
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:DYNC1LI1-GADL1 (FusionGDB2 ID:24622) |
Fusion Gene Summary for DYNC1LI1-GADL1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: DYNC1LI1-GADL1 | Fusion gene ID: 24622 | Hgene | Tgene | Gene symbol | DYNC1LI1 | GADL1 | Gene ID | 51143 | 339896 |
| Gene name | dynein cytoplasmic 1 light intermediate chain 1 | glutamate decarboxylase like 1 | |
| Synonyms | DLC-A|DNCLI1|LIC1 | ADC|CSADC|HuADC|HuCSADC | |
| Cytomap | 3p22.3 | 3p24.1-p23 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cytoplasmic dynein 1 light intermediate chain 1dynein light chain Adynein light intermediate chain 1, cytosolicdynein, cytoplasmic, light intermediate polypeptide 1 | acidic amino acid decarboxylase GADL1aspartate 1-decarboxylasecysteine sulfinic acid decarboxylaseglutamate decarboxylase-like protein 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9Y6G9 | Q6ZQY3 | |
| Ensembl transtripts involved in fusion gene | ENST00000273130, ENST00000432458, | ENST00000454381, ENST00000498387, ENST00000282538, | |
| Fusion gene scores | * DoF score | 5 X 6 X 4=120 | 7 X 7 X 6=294 |
| # samples | 6 | 7 | |
| ** MAII score | log2(6/120*10)=-1 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/294*10)=-2.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: DYNC1LI1 [Title/Abstract] AND GADL1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | DYNC1LI1(32611838)-GADL1(30903257), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across DYNC1LI1 (5'-gene) Fusion gene breakpoints across DYNC1LI1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
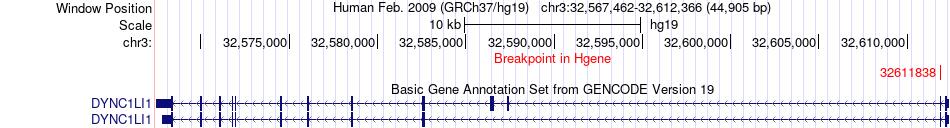 |
 Fusion gene breakpoints across GADL1 (3'-gene) Fusion gene breakpoints across GADL1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
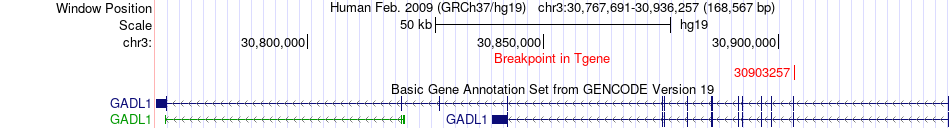 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | GBM | TCGA-14-0790-01B | DYNC1LI1 | chr3 | 32611838 | - | GADL1 | chr3 | 30903257 | - |
Top |
Fusion Gene ORF analysis for DYNC1LI1-GADL1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000273130 | ENST00000454381 | DYNC1LI1 | chr3 | 32611838 | - | GADL1 | chr3 | 30903257 | - |
