
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ECI1-GFER (FusionGDB2 ID:24901) |
Fusion Gene Summary for ECI1-GFER |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ECI1-GFER | Fusion gene ID: 24901 | Hgene | Tgene | Gene symbol | ECI1 | GFER | Gene ID | 1632 | 2671 |
| Gene name | enoyl-CoA delta isomerase 1 | growth factor, augmenter of liver regeneration | |
| Synonyms | DCI | ALR|ERV1|HERV1|HPO|HPO1|HPO2|HSS | |
| Cytomap | 16p13.3 | 16p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | enoyl-CoA delta isomerase 1, mitochondrial3,2 trans-enoyl-Coenzyme A isomerase3,2-trans-enoyl-CoA isomerase, mitochondrialD3,D2-enoyl-CoA isomeraseacetylene-allene isomerasedelta(3),Delta(2)-enoyl-CoA isomerasedodecenoyl-CoA delta isomerase (3,2 tra | FAD-linked sulfhydryl oxidase ALRERV1 homologerv1-like growth factorhepatic regenerative stimulation substancehepatopoietin protein | |
| Modification date | 20200313 | 20200322 | |
| UniProtAcc | P42126 | P55789 | |
| Ensembl transtripts involved in fusion gene | ENST00000301729, ENST00000562238, ENST00000570258, | ENST00000567719, ENST00000569451, ENST00000248114, | |
| Fusion gene scores | * DoF score | 6 X 5 X 5=150 | 2 X 2 X 2=8 |
| # samples | 6 | 3 | |
| ** MAII score | log2(6/150*10)=-1.32192809488736 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(3/8*10)=1.90689059560852 | |
| Context | PubMed: ECI1 [Title/Abstract] AND GFER [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ECI1(2293047)-GFER(2035867), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ECI1-GFER seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. ECI1-GFER seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. ECI1-GFER seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. ECI1-GFER seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across ECI1 (5'-gene) Fusion gene breakpoints across ECI1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
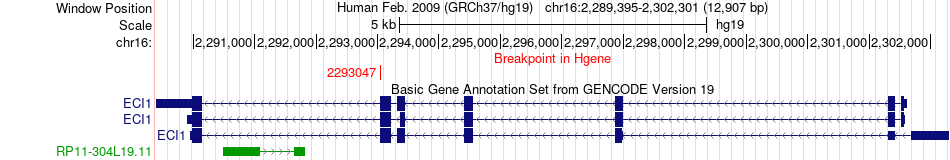 |
 Fusion gene breakpoints across GFER (3'-gene) Fusion gene breakpoints across GFER (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
