
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:EDEM3-HHAT (FusionGDB2 ID:24964) |
Fusion Gene Summary for EDEM3-HHAT |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: EDEM3-HHAT | Fusion gene ID: 24964 | Hgene | Tgene | Gene symbol | EDEM3 | HHAT | Gene ID | 80267 | 55733 |
| Gene name | ER degradation enhancing alpha-mannosidase like protein 3 | hedgehog acyltransferase | |
| Synonyms | C1orf22 | MART2|SKI1|Skn | |
| Cytomap | 1q25.3 | 1q32.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | ER degradation-enhancing alpha-mannosidase-like protein 3ER degradation enhancer, mannosidase alpha-like 3ER degradation-enhancing -mannosidase-like protein 3ER degradation-enhancing alpha-mannosidase-like 3alpha-1,2-mannosidase EDEM3 | protein-cysteine N-palmitoyltransferase HHATmelanoma antigen recognized by T-cells 2skinny hedgehog protein 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9BZQ6 | Q9HCP6 | |
| Ensembl transtripts involved in fusion gene | ENST00000318130, ENST00000367512, ENST00000466392, | ENST00000261458, ENST00000308852, ENST00000367009, ENST00000367010, ENST00000391905, ENST00000537898, ENST00000545154, ENST00000545781, ENST00000413764, ENST00000541565, | |
| Fusion gene scores | * DoF score | 5 X 5 X 4=100 | 6 X 7 X 5=210 |
| # samples | 5 | 7 | |
| ** MAII score | log2(5/100*10)=-1 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/210*10)=-1.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: EDEM3 [Title/Abstract] AND HHAT [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | EDEM3(184677287)-HHAT(210847630), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across EDEM3 (5'-gene) Fusion gene breakpoints across EDEM3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
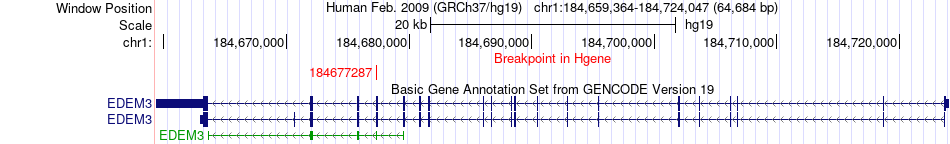 |
 Fusion gene breakpoints across HHAT (3'-gene) Fusion gene breakpoints across HHAT (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
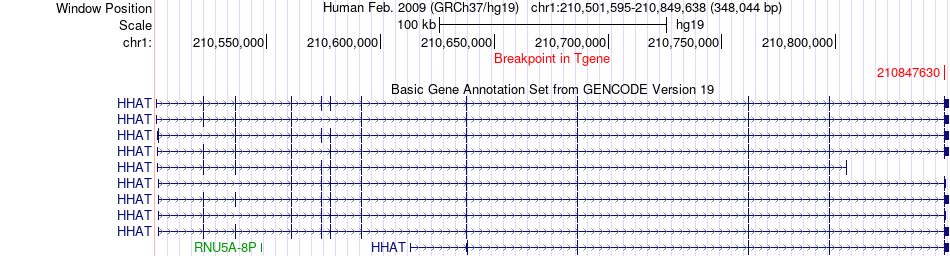 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-EJ-5518-01A | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| ChimerDB4 | PRAD | TCGA-EJ-5518 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
Top |
Fusion Gene ORF analysis for EDEM3-HHAT |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000318130 | ENST00000261458 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5CDS-intron | ENST00000318130 | ENST00000308852 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5CDS-intron | ENST00000318130 | ENST00000367009 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5CDS-intron | ENST00000318130 | ENST00000367010 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5CDS-intron | ENST00000318130 | ENST00000391905 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5CDS-intron | ENST00000318130 | ENST00000537898 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5CDS-intron | ENST00000318130 | ENST00000545154 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5CDS-intron | ENST00000318130 | ENST00000545781 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5CDS-intron | ENST00000367512 | ENST00000261458 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5CDS-intron | ENST00000367512 | ENST00000308852 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5CDS-intron | ENST00000367512 | ENST00000367009 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5CDS-intron | ENST00000367512 | ENST00000367010 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5CDS-intron | ENST00000367512 | ENST00000391905 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5CDS-intron | ENST00000367512 | ENST00000537898 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5CDS-intron | ENST00000367512 | ENST00000545154 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5CDS-intron | ENST00000367512 | ENST00000545781 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5UTR-3CDS | ENST00000466392 | ENST00000413764 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5UTR-3CDS | ENST00000466392 | ENST00000541565 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5UTR-intron | ENST00000466392 | ENST00000261458 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5UTR-intron | ENST00000466392 | ENST00000308852 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5UTR-intron | ENST00000466392 | ENST00000367009 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5UTR-intron | ENST00000466392 | ENST00000367010 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5UTR-intron | ENST00000466392 | ENST00000391905 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5UTR-intron | ENST00000466392 | ENST00000537898 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5UTR-intron | ENST00000466392 | ENST00000545154 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| 5UTR-intron | ENST00000466392 | ENST00000545781 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| In-frame | ENST00000318130 | ENST00000413764 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| In-frame | ENST00000318130 | ENST00000541565 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| In-frame | ENST00000367512 | ENST00000413764 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
| In-frame | ENST00000367512 | ENST00000541565 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000318130 | EDEM3 | chr1 | 184677287 | - | ENST00000541565 | HHAT | chr1 | 210847630 | + | 4313 | 2304 | 66 | 2318 | 750 |
| ENST00000318130 | EDEM3 | chr1 | 184677287 | - | ENST00000413764 | HHAT | chr1 | 210847630 | + | 4313 | 2304 | 66 | 2318 | 750 |
| ENST00000367512 | EDEM3 | chr1 | 184677287 | - | ENST00000541565 | HHAT | chr1 | 210847630 | + | 3967 | 1958 | 8 | 1972 | 654 |
| ENST00000367512 | EDEM3 | chr1 | 184677287 | - | ENST00000413764 | HHAT | chr1 | 210847630 | + | 3967 | 1958 | 8 | 1972 | 654 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000318130 | ENST00000541565 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + | 0.000341423 | 0.9996586 |
| ENST00000318130 | ENST00000413764 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + | 0.000341423 | 0.9996586 |
| ENST00000367512 | ENST00000541565 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + | 0.00016351 | 0.99983644 |
| ENST00000367512 | ENST00000413764 | EDEM3 | chr1 | 184677287 | - | HHAT | chr1 | 210847630 | + | 0.00016351 | 0.99983644 |
Top |
Fusion Genomic Features for EDEM3-HHAT |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| EDEM3 | chr1 | 184677286 | - | HHAT | chr1 | 210847629 | + | 2.76E-05 | 0.99997234 |
| EDEM3 | chr1 | 184677286 | - | HHAT | chr1 | 210847629 | + | 2.76E-05 | 0.99997234 |
| EDEM3 | chr1 | 184677286 | - | HHAT | chr1 | 210847629 | + | 2.76E-05 | 0.99997234 |
| EDEM3 | chr1 | 184677286 | - | HHAT | chr1 | 210847629 | + | 2.76E-05 | 0.99997234 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
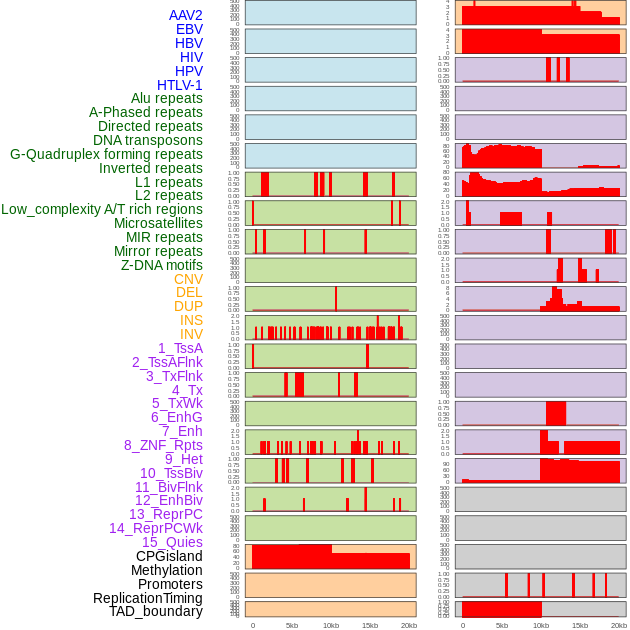 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
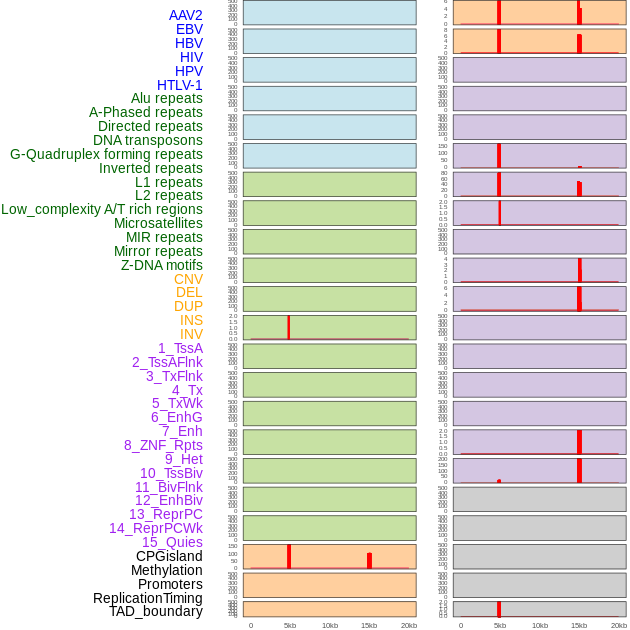 |
Top |
Fusion Protein Features for EDEM3-HHAT |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:184677287/chr1:210847630) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| EDEM3 | HHAT |
