
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:EDEM1-GPD1L (FusionGDB2 ID:24951) |
Fusion Gene Summary for EDEM1-GPD1L |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: EDEM1-GPD1L | Fusion gene ID: 24951 | Hgene | Tgene | Gene symbol | EDEM1 | GPD1L | Gene ID | 9695 | 23171 |
| Gene name | ER degradation enhancing alpha-mannosidase like protein 1 | glycerol-3-phosphate dehydrogenase 1 like | |
| Synonyms | EDEM | GPD1-L | |
| Cytomap | 3p26.1 | 3p22.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | ER degradation-enhancing alpha-mannosidase-like protein 1ER degradation enhancer, mannosidase alpha-like 1ER degradation-enhancing alpha-mannosidase-like 1 | glycerol-3-phosphate dehydrogenase 1-like protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q92611 | Q8N335 | |
| Ensembl transtripts involved in fusion gene | ENST00000256497, ENST00000445686, | ENST00000282541, | |
| Fusion gene scores | * DoF score | 4 X 4 X 4=64 | 6 X 4 X 6=144 |
| # samples | 4 | 7 | |
| ** MAII score | log2(4/64*10)=-0.678071905112638 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/144*10)=-1.04064198449735 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: EDEM1 [Title/Abstract] AND GPD1L [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | EDEM1(5229999)-GPD1L(32169568), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across EDEM1 (5'-gene) Fusion gene breakpoints across EDEM1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
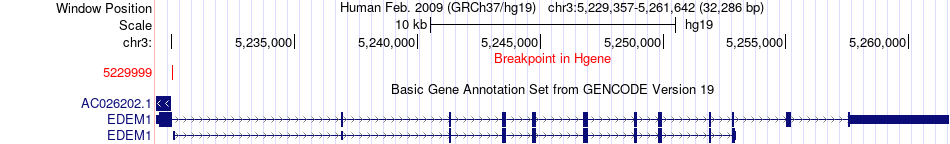 |
 Fusion gene breakpoints across GPD1L (3'-gene) Fusion gene breakpoints across GPD1L (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
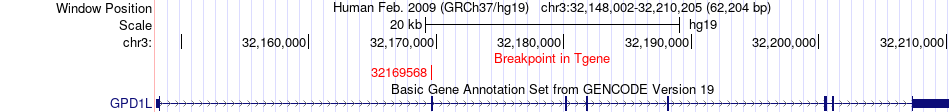 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-KK-A8I6-01A | EDEM1 | chr3 | 5229999 | - | GPD1L | chr3 | 32169568 | + |
| ChimerDB4 | PRAD | TCGA-KK-A8I6-01A | EDEM1 | chr3 | 5229999 | + | GPD1L | chr3 | 32169568 | + |
| ChimerDB4 | PRAD | TCGA-KK-A8I6 | EDEM1 | chr3 | 5229999 | + | GPD1L | chr3 | 32169568 | + |
Top |
Fusion Gene ORF analysis for EDEM1-GPD1L |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000256497 | ENST00000282541 | EDEM1 | chr3 | 5229999 | + | GPD1L | chr3 | 32169568 | + |