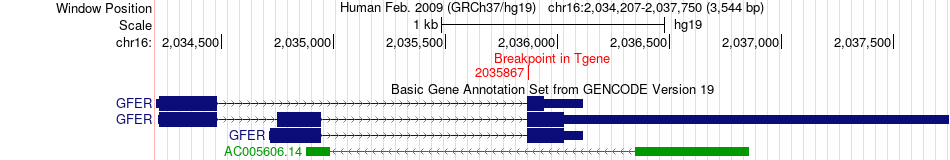 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-IN-7808-01A | ECI1 | chr16 | 2293047 | - | GFER | chr16 | 2035867 | + |
Top |
Fusion Gene ORF analysis for ECI1-GFER |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000301729 | ENST00000567719 | ECI1 | chr16 | 2293047 | - | GFER | chr16 | 2035867 | + |
| Frame-shift | ENST00000301729 | ENST00000569451 | ECI1 | chr16 | 2293047 | - | GFER | chr16 | 2035867 | + |
| Frame-shift | ENST00000562238 | ENST00000567719 | ECI1 | chr16 | 2293047 | - | GFER | chr16 | 2035867 | + |
| Frame-shift | ENST00000562238 | ENST00000569451 | ECI1 | chr16 | 2293047 | - | GFER | chr16 | 2035867 | + |
| Frame-shift | ENST00000570258 | ENST00000567719 | ECI1 | chr16 | 2293047 | - | GFER | chr16 | 2035867 | + |
| Frame-shift | ENST00000570258 | ENST00000569451 | ECI1 | chr16 | 2293047 | - | GFER | chr16 | 2035867 | + |
| In-frame | ENST00000301729 | ENST00000248114 | ECI1 | chr16 | 2293047 | - | GFER | chr16 | 2035867 | + |
| In-frame | ENST00000562238 | ENST00000248114 | ECI1 | chr16 | 2293047 | - | GFER | chr16 | 2035867 | + |
| In-frame | ENST00000570258 | ENST00000248114 | ECI1 | chr16 | 2293047 | - | GFER | chr16 | 2035867 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000301729 | ECI1 | chr16 | 2293047 | - | ENST00000248114 | GFER | chr16 | 2035867 | + | 2674 | 790 | 48 | 866 | 272 |
| ENST00000562238 | ECI1 | chr16 | 2293047 | - | ENST00000248114 | GFER | chr16 | 2035867 | + | 2581 | 697 | 6 | 773 | 255 |
| ENST00000570258 | ECI1 | chr16 | 2293047 | - | ENST00000248114 | GFER | chr16 | 2035867 | + | 3190 | 1306 | 483 | 1382 | 299 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000301729 | ENST00000248114 | ECI1 | chr16 | 2293047 | - | GFER | chr16 | 2035867 | + | 0.02865592 | 0.97134405 |
| ENST00000562238 | ENST00000248114 | ECI1 | chr16 | 2293047 | - | GFER | chr16 | 2035867 | + | 0.027196873 | 0.9728032 |
| ENST00000570258 | ENST00000248114 | ECI1 | chr16 | 2293047 | - | GFER | chr16 | 2035867 | + | 0.043911964 | 0.956088 |
Top |