| FUNCTION: Involved in endoplasmic reticulum-associated degradation (ERAD). Accelerates the glycoprotein ERAD by proteasomes, by catalyzing mannose trimming from Man8GlcNAc2 to Man7GlcNAc2 in the N-glycans. Seems to have alpha 1,2-mannosidase activity (By similarity). {ECO:0000250, ECO:0000269|PubMed:25092655}. | FUNCTION: Negatively regulates N-terminal palmitoylation of SHH by HHAT/SKN. {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 203_217 | 0 | 519.0 | Intramembrane | Ontology_term=ECO:0000255 | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 95_119 | 0 | 519.0 | Intramembrane | Ontology_term=ECO:0000255 | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 448_455 | 0 | 519.0 | Region | GTP-binding | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 448_455 | 398 | 429.0 | Region | GTP-binding | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 448_455 | 326 | 357.0 | Region | GTP-binding | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 482_493 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 482_493 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 120_131 | 0 | 519.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 149_162 | 0 | 519.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 184_202 | 0 | 519.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 1_5 | 0 | 519.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 218_243 | 0 | 519.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 23_67 | 0 | 519.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 272_281 | 0 | 519.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 311_363 | 0 | 519.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 381_383 | 0 | 519.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 400_427 | 0 | 519.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 449_462 | 0 | 519.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 482_493 | 0 | 519.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 85_94 | 0 | 519.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 482_493 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 400_427 | 398 | 429.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 449_462 | 398 | 429.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 482_493 | 398 | 429.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 381_383 | 326 | 357.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 400_427 | 326 | 357.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 449_462 | 326 | 357.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 482_493 | 326 | 357.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 482_493 | 464 | 495.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 463_481 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 463_481 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 132_148 | 0 | 519.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 163_183 | 0 | 519.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 244_271 | 0 | 519.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 282_310 | 0 | 519.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 364_380 | 0 | 519.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 384_399 | 0 | 519.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 428_448 | 0 | 519.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 463_481 | 0 | 519.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 68_84 | 0 | 519.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000391905 | 0 | 12 | 6_22 | 0 | 519.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 463_481 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 428_448 | 398 | 429.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 463_481 | 398 | 429.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 364_380 | 326 | 357.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 384_399 | 326 | 357.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 428_448 | 326 | 357.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 463_481 | 326 | 357.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 463_481 | 464 | 495.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EDEM3 | chr1:184677287 | chr1:210847630 | ENST00000318130 | - | 17 | 20 | 674_779 | 679 | 933.0 | Domain | Note=PA |
