAMSYKPICPTCQTSYGIQKGNQPEGSMVFTVSRDSLPGYESF GTIVITYSMKAGIQTEEHPNPGKRYPGIQRTAYLPDNKEGRKVLKLLYRAFDQKLIFTVGYSRVLGVSDVITWNDIHHKTSRFGGPEMLE -------------------------------------------------------------- >24413_24413_3_DTX3L-FAM162A_DTX3L_chr3_122289519_ENST00000383661_FAM162A_chr3_122126128_ENST00000232125_length(transcript)=905nt_BP=617nt ATGGCCTCCCACCTGCGCCCGCCGTCCCCGCTCCTCGTGCGGGTGTACAAGTCCGGCCCCCGAGTACGAAGGAAGCTGGAGAGCTACTTC CAGAGCTCTAAGTCCTCGGGCGGCGGGGAGTGCACGGTCAGCACCCAGGAACACGAAGCCCCGGGCACCTTCCGGGTGGAGTTCAGTGAA AGGGCAGCTAAGGAGAGAGTGTTGAAAAAAGGAGAGCACCAAATACTTGTTGACGAAAAACCTGTGCCCATTTTCCTGGTACCCACTGAA AATTCAATAAAGAAGAACACGAGACCTCAAATTTCTTCACTGACACAATCACAAGCAGAAACACCGTCTGGTGATATGCATCAACATGAA GGACATATTCCTAATGCTGTGGATTCCTGTCTCCAAAAGGAAGAACACCCAAACCCAGGAAAGAGATACCCTGGAATACAGCGAACTGCA TACTTGCCTGATAATAAGGAAGGAAGGAAGGTTTTGAAACTGCTTTATAGGGCCTTTGACCAAAAGCTGATTTTTACAGTGGGGTACTCT CGCGTATTAGGAGTCTCAGATGTCATCACTTGGAATGATATTCACCACAAAACATCCCGGTTTGGAGGACCAGAAATGTTGGAGATGCTT GATGCTGCAAAGAACAAGATGCGAGTGAAGATCAGCTATCTAATGATTGCCCTGACGGTGGTAGGATGCATCTTCATGGTTATTGAGGGC AAGAAGGCTGCCCAAAGACACGAGACTTTAACAAGCTTGAACTTAGAAAAGAAAGCTCGTCTGAAAGAGGAAGCAGCTATGAAGGCCAAA ACAGAGTAGCAGAGGTATCCGTGTTGGCTGGATTTTGAAAATCCAGGAATTATGTTATAACGTGCCTGTATTAAAAAGGATGTGGTATGA >24413_24413_3_DTX3L-FAM162A_DTX3L_chr3_122289519_ENST00000383661_FAM162A_chr3_122126128_ENST00000232125_length(amino acids)=272AA_BP=206 MASHLRPPSPLLVRVYKSGPRVRRKLESYFQSSKSSGGGECTVSTQEHEAPGTFRVEFSERAAKERVLKKGEHQILVDEKPVPIFLVPTE NSIKKNTRPQISSLTQSQAETPSGDMHQHEGHIPNAVDSCLQKEEHPNPGKRYPGIQRTAYLPDNKEGRKVLKLLYRAFDQKLIFTVGYS RVLGVSDVITWNDIHHKTSRFGGPEMLEMLDAAKNKMRVKISYLMIALTVVGCIFMVIEGKKAAQRHETLTSLNLEKKARLKEEAAMKAK -------------------------------------------------------------- >24413_24413_4_DTX3L-FAM162A_DTX3L_chr3_122289519_ENST00000383661_FAM162A_chr3_122126128_ENST00000477892_length(transcript)=3322nt_BP=617nt ATGGCCTCCCACCTGCGCCCGCCGTCCCCGCTCCTCGTGCGGGTGTACAAGTCCGGCCCCCGAGTACGAAGGAAGCTGGAGAGCTACTTC