Fusion Genomic Features for ECI1-GFER |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ECI1 | chr16 | 2293046 | - | GFER | chr16 | 2035866 | + | 0.002102152 | 0.99789786 |
| ECI1 | chr16 | 2293046 | - | GFER | chr16 | 2035866 | + | 0.002102152 | 0.99789786 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
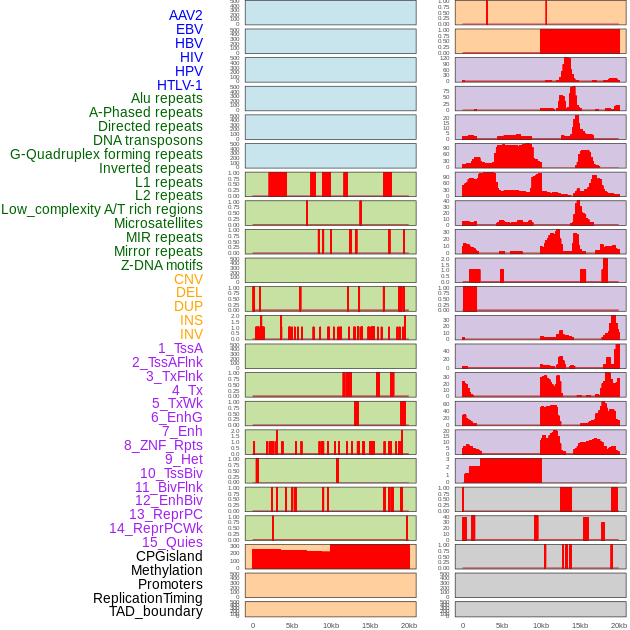 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
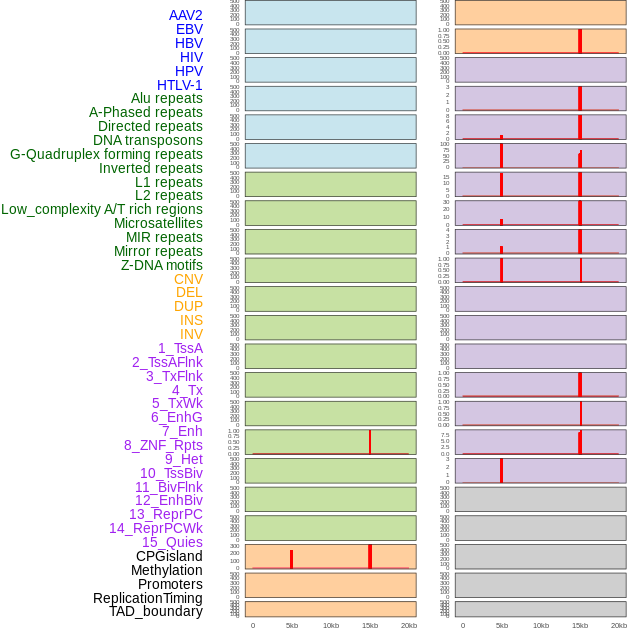 |
Top |
Fusion Protein Features for ECI1-GFER |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:2293047/chr16:2035867) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ECI1 | GFER |
| FUNCTION: Able to isomerize both 3-cis and 3-trans double bonds into the 2-trans form in a range of enoyl-CoA species. {ECO:0000269|PubMed:7818490}. | FUNCTION: [Isoform 1]: FAD-dependent sulfhydryl oxidase that regenerates the redox-active disulfide bonds in CHCHD4/MIA40, a chaperone essential for disulfide bond formation and protein folding in the mitochondrial intermembrane space. The reduced form of CHCHD4/MIA40 forms a transient intermolecular disulfide bridge with GFER/ERV1, resulting in regeneration of the essential disulfide bonds in CHCHD4/MIA40, while GFER/ERV1 becomes re-oxidized by donating electrons to cytochrome c or molecular oxygen. {ECO:0000269|PubMed:19397338, ECO:0000269|PubMed:20593814, ECO:0000269|PubMed:21383138, ECO:0000269|PubMed:22224850, ECO:0000269|PubMed:23186364, ECO:0000269|PubMed:23676665}.; FUNCTION: [Isoform 2]: May act as an autocrine hepatotrophic growth factor promoting liver regeneration. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ECI1 | chr16:2293047 | chr16:2035867 | ENST00000301729 | - | 6 | 7 | 106_110 | 247 | 303.0 | Region | Substrate binding |
| Hgene | ECI1 | chr16:2293047 | chr16:2035867 | ENST00000562238 | - | 6 | 7 | 106_110 | 230 | 286.0 | Region | Substrate binding |
| Tgene | GFER | chr16:2293047 | chr16:2035867 | ENST00000248114 | 1 | 3 | 171_183 | 151 | 206.0 | Nucleotide binding | FAD | |
| Tgene | GFER | chr16:2293047 | chr16:2035867 | ENST00000248114 | 1 | 3 | 194_195 | 151 | 206.0 | Nucleotide binding | FAD |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | GFER | chr16:2293047 | chr16:2035867 | ENST00000248114 | 1 | 3 | 95_195 | 151 | 206.0 | Domain | ERV/ALR sulfhydryl oxidase | |