| intron-3CDS | ENST00000445686 | ENST00000282541 | EDEM1 | chr3 | 5229999 | + | GPD1L | chr3 | 32169568 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000256497 | EDEM1 | chr3 | 5229999 | + | ENST00000282541 | GPD1L | chr3 | 32169568 | + | 4454 | 642 | 133 | 1650 | 505 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000256497 | ENST00000282541 | EDEM1 | chr3 | 5229999 | + | GPD1L | chr3 | 32169568 | + | 0.000110968 | 0.999889 |
Top |
Fusion Genomic Features for EDEM1-GPD1L |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| EDEM1 | chr3 | 5229999 | + | GPD1L | chr3 | 32169567 | + | 3.10E-09 | 1 |
| EDEM1 | chr3 | 5229999 | + | GPD1L | chr3 | 32169567 | + | 3.10E-09 | 1 |
| EDEM1 | chr3 | 5229999 | + | GPD1L | chr3 | 32169567 | + | 3.10E-09 | 1 |
| EDEM1 | chr3 | 5229999 | + | GPD1L | chr3 | 32169567 | + | 3.10E-09 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
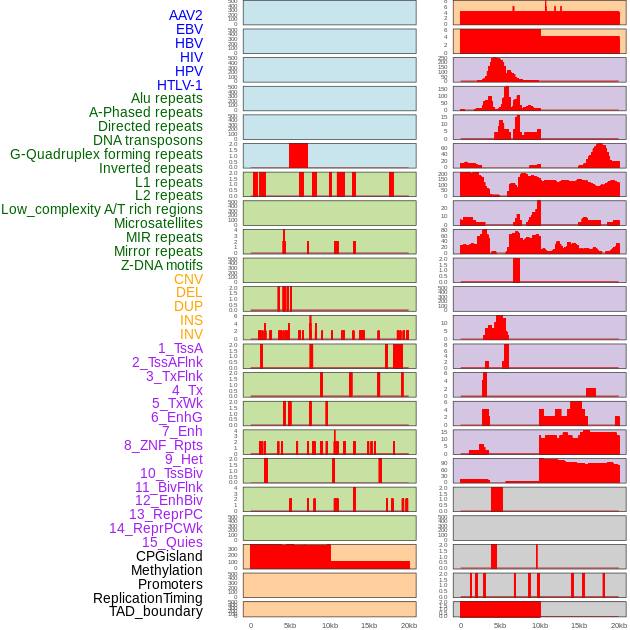 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
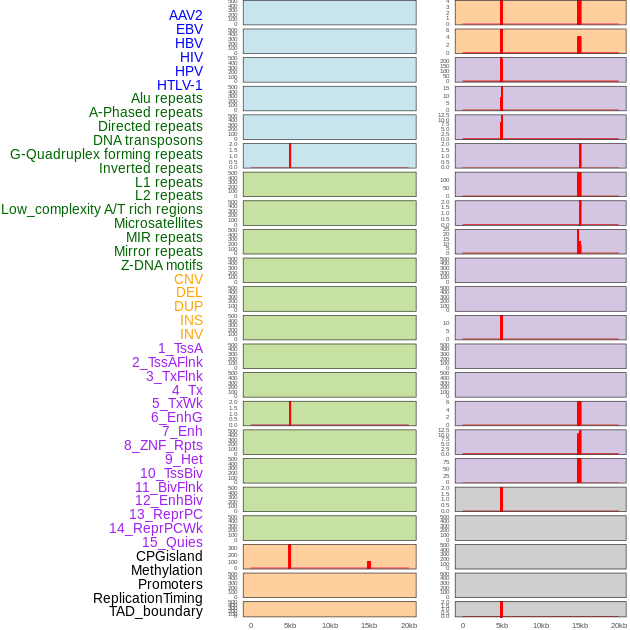 |
Top |
Fusion Protein Features for EDEM1-GPD1L |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:5229999/chr3:32169568) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| EDEM1 | GPD1L |
| FUNCTION: Extracts misfolded glycoproteins, but not glycoproteins undergoing productive folding, from the calnexin cycle. It is directly involved in endoplasmic reticulum-associated degradation (ERAD) and targets misfolded glycoproteins for degradation in an N-glycan-independent manner, probably by forming a complex with SEL1L. It has low mannosidase activity, catalyzing mannose trimming from Man8GlcNAc2 to Man7GlcNAc2. {ECO:0000269|PubMed:12610306, ECO:0000269|PubMed:19524542, ECO:0000269|PubMed:19934218, ECO:0000269|PubMed:25092655}. | FUNCTION: Plays a role in regulating cardiac sodium current; decreased enzymatic activity with resulting increased levels of glycerol 3-phosphate activating the DPD1L-dependent SCN5A phosphorylation pathway, may ultimately lead to decreased sodium current; cardiac sodium current may also be reduced due to alterations of NAD(H) balance induced by DPD1L. {ECO:0000269|PubMed:19666841, ECO:0000269|PubMed:19745168}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EDEM1 | chr3:5229999 | chr3:32169568 | ENST00000256497 | + | 1 | 12 | 1_4 | 169 | 658.0 | Topological domain | Cytoplasmic |