| 5CDS-intron | ENST00000273130 | ENST00000498387 | DYNC1LI1 | chr3 | 32611838 | - | GADL1 | chr3 | 30903257 | - |
| 5CDS-intron | ENST00000432458 | ENST00000454381 | DYNC1LI1 | chr3 | 32611838 | - | GADL1 | chr3 | 30903257 | - |
| 5CDS-intron | ENST00000432458 | ENST00000498387 | DYNC1LI1 | chr3 | 32611838 | - | GADL1 | chr3 | 30903257 | - |
| In-frame | ENST00000273130 | ENST00000282538 | DYNC1LI1 | chr3 | 32611838 | - | GADL1 | chr3 | 30903257 | - |
| In-frame | ENST00000432458 | ENST00000282538 | DYNC1LI1 | chr3 | 32611838 | - | GADL1 | chr3 | 30903257 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000273130 | DYNC1LI1 | chr3 | 32611838 | - | ENST00000282538 | GADL1 | chr3 | 30903257 | - | 3895 | 324 | 104 | 1852 | 582 |
| ENST00000432458 | DYNC1LI1 | chr3 | 32611838 | - | ENST00000282538 | GADL1 | chr3 | 30903257 | - | 3879 | 308 | 88 | 1836 | 582 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000273130 | ENST00000282538 | DYNC1LI1 | chr3 | 32611838 | - | GADL1 | chr3 | 30903257 | - | 0.000357809 | 0.99964213 |
| ENST00000432458 | ENST00000282538 | DYNC1LI1 | chr3 | 32611838 | - | GADL1 | chr3 | 30903257 | - | 0.000362424 | 0.9996376 |
Top |
Fusion Genomic Features for DYNC1LI1-GADL1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
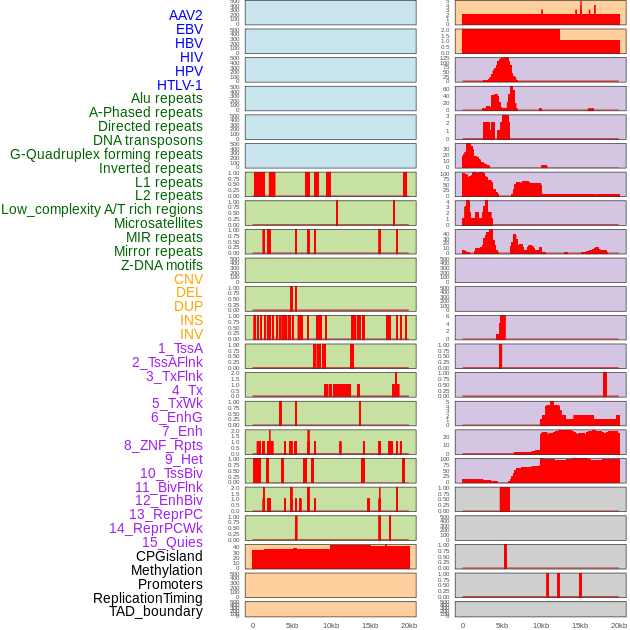 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for DYNC1LI1-GADL1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:32611838/chr3:30903257) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| DYNC1LI1 | GADL1 |
| FUNCTION: Acts as one of several non-catalytic accessory components of the cytoplasmic dynein 1 complex that are thought to be involved in linking dynein to cargos and to adapter proteins that regulate dynein function. Cytoplasmic dynein 1 acts as a motor for the intracellular retrograde motility of vesicles and organelles along microtubules. May play a role in binding dynein to membranous organelles or chromosomes. Probably involved in the microtubule-dependent transport of pericentrin. Is required for