| Tgene | GFER | chr16:2293047 | chr16:2035867 | ENST00000248114 | 1 | 3 | 99_107 | 151 | 206.0 | Nucleotide binding | FAD |
Top |
Fusion Gene Sequence for ECI1-GFER |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >24901_24901_1_ECI1-GFER_ECI1_chr16_2293047_ENST00000301729_GFER_chr16_2035867_ENST00000248114_length(transcript)=2674nt_BP=790nt AGGAGGCGAGCTCAGCCCGCGACCTTTATCCCGCGCGTTGCGGTCAAGATGGCGCTGGTGGCTTCTGTGCGAGTCCCGGCGCGCGTTCTG CTCCGCGCGGGGGCCCGGCTCCCGGGCGCGGCCCTCGGGCGGACGGAGCGGGCGGCCGGCGGCGGAGACGGCGCGCGGCGCTTCGGGAGC CAGCGGGTGCTGGTGGAGCCGGACGCGGGCGCAGGGGTCGCTGTGATGAAATTCAAGAACCCCCCAGTGAACAGCCTGAGCCTGGAGTTT CTGACGGAGCTGGTCATCAGCCTGGAGAAGCTGGAGAATGACAAGAGCTTCCGCGGTGTCATTCTGACCTCGGACCGCCCGGGTGTCTTC TCGGCCGGCCTGGACCTGACGGAGATGTGTGGGAGGAGCCCCGCCCACTACGCTGGGTACTGGAAGGCCGTTCAGGAGCTGTGGCTGCGG TTGTACCAGTCCAACCTGGTGCTGGTCTCCGCCATCAACGGAGCCTGCCCCGCTGGAGGCTGCCTGGTGGCCCTGACCTGTGACTACCGC ATCCTGGCGGACAACCCCAGGTACTGCATAGGACTCAATGAGACCCAGCTGGGCATCATCGCCCCTTTCTGGTTGAAAGACACCCTGGAG AACACCATCGGGCACCGGGCGGCGGAGCGTGCCCTGCAGCTGGGGCTGCTCTTCCCGCCGGCGGAGGCCCTGCAGGTGGGCATAGTGGAC CAGGTGGTCCCGGAGGAGCAGGTGCAGAGCACTGCGCTGTCAGCGATAGCCCAGTGGATGGCCATTCCAGGCTGTGCAGGAACCACCCAG ACACCCGCACCCGGGCATGCTTCACACAGTGGCTGTGCCACCTGCACAATGAAGTGAACCGCAAGCTGGGCAAGCCTGACTTCGACTGCT CAAAAGTGGATGAGCGCTGGCGCGACGGCTGGAAGGATGGCTCCTGTGACTAGAGGGTGGTCAGCCAGAGCTCATGGGACAGCTAGCCAG GCATGGTTGGATAGGGGCAGGGCACTCATTAAAGTGCATCACAGCCAGAGCCTGTTGTGTCTCAGTTGGGTGGTCCCCAGGACACTGCCT GTGGGGACCTGCCCTGCCCCTCTTAGGTTTGGAGCAGAAGTGGAGGTGCCCACAGCAGGTACCCACTGGCCCCCTCCTCAGTGGAGACCC CAAGGAGCTGCAGCTGAACTGCAGGGGAGGGAAGGAGGAGCAGCCTGGGCTGCCCCTTGACATTCAGGATGTAGCTTCCTGCCCACCGCA TACCCTGGCGCCTCACTCCTCACACGGGAAGACAGCGGGCCTGGCTGGGCATCCCTGTGCCTGTCCCTGGCGGCCAGGCCATTGCCTTCC CACTATGCAGCCAGGGATGCCCCTGCCCCCCATGGCTCTGTGCTGCTCACTTTAGGGGGCTCAATTCTCCACTCTGCTCAGTCCCTACAG GGAAAGCTCAGGTCGGGTCTTTCTGAGGGTCCACCAGCCATCCTACCCTCTCCCTGCCTGGCACATGCCTGCCAGCGTTGTGTCATGCCT GTCCACAGGGGATTCGTGGGGCTCACTTCATCAGAGTTTGAAGCCCAAATGAAACGCTGAAGTGACTGAGAACCTGGCTTCAGTATATTT