| Hgene | EDEM1 | chr3:5229999 | chr3:32169568 | ENST00000256497 | + | 1 | 12 | 5_25 | 169 | 658.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Tgene | GPD1L | chr3:5229999 | chr3:32169568 | ENST00000282541 | 0 | 8 | 271_272 | 15 | 352.0 | Region | Note=Substrate binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EDEM1 | chr3:5229999 | chr3:32169568 | ENST00000256497 | + | 1 | 12 | 26_657 | 169 | 658.0 | Topological domain | Lumenal |
| Tgene | GPD1L | chr3:5229999 | chr3:32169568 | ENST00000282541 | 0 | 8 | 12_17 | 15 | 352.0 | Nucleotide binding | NAD |
Top |
Fusion Gene Sequence for EDEM1-GPD1L |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >24951_24951_1_EDEM1-GPD1L_EDEM1_chr3_5229999_ENST00000256497_GPD1L_chr3_32169568_ENST00000282541_length(transcript)=4454nt_BP=642nt GGCTAGTTCCTGGTGCGTCACGGGGGAGTTCCTTAAAGGGGAAGCGAGCCGGGCTACGGGGCGAGCGCGGGGTGCGGTGGTCGGCGGGGA GGCCCCCGCGCTTTAAAATAATGCCCGCGGCGCCCGCGCGACCATGCAATGGCGAGCGCTCGTCCTGGGGCTGGTGCTCCTCCGGCTTGG CCTCCATGGAGTATTGTGGCTCGTCTTCGGGCTGGGGCCCAGCATGGGCTTCTACCAGCGCTTTCCGCTCAGCTTCGGCTTCCAGCGTCT GAGGAGCCCCGACGGCCCCGCGTCGCCCACCTCGGGGCCCGTGGGCCGGCCTGGGGGGGTATCCGGGCCGTCGTGGCTGCAGCCGCCGGG GACCGGGGCAGCGCAGAGCCCGCGCAAGGCTCCGCGGCGTCCTGGGCCGGGGATGTGCGGCCCAGCCAACTGGGGCTACGTGCTGGGCGG CCGGGGCCGCGGCCCGGACGAGTACGAGAAGCGCTACAGCGGCGCCTTCCCTCCGCAGCTGCGTGCCCAGATGCGCGACCTGGCACGGGG CATGTTCGTCTTTGGCTACGACAACTACATGGCTCACGCCTTCCCCCAGGACGAGCTCAACCCCATCCACTGCCGCGGCCGTGGGCCCGA CCGCGGGGACCCGGGTTCAGCTGTTGCAAAAATAATTGGTAATAATGTCAAGAAACTTCAGAAATTTGCCTCCACAGTCAAGATGTGGGT CTTTGAAGAAACAGTGAATGGCAGAAAACTGACAGACATCATAAATAATGACCATGAAAATGTAAAATATCTTCCTGGACACAAGCTGCC AGAAAATGTGGTTGCCATGTCAAATCTTAGCGAGGCTGTGCAGGATGCAGACCTGCTGGTGTTTGTCATTCCCCACCAGTTCATTCACAG AATCTGTGATGAGATCACTGGGAGAGTGCCCAAGAAAGCGCTGGGAATCACCCTCATCAAGGGCATAGACGAGGGCCCCGAGGGGCTGAA