progress through the spindle assembly checkpoint. The phosphorylated form appears to be involved in the selective removal of MAD1L1 and MAD1L2 but not BUB1B from kinetochores. {ECO:0000269|PubMed:19229290}. | FUNCTION: May catalyze the decarboxylation of L-aspartate, 3-sulfino-L-alanine (cysteine sulfinic acid), and L-cysteate to beta-alanine, hypotaurine and taurine, respectively. Does not exhibit any decarboxylation activity toward glutamate. {ECO:0000269|PubMed:23038267}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DYNC1LI1 | chr3:32611838 | chr3:30903257 | ENST00000273130 | - | 2 | 13 | 74_81 | 73 | 524.0 | Nucleotide binding | ATP |
Top |
Fusion Gene Sequence for DYNC1LI1-GADL1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >24622_24622_1_DYNC1LI1-GADL1_DYNC1LI1_chr3_32611838_ENST00000273130_GADL1_chr3_30903257_ENST00000282538_length(transcript)=3895nt_BP=324nt GTGCGCTGAACGCCCCATATCCGGGTTCCCGCCGCCTCCACCGCCACCGCCTCAGCCGCCTCGCACATTTAGTCTTGCCGGGAGTGGTGT GATTCCCGACCAAGATGGCGGCCGTGGGGCGAGTCGGCTCCTTCGGTTCTTCTCCGCCGGGATTATCCTCGACTTACACTGGCGGCCCCT TGGGCAACGAGATAGCGTCGGGCAACGGTGGCGCCGCGGCAGGCGACGACGAGGACGGGCAGAACCTTTGGTCCTGCATCCTCAGCGAGG TCTCCACCCGCTCGCGCTCCAAGCTCCCTGCGGGGAAGAACGTGCTACTGCTGGGAGATATTGATCAACAAGAGATGATTCCAAGTAAGA AGAATGCTGTTCTTGTGGATGGGGTTGTGCTGAATGGTCCTACAACAGATGCAAAAGCTGGAGAAAAATTTGTTGAAGAGGCCTGTAGGC TAATAATGGAAGAGGTGGTTTTGAAAGCTACAGATGTCAATGAGAAGGTGTGTGAATGGAGGCCTCCTGAACAACTGAAACAGCTTCTTG ATTTGGAGATGAGAGACTCAGGCGAGCCACCCCATAAACTATTGGAACTCTGTCGGGATGTCATACACTACAGTGTCAAAACTAACCACC CAAGATTTTTCAACCAATTGTATGCTGGACTTGATTATTACTCCTTGGTGGCCCGATTTATGACCGAAGCATTGAATCCAAGTGTTTATA CGTATGAGGTGTCCCCAGTGTTTCTGTTAGTGGAAGAAGCGGTTCTGAAGAAAATGATTGAATTTATTGGCTGGAAAGAAGGGGATGGAA TATTTAACCCAGGTGGCTCAGTGTCCAATATGTATGCAATGAATTTAGCTAGATACAAATATTGTCCTGATATTAAGGAAAAGGGGCTGT CTGGTTCGCCAAGATTAATCCTTTTCACATCTGCAGAGTGTCATTACTCTATGAAGAAGGCAGCCTCTTTTCTTGGGATTGGCACTGAGA ATGTTTGCTTTGTGGAAACAGATGGAAGAGGTAAAATGATACCTGAGGAACTGGAGAAGCAAGTCTGGCAAGCCAGAAAAGAGGGGGCAG CACCGTTTCTTGTCTGTGCCACTTCTGGTACAACTGTGTTGGGAGCTTTTGACCCTCTGGATGAAATAGCAGACATCTGCGAGAGGCACA