TCTGCTGGGGCTTAATAAAGCAGTAGACAGGGCTTGTTCCATCCCTCTGTGCTCAGCTGCATTTCCTGCTGGGGTCCTGGTTCCTCAGGA GAGAGAGACCACAGGGTGAGAGTGAGCCAGGAACAGCAAGGACGTTGATTGGTTGGGGCAGGGGGGCCAGAGTAGCTGATGTAGGAGTAC TGGGAGGCCAGACGGCACGAGGTCTCCAAGGCCCCAGCAAAGCCATGGCTTCTACCCCTAGTTCCCCTGACAGGAAGTTCTTGGCGGGTT TGGAGCCAGGGGATGGCATGGAGTGATGTGGCTTTGAAGGGTCCTCTGGCTGCTGAGCTGGGATGAGGCAGGTAAGGGTGGAACAGGAGG GGTGGGGAGGAAGCCGGGGCAGTCACCGAGTGACCACCAAGAGGAAGACCCACCCCACGGCGGGGACAGATGCGGGGTACGTTAAAGGGA GAGCCAGAGAACTCATGGGGTGAGGATGGAGTCCGAGGAGACTGCTGGGAGCCGCCGTGTGGGTCAGAGATGGAGAAGGCTGAGTGCAGC AAGGTGGGGGGTGACTGGGACCCAGCCTTTGGGCCTCCCCAGCCAGAGCAGCCCAGCAACAGTGTGTCCTGTGGTCATAAAACTCCAGGG ACCTCTATCCTCCAGGAGTCTCAGCCTTTCCCTGGGCGCAGGCCCACCTTGGCATGGCCGCCTCAGGCCTCCATGGAGGGAGCTGCTATG TCCCCACCAGATTGGCCCCGTGCGGCTGCTGGCTTCTGTAGAGGCTGCCCAGAGGGGCCAGGTGGCACAAATAAGAGAGGGGAGATGGGG GGCAGCCAGGAGAGGAGGTGTCCCTTCCTCGCCCAGACACAGCGCGCTTCTCTCTGGCCTTTCCCGAGGCCTGTGAGTGCCTCAGGAAGC AGCTGGGCCCTCTGGGAAGGCTGTGTTCAGCTTAGGAACATACCGCCTGTATCTGCTGTCCCTCCCCTGCCCCCCTGCCCCCCCCACCGC >24901_24901_1_ECI1-GFER_ECI1_chr16_2293047_ENST00000301729_GFER_chr16_2035867_ENST00000248114_length(amino acids)=272AA_BP=247 MALVASVRVPARVLLRAGARLPGAALGRTERAAGGGDGARRFGSQRVLVEPDAGAGVAVMKFKNPPVNSLSLEFLTELVISLEKLENDKS FRGVILTSDRPGVFSAGLDLTEMCGRSPAHYAGYWKAVQELWLRLYQSNLVLVSAINGACPAGGCLVALTCDYRILADNPRYCIGLNETQ LGIIAPFWLKDTLENTIGHRAAERALQLGLLFPPAEALQVGIVDQVVPEEQVQSTALSAIAQWMAIPGCAGTTQTPAPGHASHSGCATCT -------------------------------------------------------------- >24901_24901_2_ECI1-GFER_ECI1_chr16_2293047_ENST00000562238_GFER_chr16_2035867_ENST00000248114_length(transcript)=2581nt_BP=697nt GTCAAGATGGCGCTGGTGGCTTCTGTGCGAGTCCCGGCGCGCGTTCTGCTCCGCGCGGGGGCCCGGCTCCCGGGCGCGGCCCTCGGGCGG ACGGAGCGGGCGGCCGGCGGCGGAGACGGCGCGCGGCGCTTCGGGAGCCAGCGGGTGCTGGTGGAGCCGGACGCGGGCGCAGGGGTCGCT GTGATGAAATTCAAGAACCCCCCAGTGAACAGCCTGAGCCTGGAGTTTCTGACGGAGCTGGTCATCAGCCTGGAGAAGCTGGAGAATGAC AAGAGCTTCCGCGGTGTCATTCTGACCTCGGACCGCCCGGGTGTCTTCTCGGCCGGCCTGGACCTGACGGAGATGTGTGGGAGGAGCCCC GCCCACTACGCTGGGTACTGGAAGGCCGTTCAGGAGCTGTGGCTGCGGTTGTACCAGTCCAACCTGGTGCTGGTCTCCGCCATCAACGGA GCCTGCCCCGCTGGAGGCTGCCTGGTGGCCCTGACCTGTGACTACCGCATCCTGGCGGACAACCCCAGGTTGAAAGACACCCTGGAGAAC