GCTCATTTCTGACATCATCCGTGAGAAGATGGGTATTGACATCAGTGTGCTGATGGGAGCCAACATTGCCAATGAGGTGGCTGCAGAGAA GTTCTGTGAGACCACCATCGGCAGCAAAGTAATGGAGAACGGCCTTCTCTTCAAAGAACTTCTGCAGACTCCAAATTTTCGAATTACCGT GGTTGATGATGCAGACACTGTTGAACTCTGTGGTGCGCTTAAGAACATCGTAGCTGTGGGAGCTGGGTTCTGCGACGGCCTCCGCTGTGG AGACAACACCAAAGCGGCCGTCATCCGCCTGGGACTCATGGAAATGATTGCTTTTGCCAGGATCTTCTGCAAAGGCCAAGTGTCTACAGC CACCTTCCTAGAGAGCTGCGGGGTGGCCGACCTGATCACCACCTGTTACGGAGGGCGGAACCGCAGGGTGGCCGAGGCCTTCGCCAGAAC TGGGAAGACCATTGAAGAGTTGGAGAAGGAGATGCTGAATGGGCAAAAGCTCCAAGGACCGCAGACTTCTGCTGAAGTGTACCGCATCCT CAAACAGAAGGGACTACTGGACAAGTTTCCATTGTTTACTGCAGTGTATCAGATCTGCTACGAAAGCAGACCAGTTCAAGAGATGTTGTC TTGTCTTCAGAGCCATCCAGAGCATACATAAAGTGAATCATGCAACGTGTTGGGGGAAGTTCTGCCTTTCTGATCAATCTTTTGGGTTCA CGTGGAAACCAGGACTTGGCAACATGATGTTTGACTGTAATCTCATCACGGATATGTATGAATTTTTACAGGTTCGTTTTTGAATTGTGA GAGGCAGTTCATTAGCAAAGATGTACTGGGCAGTAACTAAACACACATGCAAACATGTGAATGGTGGTTTATTCCTCATTCTGTGGATGT TTCTATGAGCCAAAATTTGATGTCTTTTTTTCAAAATTGCTTATGAAATTTCCACACAATCGTAGCTTATAAGATTGGAACGATCTCAGC CAAATATTTTAGGTGTAATTCATATGTATTTGAGTGGAGGATTTTTTTTCTCATTTTTCTAGTGTTAAATTTTAACCAGCATTAACATGG TAGAGTGGAGGAGTGAGTGTGTTCAAAGATCAACATATTTAACTTTTAAACACTATCTCAAAGCCAGCATAATTAACTACTTTGATTGTG GGCTGACCTTTGTTTTTTTAACAATCAGGCATTTTTAATTAGATAATCCACTCATGTATTTCCCCCTCACTGCAGTTGTCTGCATTTTTA GCCTCTTTTCTCTTCGTTAGTTGTCAGAATATGCCTTCGTCAAGGCTCAGAGGTAACAAGACAGAAAATTCATCTGGGATTTTCCTGCTG TGGCTGGCACATTCTTCTGATTAACAGACACTTGTATGATGCTTTAGGCTAGTTAGTGCATTTTTTAGCAAACATTTATCTTAAACATCA CAGATCCACTGGGGGGTGCAAGGGGCTACTGTTAGTCCTCTTGTTAGATGCAGTCACTCCTCCTGGTCACCTAGTGAGCAGGGACAGAGC CAGGAGTCAAGTGCAGTGCCAAGGTGCATGACCCTCTGAGAAGTCACTGGGCTGATTTGACCTCCGACTCATTGGTTGTGCAAATGCCAT GTGCAGCCTTTCCTGAGGCCATAGGAGGGCTTCCTGCAGCTGAGATCTATGCAGGCCATCCTCTCAACAAGTGCCACTCCAAGGGCGGTC CTCGGTGCAGCAGCATCAGCTTCACTTGTGGGGGGGTGGGGGAAGGGGCGGTCTCAGAAATGCAGGTTCCCAGGTCCCACCCTGGACTTC TGAAGGGGTGTGGCATCTGTGTTTCTGATGCTTACTACAATATGTGAACCACTACTTTAGAAAATCTGCTTTAACTTGGTATTCCTCTAA TTGTGTTCCCTAGGAAATGACTGTCCCAAGAGCCAGTGATTATTCCAGGTGTTCCCTGGAAAGGTCAAGTGAGTCTGGGAAACACTATGT CTGTACACCTCTTGAAGGTGTCGAATGTATGTTTATACATCAGTGGAACCCATTTTTCTAGCCTAGCAAGTCCCAAACACATTACACTGA AGAGATTTTGGTGAGGAAACTTGCTGGAGTTTTCAGGGAACACTGTTCTAGGCTTAGGTGACCTTAGGATCACTCAAGTAGACCCTTCAC