GCCTCTGGCTTCATGTAGATGCTTCTTGGGGTGGCTCAGCTTTGATGTCGAGGAAGCACCGCAAGCTTCTGCATGGCATCCACAGGGCTG ACTCTGTGGCCTGGAACCCACACAAGATGCTGATGGCTGGGATCCAGTGCTGTGCTCTCCTTGTGAAAGACAAATCTGATCTTCTTAAAA AATGCTACTCTGCCAAGGCATCTTACCTCTTCCAGCAGGATAAATTCTATGATGTGAGCTATGACACAGGAGACAAGTCTATCCAGTGTA GCAGAAGACCAGATGCATTCAAGTTCTGGATGACCTGGAAGGCCCTGGGTACATTAGGCCTTGAAGAAAGAGTTAATCGTGCTCTTGCTT TATCTAGGTACCTAGTAGATGAAATCAAGAAAAGAGAAGGATTCAAGTTACTGATGGAACCTGAATATGCCAATATTTGCTTTTGGTACA TTCCACCGAGCCTCAGAGAGATGGAAGAAGGACCCGAGTTCTGGGCAAAACTTAATTTGGTGGCCCCAGCCATTAAGGAGAGGATGATGA AGAAGGGAAGCTTGATGCTGGGCTACCAGCCGCACCGGGGAAAGGTCAACTTCTTCCGCCAGGTGGTGATCAGCCCTCAAGTGAGCCGGG AGGACATGGACTTCCTCCTGGATGAGATAGACTTACTGGGTAAAGACATGTAGCTGTGGCTTTGGTCCCCCAGAGGCATAGATCCTATCC TGGGAGAGTTTAGATCCAGAACATCTTGGAGATACACAGTAGATTGCAGCCCTTCTGATGAGAAATAGGGAATACTCCCAGTCCAGGCCC AGCAAAACCAAAATGCTAAGCAATGAATATTAAGGACTCTCTAGCTGCCTGGGCATTACTGTTGCTAAAAGAAGAAAGTTTAAAAAAAAA AATGATTTTCTCAAGGAATGCCCCTGGAACACAGCTCTGAAGAGAGTTTAGTAAGTACCATGTAGGTTCTGGATTCTAAGCTTACATTGC TCTTTAAAGAACTTATAAACTAACGGTTTAAAGCAGTGGTTCTCAAAGTGTGGTCCCTGGACTATCAGCATCAAAGCATCACCTGGGAAC TTGCTAAAAATGCAGATTCTCAGGCTTTCTCTAGACCAACTGGATCAGAAGCTCTGGGGGTGAGGCCCAGTATTCTGTGTTTTAACAAGC CCGTCAGGGAATTCTGATGCACAGTAAAATCCGAGAAACACTGGTTTAAGAAAAACCTTGTAATGATCGAATACCCACTCTGATGTTTTG CCAGCAAAGGGATATCTAATATTTCAGAAGCCTCTGAGCCAGTCTTTGAAAAAATACAACTATGGCATCTGCAGCACAAATATTTAAGGA CATCAGAAGCATGTCAAAGCTATTTTTAAAGAGAGAAACTGTATAAGATGTTTACTTCATAGAGATTTATGTTTTATGCAGGCTGAATGT TTATCTCAAAAGTTAAAATTATCCATTCTCAAAAGTTAAAATTATATATATATATATATATACACACACACACATATATATATATATATA ATTCAAAGCACAATAATTGAAAGCACAATAATTGACAGAAAAATACAGGTTCTATTAATAAATTAATAAACTGTTGGTCTTCAAAATAGA AATGCATGTAATATCCATATTAGTTTTTTCTTGGTAGACAACTGGAAGGTTTTCTTTTTTTTCGTCTATGACTAATTTTCTTTATTCAAG ATACCTGAACTGGGGTGCTTTTTAAGAAAAATTTGGGAAATATATATGTTTCTGTGATATACATATATACATATATATGTATATATATAC ACACACATACATATGTGTGTGTATAGTATATATATATATACACACATATATGTTTCTGTGTTCCTCTTTTAGCTTGAGGGGCTTGTTTAT TATCTTGCTCTGTGCCTCATAGGGAATAAACACAATGAAGTCCAGGGTTGTACAACATTCCCTTTCCTAAGCTTTGAAATGTCAGTATAG ATTATTAAGTGGTTTATATTACAGAATCTGGGATTCAGCAGACTTTCAGTGTAAATGCTTCCTCCATTTCTCCTGAGAGTGGGTGATTTT AATTCTATCTCTGACCCTGGTCCTAGGTTTCTAGGAGAGTTTTGTTTAACTAAGAAATTGACAGAATTCATAGGTGTGGGTGTAGAGTTC