CAGAGCTCTAAGTCCTCGGGCGGCGGGGAGTGCACGGTCAGCACCCAGGAACACGAAGCCCCGGGCACCTTCCGGGTGGAGTTCAGTGAA AGGGCAGCTAAGGAGAGAGTGTTGAAAAAAGGAGAGCACCAAATACTTGTTGACGAAAAACCTGTGCCCATTTTCCTGGTACCCACTGAA AATTCAATAAAGAAGAACACGAGACCTCAAATTTCTTCACTGACACAATCACAAGCAGAAACACCGTCTGGTGATATGCATCAACATGAA GGACATATTCCTAATGCTGTGGATTCCTGTCTCCAAAAGGAAGAACACCCAAACCCAGGAAAGAGATACCCTGGAATACAGCGAACTGCA TACTTGCCTGATAATAAGGAAGGAAGGAAGGTTTTGAAACTGCTTTATAGGGCCTTTGACCAAAAGCTGATTTTTACAGTGGGGTACTCT CGCGTATTAGGAGTCTCAGATGTCATCACTTGGAATGATATTCACCACAAAACATCCCGGTTTGGAGGACCAGAAATGTTGGAGATGCTT GATGCTGCAAAGAACAAGATGCGAGTGAAGATCAGCTATCTAATGATTGCCCTGACGGTGGTAGGATGCATCTTCATGGTTATTGAGGGC AAGAAGGCTGCCCAAAGACACGAGACTTTAACAAGCTTGAACTTAGAAAAGAAAGCTCGTCTGAAAGAGGAAGCAGCTATGAAGGCCAAA ACAGAGTAGCAGAGGTATCCGTGTTGGCTGGATTTTGAAAATCCAGGAATTATGTTATAACGTGCCTGTATTAAAAAGGATGTGGTATGA GGATCCATTTCATAAAGTATGATTTGCCCAAACCTGTACCATTTCCGTATTTCTGCTGTAGAAGTAGAAATAAATTTTCTTAAATAATGA CTGTGTTTTATTGTTTTGATCCAAGTCAAGTGTAGCCTCTCAGCCTTTTGGAGAAAAATGAAACAATTTATTTCAACTTGAATACCAACT CTTCAGAGAAGCAGGTACCATATCTTACTTCTCTTCCCATTATCAAAGTAACCTCTCCACAGGCGAACTCATTTTGAGGAGATAAGGAAG GCCAGTTGAGTATATGGCCCAGGTGCAGCTTGGAGGGCCAGCCTTCTCAAATGTCCTGGTGTACTGGTGGGGAGGCTGCCCCCTAATAGA GCGAGCGCTGAGAAGCAGCCAGACCACATTCTATAATCTTTGAACATGCTGGCAGATGGAGGCTTTTAGGAGAATAGTTTTTTAAACATT CTGACAAGGTCCAGCCAGGAATTTCTAAGGACTTAGAAATCCCAGCGTCATGGAGTTGCTTGGGGTCAACACAAGTCACCTCTTAGCTTC ATGCCTTAGAATTCTCAACTGGCTGTTTTCTTACCACCTGAGAAACTAAATTTCTCAATTATAAAAATCTCATGTATTATTACTGTTAGA AGTATTTGCACCTCATATATATCTCATTTACAGTTTGACCTTGGGATATCGAGTTGTCTCATATTTAGGTATGAATACAGGTTTCTATTT TAAACATTGATTCTCACATGCTTCTTAATTATTCATGGATGACAAGCCATTATAGTAGTCCCTTCTTATCCACGGCTTTGTTTTCTGTGG TTTCAGTTACCTGAGGTCTGAAAATATTAAATGGAAAATTCCTATAACAATGCATAAGTTATAGATTACATACCATTCAGTGTAGCATGA TGAAATCTCTTGCCAATCTGTTCCATCCACCTGGGATGTGAATCATCCCTCTCTCTGGTGTATTCACACTGTATGTGCTAGCCGTCTCTG TTACGAGATCAGCTGTCGTGGAATCAGTGCTAGTGTTCAGGTAACTCTTATTTAATAATTGCCTCAAAGTGCATTGTGCTGTGCCTAATT