| Hgene | EDEM3 | chr1:184677287 | chr1:210847630 | ENST00000318130 | - | 17 | 20 | 929_932 | 679 | 933.0 | Motif | Prevents secretion from ER |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 203_217 | 463 | 494.0 | Intramembrane | Ontology_term=ECO:0000255 | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 95_119 | 463 | 494.0 | Intramembrane | Ontology_term=ECO:0000255 | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 203_217 | 463 | 494.0 | Intramembrane | Ontology_term=ECO:0000255 | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 95_119 | 463 | 494.0 | Intramembrane | Ontology_term=ECO:0000255 | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 203_217 | 463 | 494.0 | Intramembrane | Ontology_term=ECO:0000255 | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 95_119 | 463 | 494.0 | Intramembrane | Ontology_term=ECO:0000255 | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 203_217 | 398 | 429.0 | Intramembrane | Ontology_term=ECO:0000255 | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 95_119 | 398 | 429.0 | Intramembrane | Ontology_term=ECO:0000255 | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 203_217 | 326 | 357.0 | Intramembrane | Ontology_term=ECO:0000255 | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 95_119 | 326 | 357.0 | Intramembrane | Ontology_term=ECO:0000255 | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 203_217 | 464 | 495.0 | Intramembrane | Ontology_term=ECO:0000255 | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 95_119 | 464 | 495.0 | Intramembrane | Ontology_term=ECO:0000255 | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 448_455 | 463 | 494.0 | Region | GTP-binding | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 448_455 | 463 | 494.0 | Region | GTP-binding | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 448_455 | 463 | 494.0 | Region | GTP-binding | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 448_455 | 464 | 495.0 | Region | GTP-binding | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 120_131 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 149_162 | 463 | 494.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 184_202 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 1_5 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 218_243 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 23_67 | 463 | 494.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 272_281 | 463 | 494.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 311_363 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 381_383 | 463 | 494.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 400_427 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 449_462 | 463 | 494.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 85_94 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 120_131 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 149_162 | 463 | 494.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 184_202 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 1_5 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 218_243 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 23_67 | 463 | 494.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 272_281 | 463 | 494.