ACCATCGGGCACCGGGCGGCGGAGCGTGCCCTGCAGCTGGGGCTGCTCTTCCCGCCGGCGGAGGCCCTGCAGGTGGGCATAGTGGACCAG GTGGTCCCGGAGGAGCAGGTGCAGAGCACTGCGCTGTCAGCGATAGCCCAGTGGATGGCCATTCCAGGCTGTGCAGGAACCACCCAGACA CCCGCACCCGGGCATGCTTCACACAGTGGCTGTGCCACCTGCACAATGAAGTGAACCGCAAGCTGGGCAAGCCTGACTTCGACTGCTCAA AAGTGGATGAGCGCTGGCGCGACGGCTGGAAGGATGGCTCCTGTGACTAGAGGGTGGTCAGCCAGAGCTCATGGGACAGCTAGCCAGGCA TGGTTGGATAGGGGCAGGGCACTCATTAAAGTGCATCACAGCCAGAGCCTGTTGTGTCTCAGTTGGGTGGTCCCCAGGACACTGCCTGTG GGGACCTGCCCTGCCCCTCTTAGGTTTGGAGCAGAAGTGGAGGTGCCCACAGCAGGTACCCACTGGCCCCCTCCTCAGTGGAGACCCCAA GGAGCTGCAGCTGAACTGCAGGGGAGGGAAGGAGGAGCAGCCTGGGCTGCCCCTTGACATTCAGGATGTAGCTTCCTGCCCACCGCATAC CCTGGCGCCTCACTCCTCACACGGGAAGACAGCGGGCCTGGCTGGGCATCCCTGTGCCTGTCCCTGGCGGCCAGGCCATTGCCTTCCCAC TATGCAGCCAGGGATGCCCCTGCCCCCCATGGCTCTGTGCTGCTCACTTTAGGGGGCTCAATTCTCCACTCTGCTCAGTCCCTACAGGGA AAGCTCAGGTCGGGTCTTTCTGAGGGTCCACCAGCCATCCTACCCTCTCCCTGCCTGGCACATGCCTGCCAGCGTTGTGTCATGCCTGTC CACAGGGGATTCGTGGGGCTCACTTCATCAGAGTTTGAAGCCCAAATGAAACGCTGAAGTGACTGAGAACCTGGCTTCAGTATATTTTCT GCTGGGGCTTAATAAAGCAGTAGACAGGGCTTGTTCCATCCCTCTGTGCTCAGCTGCATTTCCTGCTGGGGTCCTGGTTCCTCAGGAGAG AGAGACCACAGGGTGAGAGTGAGCCAGGAACAGCAAGGACGTTGATTGGTTGGGGCAGGGGGGCCAGAGTAGCTGATGTAGGAGTACTGG GAGGCCAGACGGCACGAGGTCTCCAAGGCCCCAGCAAAGCCATGGCTTCTACCCCTAGTTCCCCTGACAGGAAGTTCTTGGCGGGTTTGG AGCCAGGGGATGGCATGGAGTGATGTGGCTTTGAAGGGTCCTCTGGCTGCTGAGCTGGGATGAGGCAGGTAAGGGTGGAACAGGAGGGGT GGGGAGGAAGCCGGGGCAGTCACCGAGTGACCACCAAGAGGAAGACCCACCCCACGGCGGGGACAGATGCGGGGTACGTTAAAGGGAGAG CCAGAGAACTCATGGGGTGAGGATGGAGTCCGAGGAGACTGCTGGGAGCCGCCGTGTGGGTCAGAGATGGAGAAGGCTGAGTGCAGCAAG GTGGGGGGTGACTGGGACCCAGCCTTTGGGCCTCCCCAGCCAGAGCAGCCCAGCAACAGTGTGTCCTGTGGTCATAAAACTCCAGGGACC TCTATCCTCCAGGAGTCTCAGCCTTTCCCTGGGCGCAGGCCCACCTTGGCATGGCCGCCTCAGGCCTCCATGGAGGGAGCTGCTATGTCC CCACCAGATTGGCCCCGTGCGGCTGCTGGCTTCTGTAGAGGCTGCCCAGAGGGGCCAGGTGGCACAAATAAGAGAGGGGAGATGGGGGGC AGCCAGGAGAGGAGGTGTCCCTTCCTCGCCCAGACACAGCGCGCTTCTCTCTGGCCTTTCCCGAGGCCTGTGAGTGCCTCAGGAAGCAGC TGGGCCCTCTGGGAAGGCTGTGTTCAGCTTAGGAACATACCGCCTGTATCTGCTGTCCCTCCCCTGCCCCCCTGCCCCCCCCACCGCCTT >24901_24901_2_ECI1-GFER_ECI1_chr16_2293047_ENST00000562238_GFER_chr16_2035867_ENST00000248114_length(amino