TCCCTGCGAGAAATTAGGATGAATAACTACCTGTGGCATTGTTGGTTCTGAACTTTTACAGTTCAGGCCTGCTGTGAATCTTTGATGAAG CTTTAAGGTGACACTGTTGTACAAGATGTCAGCTTTGCTGAAACGCACATTACCTGGAATAAGTGCTTTAATTGTAGAATTAGAATGGGA TTTACTGTACTGTTTTAAATGAGATTGGCTTCAGAATCCATTACAGTTACCTTACATAGCACTTGATACGTGTTAAATGAACATATGAAT GTAATTTATATATTCCTAGAATTTAAGTTACTTTGTGAGATTTGGGCCTGTCCCTCAATGCCAGTTTAGGATTTCTTTTTTTCTATACCT TGAAATGATTATAAAATAGATTTTCATGGGAATTTTAAAAACTCTATCCAAAACATTTTTGGAGCATTTTAAAGCCCCATACACAGAAGT ATACGAAAGCACACAAAACACTCCAAGTTTCAGCAGTTTTAGCGCCACCATTAACCCACTTTGCTTGTCTCATGAAAAATCTTTGTTAAA GTTTGTACACAGGTAACAAAAAGTTACTTTAAAAGATATATAAAGGGCTGTAAGCTAATTGTGGTGTCTAGTAAGTAGCATAATGAGATG TGAGGAGTTGGAACTTTGCGTGTTTTGCGTATTTTCATCTGCATTCAGCTTCTTACTCTGGGTTTGTACTCGAGTGTTATTTCTTTACAA ATGCCCTTGTAATTACCACTCTGAAGTCTGCTGACTGTGTCTCTTGAACATACTTAGGATATTCTGCACATTATGGAAAAAGGTAAATTT TAGAAGTTTCTGCTCTACTAACTGTAGATATTTATGACTCTGCGAGTTATCTATTTTTATAACCACCTGTGGTCCATTGTTCATTTTAAT TCACATTTCTTATGAAGTATGGTAACAGGGAGGGAGACACCTAGATTAGCAGCTCAATTTGTACTACTTCAGCCAATCTGTGAATGTAAA AACTACACTGTTGCCTTGCTAGGATCCACCCTCCTATAATATGGAACAAATATCTGAATGAAATCCACCCTAGGAGACGGAGTCAAACTA AACTTGTGGTTTTTCATTTAACTTTTGACTACAGCATGGCCCCATGGCATCCACACCAAGAGGGTGTTGTGATGAGGTGCCGGTGTGCAA AGGGAACTTTAGTTTTTCCACTGGTTCTTATCTGCTAGCCTTTTACATACATGTGTACTATATTTGTTTATAGACTGTAGGTGGATATAT >24951_24951_1_EDEM1-GPD1L_EDEM1_chr3_5229999_ENST00000256497_GPD1L_chr3_32169568_ENST00000282541_length(amino acids)=505AA_BP=170 MQWRALVLGLVLLRLGLHGVLWLVFGLGPSMGFYQRFPLSFGFQRLRSPDGPASPTSGPVGRPGGVSGPSWLQPPGTGAAQSPRKAPRRP GPGMCGPANWGYVLGGRGRGPDEYEKRYSGAFPPQLRAQMRDLARGMFVFGYDNYMAHAFPQDELNPIHCRGRGPDRGDPGSAVAKIIGN NVKKLQKFASTVKMWVFEETVNGRKLTDIINNDHENVKYLPGHKLPENVVAMSNLSEAVQDADLLVFVIPHQFIHRICDEITGRVPKKAL GITLIKGIDEGPEGLKLISDIIREKMGIDISVLMGANIANEVAAEKFCETTIGSKVMENGLLFKELLQTPNFRITVVDDADTVELCGALK NIVAVGAGFCDGLRCGDNTKAAVIRLGLMEMIAFARIFCKGQVSTATFLESCGVADLITTCYGGRNRRVAEAFARTGKTIEELEKEMLNG -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for EDEM1-GPD1L |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for EDEM1-GPD1L |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for EDEM1-GPD1L |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