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 311_363 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 381_383 | 463 | 494.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 400_427 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 449_462 | 463 | 494.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 85_94 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 120_131 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 149_162 | 463 | 494.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 184_202 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 1_5 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 218_243 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 23_67 | 463 | 494.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 272_281 | 463 | 494.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 311_363 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 381_383 | 463 | 494.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 400_427 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 449_462 | 463 | 494.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 85_94 | 463 | 494.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 120_131 | 398 | 429.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 149_162 | 398 | 429.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 184_202 | 398 | 429.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 1_5 | 398 | 429.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 218_243 | 398 | 429.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 23_67 | 398 | 429.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 272_281 | 398 | 429.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 311_363 | 398 | 429.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 381_383 | 398 | 429.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 85_94 | 398 | 429.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 120_131 | 326 | 357.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 149_162 | 326 | 357.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 184_202 | 326 | 357.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 1_5 | 326 | 357.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 218_243 | 326 | 357.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 23_67 | 326 | 357.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 272_281 | 326 | 357.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 311_363 | 326 | 357.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 85_94 | 326 | 357.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 120_131 | 464 | 495.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 149_162 | 464 | 495.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 184_202 | 464 | 495.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 1_5 | 464 | 495.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 218_243 | 464 | 495.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 23_67 | 464 | 495.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 272_281 | 464 | 495.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 311_363 | 464 | 495.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 381_383 | 464 | 495.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 400_427 | 464 | 495.