acids)=255AA_BP=230 MALVASVRVPARVLLRAGARLPGAALGRTERAAGGGDGARRFGSQRVLVEPDAGAGVAVMKFKNPPVNSLSLEFLTELVISLEKLENDKS FRGVILTSDRPGVFSAGLDLTEMCGRSPAHYAGYWKAVQELWLRLYQSNLVLVSAINGACPAGGCLVALTCDYRILADNPRLKDTLENTI -------------------------------------------------------------- >24901_24901_3_ECI1-GFER_ECI1_chr16_2293047_ENST00000570258_GFER_chr16_2035867_ENST00000248114_length(transcript)=3190nt_BP=1306nt AACACAGCTGGGACCTTACTGCCCAGTGCTCCCAGTGGTCTGACCAAAACCCCACACCAGACAGTACAAAGGCCCACTCCCGAGCTGCCG CTTACGGCCCAGCGGGAACAGACTGTATTCTTATCTTTCGGCATTTCTCTGCAAATAAAAAGGTGCCACTGCGGTCCGGCCTTTGTGCAC TTGCCCTTGTGCTGAGCAAGGCGGGTGGCCTGTTCTTACCGCTGGGTTCTGCTGCCCCCACCCCCATTTCGGTGACTAAACCTGCAGCTC TGATCCTGTTCCCAAAGGGGCAGGAGGCCGGGCTCACCCCAGGCAGCCCCTGGCCTCCCTGACGGCCCCCTCGGGAGGGGTTCCGTGCTT TGATCGCCTTTCCAGCGCCGATACTGCAGTAGCTGGGCCCGGCCGAGAGCTCGCGGTCACGAACACCGGCAGCCTCTTGCCTACAGTGTC CCGCGCTGTAGAAGAGCCTGCCCTGGCGCTGAGCTGGTCGTGGCTGCTTCAGCAGTGGGCCTGCGCCTCCTTGTAGGCCAGCCCTGGCCC AGATCCAGGTGCCTGCCCCGGAAGCGCGCGGCTGCCCGGGGTGAAGCCGGAGGGCGGCGAGCGGCTGTTTCGGCCGGGGCCCGGCTCCCG GGCGCGGCCCTCGGGCGGACGGAGCGGGCGGCCGGCGGCGGAGACGGCGCGCGGCGCTTCGGGAGCCAGCGGGTGCTGGTGGAGCCGGAC GCGGGCGCAGGGGTCGCTGTGATGAAATTCAAGAACCCCCCAGTGAACAGCCTGAGCCTGGAGTTTCTGACGGAGCTGGTCATCAGCCTG GAGAAGCTGGAGAATGACAAGAGCTTCCGCGGTGTCATTCTGACCTCGGACCGCCCGGGTGTCTTCTCGGCCGGCCTGGACCTGACGGAG ATGTGTGGGAGGAGCCCCGCCCACTACGCTGGGTACTGGAAGGCCGTTCAGGAGCTGTGGCTGCGGTTGTACCAGTCCAACCTGGTGCTG GTCTCCGCCATCAACGGAGCCTGCCCCGCTGGAGGCTGCCTGGTGGCCCTGACCTGTGACTACCGCATCCTGGCGGACAACCCCAGGTAC TGCATAGGACTCAATGAGACCCAGCTGGGCATCATCGCCCCTTTCTGGTTGAAAGACACCCTGGAGAACACCATCGGGCACCGGGCGGCG GAGCGTGCCCTGCAGCTGGGGCTGCTCTTCCCGCCGGCGGAGGCCCTGCAGGTGGGCATAGTGGACCAGGTGGTCCCGGAGGAGCAGGTG CAGAGCACTGCGCTGTCAGCGATAGCCCAGTGGATGGCCATTCCAGGCTGTGCAGGAACCACCCAGACACCCGCACCCGGGCATGCTTCA CACAGTGGCTGTGCCACCTGCACAATGAAGTGAACCGCAAGCTGGGCAAGCCTGACTTCGACTGCTCAAAAGTGGATGAGCGCTGGCGCG ACGGCTGGAAGGATGGCTCCTGTGACTAGAGGGTGGTCAGCCAGAGCTCATGGGACAGCTAGCCAGGCATGGTTGGATAGGGGCAGGGCA CTCATTAAAGTGCATCACAGCCAGAGCCTGTTGTGTCTCAGTTGGGTGGTCCCCAGGACACTGCCTGTGGGGACCTGCCCTGCCCCTCTT AGGTTTGGAGCAGAAGTGGAGGTGCCCACAGCAGGTACCCACTGGCCCCCTCCTCAGTGGAGACCCCAAGGAGCTGCAGCTGAACTGCAG