ACCAAGATAAGATTATGAATATAATTAAAGGTCTGCATTAAAAGGTGAATGATTGAAGAGTGTTAAAGCATTAGACTTAGCACATTCAAT AACCTTTTCGTACTCCATTGTTAACCAATGTCATTTAAATTTTGAGTACTATTTGCTTTTATTGCTTATTTTCATTTTAGTGTGCACAGT TTCTCGGTATCTCTATTGGTCAAAGAATATTAAATCTGTCTCTGAATTACTTCAAATTCTCAGGTGAAACCTATTGGTGTGTGTGTGTGT GTGTGTGTGTTTATTTTGCATTTCTTGTTGCCTTTTTGTTTTAATGTCTACATAAAATATTTCTAAAATTGATGTTTGTAACAATTTGGG TTTCATGAAACAAAAAGGAACATTACTATACTTAGTGTTGTTGACTTTTCTTTTCCTGTCATCTCCTCTTTACTGGATTGTACCAATACA TTTTAGAAGTGAACTGGACTTGGTTGGCATTTTAGTTTAATGACTGAAAAAGTAGGTTGAAAGCTCTCTGTATTTTAGTTAACACCTTGA >24622_24622_1_DYNC1LI1-GADL1_DYNC1LI1_chr3_32611838_ENST00000273130_GADL1_chr3_30903257_ENST00000282538_length(amino acids)=582AA_BP=70 MAAVGRVGSFGSSPPGLSSTYTGGPLGNEIASGNGGAAAGDDEDGQNLWSCILSEVSTRSRSKLPAGKNVLLLGDIDQQEMIPSKKNAVL VDGVVLNGPTTDAKAGEKFVEEACRLIMEEVVLKATDVNEKVCEWRPPEQLKQLLDLEMRDSGEPPHKLLELCRDVIHYSVKTNHPRFFN QLYAGLDYYSLVARFMTEALNPSVYTYEVSPVFLLVEEAVLKKMIEFIGWKEGDGIFNPGGSVSNMYAMNLARYKYCPDIKEKGLSGSPR LILFTSAECHYSMKKAASFLGIGTENVCFVETDGRGKMIPEELEKQVWQARKEGAAPFLVCATSGTTVLGAFDPLDEIADICERHSLWLH VDASWGGSALMSRKHRKLLHGIHRADSVAWNPHKMLMAGIQCCALLVKDKSDLLKKCYSAKASYLFQQDKFYDVSYDTGDKSIQCSRRPD AFKFWMTWKALGTLGLEERVNRALALSRYLVDEIKKREGFKLLMEPEYANICFWYIPPSLREMEEGPEFWAKLNLVAPAIKERMMKKGSL -------------------------------------------------------------- >24622_24622_2_DYNC1LI1-GADL1_DYNC1LI1_chr3_32611838_ENST00000432458_GADL1_chr3_30903257_ENST00000282538_length(transcript)=3879nt_BP=308nt ATATCCGGGTTCCCGCCGCCTCCACCGCCACCGCCTCAGCCGCCTCGCACATTTAGTCTTGCCGGGAGTGGTGTGATTCCCGACCAAGAT GGCGGCCGTGGGGCGAGTCGGCTCCTTCGGTTCTTCTCCGCCGGGATTATCCTCGACTTACACTGGCGGCCCCTTGGGCAACGAGATAGC GTCGGGCAACGGTGGCGCCGCGGCAGGCGACGACGAGGACGGGCAGAACCTTTGGTCCTGCATCCTCAGCGAGGTCTCCACCCGCTCGCG CTCCAAGCTCCCTGCGGGGAAGAACGTGCTACTGCTGGGAGATATTGATCAACAAGAGATGATTCCAAGTAAGAAGAATGCTGTTCTTGT GGATGGGGTTGTGCTGAATGGTCCTACAACAGATGCAAAAGCTGGAGAAAAATTTGTTGAAGAGGCCTGTAGGCTAATAATGGAAGAGGT GGTTTTGAAAGCTACAGATGTCAATGAGAAGGTGTGTGAATGGAGGCCTCCTGAACAACTGAAACAGCTTCTTGATTTGGAGATGAGAGA CTCAGGCGAGCCACCCCATAAACTATTGGAACTCTGTCGGGATGTCATACACTACAGTGTCAAAACTAACCACCCAAGATTTTTCAACCA