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 449_462 | 464 | 495.0 | Topological domain | Lumenal | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 85_94 | 464 | 495.0 | Topological domain | Cytoplasmic | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 132_148 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 163_183 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 244_271 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 282_310 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 364_380 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 384_399 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 428_448 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 68_84 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000261458 | 10 | 12 | 6_22 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 132_148 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 163_183 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 244_271 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 282_310 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 364_380 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 384_399 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 428_448 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 68_84 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000367010 | 10 | 12 | 6_22 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 132_148 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 163_183 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 244_271 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 282_310 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 364_380 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 384_399 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 428_448 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 68_84 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000413764 | 10 | 12 | 6_22 | 463 | 494.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 132_148 | 398 | 429.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 163_183 | 398 | 429.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 244_271 | 398 | 429.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 282_310 | 398 | 429.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 364_380 | 398 | 429.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 384_399 | 398 | 429.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 68_84 | 398 | 429.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000537898 | 9 | 11 | 6_22 | 398 | 429.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 132_148 | 326 | 357.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 163_183 | 326 | 357.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 244_271 | 326 | 357.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 282_310 | 326 | 357.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 68_84 | 326 | 357.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000541565 | 8 | 10 | 6_22 | 326 | 357.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 132_148 | 464 | 495.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 163_183 | 464 | 495.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 244_271 | 464 | 495.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 282_310 | 464 | 495.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 364_380 | 464 | 495.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 384_399 | 464 | 495.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 428_448 | 464 | 495.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 68_84 | 464 | 495.0 | Transmembrane | Helical | |