TATAAAATAAACGGTATCATAGGTATGTATGTATACGAAAAACATAGTATATGCAGGGTTGGGTACTATCCACAGTTTCAGGCATCTACT GGGGGGTCTTGGAATATATCCCCAGAGGGTAAGGGGTGACTACTGTATACAGCCATGCCAACATGCTGGTTCTCAGAGAGTTATAAGAGA GATTAAAAATACATGCAGCTGTTTGGGCACTTTCTTCCCAAATAAAATACCCAGTAAAATGTAACATACTACCTATAGCTTGATAATCTA TCTGAGGATGTTTATGATGTCTGTCTATATACTTTAATTTGGCCTTAGCTTCTTTGTAGGTATCAAGTTATCACTTGCAAATTTATTTTC AAAAAGCATAGTTTCTTGTACATTTTAGAAGATTAGTTTTTCTTATTTTGGGACACCTACACCAATGTTTTAAATGTGGATAAATTTTAA TGTGGAATTTCTTTTAAAAATAATCCTCTTCATAGAGCAATACAACAAAAATGTGTTGTATCTTGGAGATTTACATTATAAATGTAGTAA CATTTATGAGCTTAAATTTAATGAAACCTTTTTTTTTTTTTTGAGACAGTCTTGCACTGTCGCCCAGGCTAGAGTGCAGGGCATGATCTC AGCTCACTGCAACCTCCGTCTCCCGGGTTCAAGCAATTCTCGTGCCTCAGCCTCCTCAGTAACTGGCATTATAGGTGTGCACCACCACAC CTGGCTAATTTTTGTATTTTTCATAAAGACGGGGGTTTCACCATGTTGGCCAGGCTGGTCTCAAACTCCTGACCTCAGGTGATCAGCCTC CCTCGGCCTCCCAAAGTGCTGGGATTACAGGCATGAGCCATGGTGCCTGGCCAAACGTTTTTATTACAAATATTACATTAAACTAAAGCT CACAACTGTTATTATGCATCAGGAATTAACAGTCATTTCTCATTTGTGTTATTCTCCTATTTGATAACATATAGTGACTATCTCTTTATG GGTTAAATATCAGTTTCTTGGCCAGAATTCAAGATCTGCCCCTAACTAATCCTACTTCTCCCAGTACCCTTACAGCACTCCTACCCTACT AGTAACTCTGATATCTCATAGTCCTCAGATTTATCAAGCAAACGAAACTGAATGAGCACTACTATAAGGTACCACAAACTACAAAAGGAG ACAGTTTTGCCCTTGTTGCTTTCAGTTAAATTATTGTGCTATTTTTTGCTTAATTTCTAGGTTTTTTTTGCGTGTTTATCATGTCTCCGT >24413_24413_4_DTX3L-FAM162A_DTX3L_chr3_122289519_ENST00000383661_FAM162A_chr3_122126128_ENST00000477892_length(amino acids)=272AA_BP=206 MASHLRPPSPLLVRVYKSGPRVRRKLESYFQSSKSSGGGECTVSTQEHEAPGTFRVEFSERAAKERVLKKGEHQILVDEKPVPIFLVPTE NSIKKNTRPQISSLTQSQAETPSGDMHQHEGHIPNAVDSCLQKEEHPNPGKRYPGIQRTAYLPDNKEGRKVLKLLYRAFDQKLIFTVGYS RVLGVSDVITWNDIHHKTSRFGGPEMLEMLDAAKNKMRVKISYLMIALTVVGCIFMVIEGKKAAQRHETLTSLNLEKKARLKEEAAMKAK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for DTX3L-FAM162A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for DTX3L-FAM162A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for DTX3L-FAM162A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