ATTGTATGCTGGACTTGATTATTACTCCTTGGTGGCCCGATTTATGACCGAAGCATTGAATCCAAGTGTTTATACGTATGAGGTGTCCCC AGTGTTTCTGTTAGTGGAAGAAGCGGTTCTGAAGAAAATGATTGAATTTATTGGCTGGAAAGAAGGGGATGGAATATTTAACCCAGGTGG CTCAGTGTCCAATATGTATGCAATGAATTTAGCTAGATACAAATATTGTCCTGATATTAAGGAAAAGGGGCTGTCTGGTTCGCCAAGATT AATCCTTTTCACATCTGCAGAGTGTCATTACTCTATGAAGAAGGCAGCCTCTTTTCTTGGGATTGGCACTGAGAATGTTTGCTTTGTGGA AACAGATGGAAGAGGTAAAATGATACCTGAGGAACTGGAGAAGCAAGTCTGGCAAGCCAGAAAAGAGGGGGCAGCACCGTTTCTTGTCTG TGCCACTTCTGGTACAACTGTGTTGGGAGCTTTTGACCCTCTGGATGAAATAGCAGACATCTGCGAGAGGCACAGCCTCTGGCTTCATGT AGATGCTTCTTGGGGTGGCTCAGCTTTGATGTCGAGGAAGCACCGCAAGCTTCTGCATGGCATCCACAGGGCTGACTCTGTGGCCTGGAA CCCACACAAGATGCTGATGGCTGGGATCCAGTGCTGTGCTCTCCTTGTGAAAGACAAATCTGATCTTCTTAAAAAATGCTACTCTGCCAA GGCATCTTACCTCTTCCAGCAGGATAAATTCTATGATGTGAGCTATGACACAGGAGACAAGTCTATCCAGTGTAGCAGAAGACCAGATGC ATTCAAGTTCTGGATGACCTGGAAGGCCCTGGGTACATTAGGCCTTGAAGAAAGAGTTAATCGTGCTCTTGCTTTATCTAGGTACCTAGT AGATGAAATCAAGAAAAGAGAAGGATTCAAGTTACTGATGGAACCTGAATATGCCAATATTTGCTTTTGGTACATTCCACCGAGCCTCAG AGAGATGGAAGAAGGACCCGAGTTCTGGGCAAAACTTAATTTGGTGGCCCCAGCCATTAAGGAGAGGATGATGAAGAAGGGAAGCTTGAT GCTGGGCTACCAGCCGCACCGGGGAAAGGTCAACTTCTTCCGCCAGGTGGTGATCAGCCCTCAAGTGAGCCGGGAGGACATGGACTTCCT CCTGGATGAGATAGACTTACTGGGTAAAGACATGTAGCTGTGGCTTTGGTCCCCCAGAGGCATAGATCCTATCCTGGGAGAGTTTAGATC CAGAACATCTTGGAGATACACAGTAGATTGCAGCCCTTCTGATGAGAAATAGGGAATACTCCCAGTCCAGGCCCAGCAAAACCAAAATGC TAAGCAATGAATATTAAGGACTCTCTAGCTGCCTGGGCATTACTGTTGCTAAAAGAAGAAAGTTTAAAAAAAAAAATGATTTTCTCAAGG AATGCCCCTGGAACACAGCTCTGAAGAGAGTTTAGTAAGTACCATGTAGGTTCTGGATTCTAAGCTTACATTGCTCTTTAAAGAACTTAT AAACTAACGGTTTAAAGCAGTGGTTCTCAAAGTGTGGTCCCTGGACTATCAGCATCAAAGCATCACCTGGGAACTTGCTAAAAATGCAGA TTCTCAGGCTTTCTCTAGACCAACTGGATCAGAAGCTCTGGGGGTGAGGCCCAGTATTCTGTGTTTTAACAAGCCCGTCAGGGAATTCTG ATGCACAGTAAAATCCGAGAAACACTGGTTTAAGAAAAACCTTGTAATGATCGAATACCCACTCTGATGTTTTGCCAGCAAAGGGATATC TAATATTTCAGAAGCCTCTGAGCCAGTCTTTGAAAAAATACAACTATGGCATCTGCAGCACAAATATTTAAGGACATCAGAAGCATGTCA AAGCTATTTTTAAAGAGAGAAACTGTATAAGATGTTTACTTCATAGAGATTTATGTTTTATGCAGGCTGAATGTTTATCTCAAAAGTTAA AATTATCCATTCTCAAAAGTTAAAATTATATATATATATATATATACACACACACACATATATATATATATATAATTCAAAGCACAATAA