| Tgene | HHAT | chr1:184677287 | chr1:210847630 | ENST00000545154 | 9 | 11 | 6_22 | 464 | 495.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for EDEM3-HHAT |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >24964_24964_1_EDEM3-HHAT_EDEM3_chr1_184677287_ENST00000318130_HHAT_chr1_210847630_ENST00000413764_length(transcript)=4313nt_BP=2304nt AGTAACGTTGGGGAGGCGGTTTCCTAGACTACGACACCGGAAAGCTGGGGGAGGCGCTCCGGGATACTGAGGGCGGAGGGCGGTGGCAGC GCTGGCGCTGGGGACCGGCTTGGTGGCTTCGGGAAACAGTTTGGCGCCGGCGGCCGTCCGTGTTACTCCGCATCCCGCCCCGTCTCGGCA CGGCTAGCAGCCCCCTGGCCACCAGCGTCCAGCAATGTGTCTCACCGGCCGGGCGTAGCAGCTGCGCGTGCGCGGAACCGCGGGGCCATG AGCGAAGCCGGCGGCCGGGGCTGTGGGTCCCCGGTTCCCCAGCGAGCGCGATGGAGACTAGTGGCGGCGACGGCCGCGTTCTGCCTGGTG TCGGCCACCTCCGTGTGGACGGCGGGGGCCGAGCCCATGAGTAGGGAGGAGAAACAGAAGCTTGGGAATCAAGTACTGGAAATGTTTGAT CATGCTTATGGTAACTATATGGAACATGCTTACCCTGCTGATGAACTCATGCCTTTAACCTGTAGAGGTCGAGTTAGAGGCCAAGAGCCA AGTCGCGGTGACGTTGATGATGCCTTGGGAAAATTTTCTCTGACACTGATTGATTCTTTGGACACTCTTGTGGTTTTAAATAAAACTAAA GAATTTGAAGATGCAGTGAGAAAAGTTTTAAGAGATGTTAATTTAGATAACGATGTAGTCGTATCAGTCTTTGAAACAAACATCAGAGTT CTTGGGGGTCTTTTGGGTGGGCACTCCCTGGCAATCATGCTGAAAGAAAAAGGTGAATATATGCAGTGGTACAATGATGAACTTCTCCAA ATGGCAAAGCAGTTAGGTTACAAACTTTTACCGGCTTTCAACACTACCAGTGGCCTTCCTTATCCAAGAATTAATTTAAAGTTTGGCATC AGAAAACCAGAAGCTCGGACAGGAACTGAGACAGATACCTGTACAGCTTGTGCAGGTACCTTGATCCTTGAATTTGCTGCTTTAAGTCGA TTCACAGGAGCAACAATATTTGAGGAATATGCCAGAAAAGCTCTTGATTTTCTCTGGGAAAAAAGACAGCGAAGTAGTAATTTAGTGGGC GTGACTATAAATATTCATACTGGAGATTGGGTACGAAAAGATAGTGGAGTTGGAGCAGGGATTGATTCATATTATGAATATCTGTTGAAA GCCTATGTCTTGCTTGGAGATGACAGTTTTCTGGAAAGATTTAACACACACTATGATGCCATAATGAGGTATATTAGCCAGCCACCTCTT CTACTTGATGTGCATATCCACAAACCAATGCTGAATGCTCGGACTTGGATGGATGCTTTGCTTGCCTTCTTCCCAGGCTTGCAGGTGTTA AAGGGGGATATTAGACCTGCTATTGAAACTCATGAAATGTTATATCAGGTGATTAAAAAACACAATTTTCTACCAGAGGCATTTACCACA GATTTCAGAGTACACTGGGCTCAACATCCTTTAAGGCCAGAATTTGCAGAAAGTACCTACTTCTTATATAAAGCTACAGGAGATCCTTAC TACCTTGAAGTAGGGAAGACACTTATTGAAAATTTAAATAAATATGCTAGAGTGCCTTGCGGATTTGCTGCCATGAAGGATGTTCGTACT GGAAGTCATGAGGACAGAATGGATTCTTTCTTCTTGGCTGAAATGTTTAAATATCTTTACCTGTTATTTGCTGATAAAGAAGACATTATT TTTGACATAGAAGATTACATCTTTACAACAGAAGCTCATCTGTTACCTCTTTGGCTCTCTACTACAAATCAAAGCATCTCTAAAAAGAAC ACAACCTCGGAATATACAGAACTGGATGACAGTAACTTCGATTGGACTTGTCCAAATACTCAGATCCTCTTTCCTAATGACCCATTGTAT GCTCAAAGTATTCGTGAGCCCTTGAAAAATGTGGTGGATAAGAGCTGTCCTAGAGGCATCATCAGAGTAGAGGAGAGTTTCAGGAGTGGA GCTAAACCCCCTCTGAGAGCCAGAGATTTCATGGCCACTAACCCTGAGCATTTAGAAATCCTGAAGAAGATGGGGGTGAGTTTGATTCAC CTCAAAGATGGGAGAGTCCAGTTGGTCCAACATGCAATCCAAGCTGCTAGTTCAATCGACGCTGAAGATGGGTTGAGGTTCATGCAGGAG ATGATTGAATTGTCAAGTCAGCAACAAAAAGAACAGCAGCTGCCTCCACGAGCTGTACAAATTGTTTCCCACCCATTTTTTGGCAGGGTA GTATTGACTGCTGGACCAGCTCAGTTTGGGCTGGATCTGTCTAAACATAAAGAGGCTGGCCTTGGGTGACCCTCTCTGTCCTGGGATTCC TGTACTGCTACTCCCACGTGGGCATTGCCTGGGCCCAGACCTACGCCACGGACTAATGCTGTTGGGCCCAGGCCAGTCCTTGTTGCTGGC CTCCAAGGCAAATAGTGCTTCACCCTGACCTCTCACTCCAGGACAGCCTCTAAGGGATTTGATCTGCTCATCTTCAGTTGAATGCCCTCA CTCCAAGACTGGATGCTGGATCTCATAGAAAATTCACAGCCAGACAATCTTCTAATCTGGAGTCTTTGAGATCTTCTACCCCAACTCATC ATTTTCCTATTGAGGAAACGGGTCCAGGGCAGTCGTGTGTCTTACCCAGCTACACAGGGTGACATTTTGGTCTAGAACCTAGTCTCATGA GCCCTTTGCATTCTTTCCCCTTAAGCAAGAATAGAATGTAGTGGAAATTTATTGATTGAGACACAGAAATCCTGATTAGTGATAGGCCTG TCTTCTGGGCAATATTTCTAAAATATTTGAGTGACATTGATGTTTAAGTGACCTCCTTCATCACATCCTGCAGTATCTCCAGAAGCAGCA CTAGGGTTTGAGTTTCATACTTCAGCCCCTCTGTAGGAATGAGACCAAGCCACAGCTGTCTTTTGAATTACGTAGTCGAAGAAGACGGTA GCAGCCCTGTAGCATTCTAAGGCATCTATACCCAAGGAGTCCTGTGATTTGAGCTTCAGCAGGGTGATCTACATTTGGGTGCTTGTTTCT GAGATTTGCAGAGAAGTATAACATGGTAGTTCCTCTACCTTACAGTTAATCGTTTCTTAATAAAGAAGCAGAATTTAGAAACCACAGGAT AGTGTACCCACAGATGGGTGTTATCAAGGCCAGTCATGAGGATGGTGTCCTGGAGTCTTGTCCACCCTCTCCATACAAGTCTCAAAAGTC ATCCTCCTACTCAGTGATTCACGTTTAGTGGTTTATATTATTAAGGTTTGATTCAAACAGAGCCTTTTCTGTCCTGTAGATAATCTACAT GTTTGTAGAATTATTTTGAATATGTTTGAGGAAAATGTTTAAAATCTAAATATACTCACATAACTTGATTATTCACTCCTCTGAAAAGAT GCTGGATAGGCTACCAAAGTTCCCAAGTGGTAGATAATTCAGAAGACTTGTTTGAATTTGGATTTTTTTTTTTTTTGGAGTGGGGAAGGG TATAAAGGAGGCTTAAAATTTGAATCCATAATATATCTAATTACAGGAGAATTTACAACATCTCAAGTACGTAAATTAAGTTGTCATTGA GTGAAAGGTTCACTTGGACCTAGTGCTGCCTCCTGTTTATTACATAGCATGGCCCTTATGTCTTGAGTTGAGGTTATCATCTCAATGAGG CTTAAGCTCCTAGAGTACAGGACCATTTTGTTGATTGTCTTTCTTCATAGCTTCTCTGCTTGGCAAAGAGATGGGAGGGGGCCAGATACT