GGGAGGGAAGGAGGAGCAGCCTGGGCTGCCCCTTGACATTCAGGATGTAGCTTCCTGCCCACCGCATACCCTGGCGCCTCACTCCTCACA CGGGAAGACAGCGGGCCTGGCTGGGCATCCCTGTGCCTGTCCCTGGCGGCCAGGCCATTGCCTTCCCACTATGCAGCCAGGGATGCCCCT GCCCCCCATGGCTCTGTGCTGCTCACTTTAGGGGGCTCAATTCTCCACTCTGCTCAGTCCCTACAGGGAAAGCTCAGGTCGGGTCTTTCT GAGGGTCCACCAGCCATCCTACCCTCTCCCTGCCTGGCACATGCCTGCCAGCGTTGTGTCATGCCTGTCCACAGGGGATTCGTGGGGCTC ACTTCATCAGAGTTTGAAGCCCAAATGAAACGCTGAAGTGACTGAGAACCTGGCTTCAGTATATTTTCTGCTGGGGCTTAATAAAGCAGT AGACAGGGCTTGTTCCATCCCTCTGTGCTCAGCTGCATTTCCTGCTGGGGTCCTGGTTCCTCAGGAGAGAGAGACCACAGGGTGAGAGTG AGCCAGGAACAGCAAGGACGTTGATTGGTTGGGGCAGGGGGGCCAGAGTAGCTGATGTAGGAGTACTGGGAGGCCAGACGGCACGAGGTC TCCAAGGCCCCAGCAAAGCCATGGCTTCTACCCCTAGTTCCCCTGACAGGAAGTTCTTGGCGGGTTTGGAGCCAGGGGATGGCATGGAGT GATGTGGCTTTGAAGGGTCCTCTGGCTGCTGAGCTGGGATGAGGCAGGTAAGGGTGGAACAGGAGGGGTGGGGAGGAAGCCGGGGCAGTC ACCGAGTGACCACCAAGAGGAAGACCCACCCCACGGCGGGGACAGATGCGGGGTACGTTAAAGGGAGAGCCAGAGAACTCATGGGGTGAG GATGGAGTCCGAGGAGACTGCTGGGAGCCGCCGTGTGGGTCAGAGATGGAGAAGGCTGAGTGCAGCAAGGTGGGGGGTGACTGGGACCCA GCCTTTGGGCCTCCCCAGCCAGAGCAGCCCAGCAACAGTGTGTCCTGTGGTCATAAAACTCCAGGGACCTCTATCCTCCAGGAGTCTCAG CCTTTCCCTGGGCGCAGGCCCACCTTGGCATGGCCGCCTCAGGCCTCCATGGAGGGAGCTGCTATGTCCCCACCAGATTGGCCCCGTGCG GCTGCTGGCTTCTGTAGAGGCTGCCCAGAGGGGCCAGGTGGCACAAATAAGAGAGGGGAGATGGGGGGCAGCCAGGAGAGGAGGTGTCCC TTCCTCGCCCAGACACAGCGCGCTTCTCTCTGGCCTTTCCCGAGGCCTGTGAGTGCCTCAGGAAGCAGCTGGGCCCTCTGGGAAGGCTGT GTTCAGCTTAGGAACATACCGCCTGTATCTGCTGTCCCTCCCCTGCCCCCCTGCCCCCCCCACCGCCTTCCCTTTTTCCCTGTCTTCCTT >24901_24901_3_ECI1-GFER_ECI1_chr16_2293047_ENST00000570258_GFER_chr16_2035867_ENST00000248114_length(amino acids)=299AA_BP=274 MVVAASAVGLRLLVGQPWPRSRCLPRKRAAARGEAGGRRAAVSAGARLPGAALGRTERAAGGGDGARRFGSQRVLVEPDAGAGVAVMKFK NPPVNSLSLEFLTELVISLEKLENDKSFRGVILTSDRPGVFSAGLDLTEMCGRSPAHYAGYWKAVQELWLRLYQSNLVLVSAINGACPAG GCLVALTCDYRILADNPRYCIGLNETQLGIIAPFWLKDTLENTIGHRAAERALQLGLLFPPAEALQVGIVDQVVPEEQVQSTALSAIAQW -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ECI1-GFER |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ECI1-GFER |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ECI1-GFER |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