TTGAAAGCACAATAATTGACAGAAAAATACAGGTTCTATTAATAAATTAATAAACTGTTGGTCTTCAAAATAGAAATGCATGTAATATCC ATATTAGTTTTTTCTTGGTAGACAACTGGAAGGTTTTCTTTTTTTTCGTCTATGACTAATTTTCTTTATTCAAGATACCTGAACTGGGGT GCTTTTTAAGAAAAATTTGGGAAATATATATGTTTCTGTGATATACATATATACATATATATGTATATATATACACACACATACATATGT GTGTGTATAGTATATATATATATACACACATATATGTTTCTGTGTTCCTCTTTTAGCTTGAGGGGCTTGTTTATTATCTTGCTCTGTGCC TCATAGGGAATAAACACAATGAAGTCCAGGGTTGTACAACATTCCCTTTCCTAAGCTTTGAAATGTCAGTATAGATTATTAAGTGGTTTA TATTACAGAATCTGGGATTCAGCAGACTTTCAGTGTAAATGCTTCCTCCATTTCTCCTGAGAGTGGGTGATTTTAATTCTATCTCTGACC CTGGTCCTAGGTTTCTAGGAGAGTTTTGTTTAACTAAGAAATTGACAGAATTCATAGGTGTGGGTGTAGAGTTCACCAAGATAAGATTAT GAATATAATTAAAGGTCTGCATTAAAAGGTGAATGATTGAAGAGTGTTAAAGCATTAGACTTAGCACATTCAATAACCTTTTCGTACTCC ATTGTTAACCAATGTCATTTAAATTTTGAGTACTATTTGCTTTTATTGCTTATTTTCATTTTAGTGTGCACAGTTTCTCGGTATCTCTAT TGGTCAAAGAATATTAAATCTGTCTCTGAATTACTTCAAATTCTCAGGTGAAACCTATTGGTGTGTGTGTGTGTGTGTGTGTGTTTATTT TGCATTTCTTGTTGCCTTTTTGTTTTAATGTCTACATAAAATATTTCTAAAATTGATGTTTGTAACAATTTGGGTTTCATGAAACAAAAA GGAACATTACTATACTTAGTGTTGTTGACTTTTCTTTTCCTGTCATCTCCTCTTTACTGGATTGTACCAATACATTTTAGAAGTGAACTG GACTTGGTTGGCATTTTAGTTTAATGACTGAAAAAGTAGGTTGAAAGCTCTCTGTATTTTAGTTAACACCTTGAATAAAATGGAAAAAGC >24622_24622_2_DYNC1LI1-GADL1_DYNC1LI1_chr3_32611838_ENST00000432458_GADL1_chr3_30903257_ENST00000282538_length(amino acids)=582AA_BP=70 MAAVGRVGSFGSSPPGLSSTYTGGPLGNEIASGNGGAAAGDDEDGQNLWSCILSEVSTRSRSKLPAGKNVLLLGDIDQQEMIPSKKNAVL VDGVVLNGPTTDAKAGEKFVEEACRLIMEEVVLKATDVNEKVCEWRPPEQLKQLLDLEMRDSGEPPHKLLELCRDVIHYSVKTNHPRFFN QLYAGLDYYSLVARFMTEALNPSVYTYEVSPVFLLVEEAVLKKMIEFIGWKEGDGIFNPGGSVSNMYAMNLARYKYCPDIKEKGLSGSPR LILFTSAECHYSMKKAASFLGIGTENVCFVETDGRGKMIPEELEKQVWQARKEGAAPFLVCATSGTTVLGAFDPLDEIADICERHSLWLH VDASWGGSALMSRKHRKLLHGIHRADSVAWNPHKMLMAGIQCCALLVKDKSDLLKKCYSAKASYLFQQDKFYDVSYDTGDKSIQCSRRPD AFKFWMTWKALGTLGLEERVNRALALSRYLVDEIKKREGFKLLMEPEYANICFWYIPPSLREMEEGPEFWAKLNLVAPAIKERMMKKGSL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for DYNC1LI1-GADL1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for DYNC1LI1-GADL1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for DYNC1LI1-GADL1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