GACTACCTGGGGTAGGCACATTATGTGTTAAAGCAAGACAGAGGCCAGAGAGGGGCAGGTAGACCTGCATAGCAGCAGCCTCAGCAGCTG TCTTGGTAAAGGAGAGAGAGAGACATGGGGCCAGTAATTCCGGGGTGCTCAGAAGTTTTAGGAGGGAATGAGCCTCAGGGAGGAGTGAGC ACCTAAATGAACGCAGTAAACCTTCATGGACCAACAGTGATTGAGGATTTGTGGGCAGCCAGAGGGAGTCTGACTGAAGTTTACTTGGAA AGAAAGGGCTTGCTAAGAAAAAAGGGAGTAAAAATGATGATAGGGAAGTGTCTAATGTATGTGCACATATAAGTAATACAAAAGTTTTGA GCTCTTCCAAGTATACCATTTATATACAAACAAATAGGTTTATTCATTCATTAAACTACTTTGGAAGCGTCAGTGGATATATTTGAAAGT >24964_24964_1_EDEM3-HHAT_EDEM3_chr1_184677287_ENST00000318130_HHAT_chr1_210847630_ENST00000413764_length(amino acids)=750AA_BP= MRAEGGGSAGAGDRLGGFGKQFGAGGRPCYSASRPVSARLAAPWPPASSNVSHRPGVAAARARNRGAMSEAGGRGCGSPVPQRARWRLVA ATAAFCLVSATSVWTAGAEPMSREEKQKLGNQVLEMFDHAYGNYMEHAYPADELMPLTCRGRVRGQEPSRGDVDDALGKFSLTLIDSLDT LVVLNKTKEFEDAVRKVLRDVNLDNDVVVSVFETNIRVLGGLLGGHSLAIMLKEKGEYMQWYNDELLQMAKQLGYKLLPAFNTTSGLPYP RINLKFGIRKPEARTGTETDTCTACAGTLILEFAALSRFTGATIFEEYARKALDFLWEKRQRSSNLVGVTINIHTGDWVRKDSGVGAGID SYYEYLLKAYVLLGDDSFLERFNTHYDAIMRYISQPPLLLDVHIHKPMLNARTWMDALLAFFPGLQVLKGDIRPAIETHEMLYQVIKKHN FLPEAFTTDFRVHWAQHPLRPEFAESTYFLYKATGDPYYLEVGKTLIENLNKYARVPCGFAAMKDVRTGSHEDRMDSFFLAEMFKYLYLL FADKEDIIFDIEDYIFTTEAHLLPLWLSTTNQSISKKNTTSEYTELDDSNFDWTCPNTQILFPNDPLYAQSIREPLKNVVDKSCPRGIIR VEESFRSGAKPPLRARDFMATNPEHLEILKKMGVSLIHLKDGRVQLVQHAIQAASSIDAEDGLRFMQEMIELSSQQQKEQQLPPRAVQIV -------------------------------------------------------------- >24964_24964_2_EDEM3-HHAT_EDEM3_chr1_184677287_ENST00000318130_HHAT_chr1_210847630_ENST00000541565_length(transcript)=4313nt_BP=2304nt AGTAACGTTGGGGAGGCGGTTTCCTAGACTACGACACCGGAAAGCTGGGGGAGGCGCTCCGGGATACTGAGGGCGGAGGGCGGTGGCAGC GCTGGCGCTGGGGACCGGCTTGGTGGCTTCGGGAAACAGTTTGGCGCCGGCGGCCGTCCGTGTTACTCCGCATCCCGCCCCGTCTCGGCA CGGCTAGCAGCCCCCTGGCCACCAGCGTCCAGCAATGTGTCTCACCGGCCGGGCGTAGCAGCTGCGCGTGCGCGGAACCGCGGGGCCATG AGCGAAGCCGGCGGCCGGGGCTGTGGGTCCCCGGTTCCCCAGCGAGCGCGATGGAGACTAGTGGCGGCGACGGCCGCGTTCTGCCTGGTG TCGGCCACCTCCGTGTGGACGGCGGGGGCCGAGCCCATGAGTAGGGAGGAGAAACAGAAGCTTGGGAATCAAGTACTGGAAATGTTTGAT CATGCTTATGGTAACTATATGGAACATGCTTACCCTGCTGATGAACTCATGCCTTTAACCTGTAGAGGTCGAGTTAGAGGCCAAGAGCCA AGTCGCGGTGACGTTGATGATGCCTTGGGAAAATTTTCTCTGACACTGATTGATTCTTTGGACACTCTTGTGGTTTTAAATAAAACTAAA GAATTTGAAGATGCAGTGAGAAAAGTTTTAAGAGATGTTAATTTAGATAACGATGTAGTCGTATCAGTCTTTGAAACAAACATCAGAGTT CTTGGGGGTCTTTTGGGTGGGCACTCCCTGGCAATCATGCTGAAAGAAAAAGGTGAATATATGCAGTGGTACAATGATGAACTTCTCCAA ATGGCAAAGCAGTTAGGTTACAAACTTTTACCGGCTTTCAACACTACCAGTGGCCTTCCTTATCCAAGAATTAATTTAAAGTTTGGCATC AGAAAACCAGAAGCTCGGACAGGAACTGAGACAGATACCTGTACAGCTTGTGCAGGTACCTTGATCCTTGAATTTGCTGCTTTAAGTCGA TTCACAGGAGCAACAATATTTGAGGAATATGCCAGAAAAGCTCTTGATTTTCTCTGGGAAAAAAGACAGCGAAGTAGTAATTTAGTGGGC GTGACTATAAATATTCATACTGGAGATTGGGTACGAAAAGATAGTGGAGTTGGAGCAGGGATTGATTCATATTATGAATATCTGTTGAAA GCCTATGTCTTGCTTGGAGATGACAGTTTTCTGGAAAGATTTAACACACACTATGATGCCATAATGAGGTATATTAGCCAGCCACCTCTT CTACTTGATGTGCATATCCACAAACCAATGCTGAATGCTCGGACTTGGATGGATGCTTTGCTTGCCTTCTTCCCAGGCTTGCAGGTGTTA AAGGGGGATATTAGACCTGCTATTGAAACTCATGAAATGTTATATCAGGTGATTAAAAAACACAATTTTCTACCAGAGGCATTTACCACA GATTTCAGAGTACACTGGGCTCAACATCCTTTAAGGCCAGAATTTGCAGAAAGTACCTACTTCTTATATAAAGCTACAGGAGATCCTTAC TACCTTGAAGTAGGGAAGACACTTATTGAAAATTTAAATAAATATGCTAGAGTGCCTTGCGGATTTGCTGCCATGAAGGATGTTCGTACT GGAAGTCATGAGGACAGAATGGATTCTTTCTTCTTGGCTGAAATGTTTAAATATCTTTACCTGTTATTTGCTGATAAAGAAGACATTATT TTTGACATAGAAGATTACATCTTTACAACAGAAGCTCATCTGTTACCTCTTTGGCTCTCTACTACAAATCAAAGCATCTCTAAAAAGAAC ACAACCTCGGAATATACAGAACTGGATGACAGTAACTTCGATTGGACTTGTCCAAATACTCAGATCCTCTTTCCTAATGACCCATTGTAT GCTCAAAGTATTCGTGAGCCCTTGAAAAATGTGGTGGATAAGAGCTGTCCTAGAGGCATCATCAGAGTAGAGGAGAGTTTCAGGAGTGGA GCTAAACCCCCTCTGAGAGCCAGAGATTTCATGGCCACTAACCCTGAGCATTTAGAAATCCTGAAGAAGATGGGGGTGAGTTTGATTCAC CTCAAAGATGGGAGAGTCCAGTTGGTCCAACATGCAATCCAAGCTGCTAGTTCAATCGACGCTGAAGATGGGTTGAGGTTCATGCAGGAG ATGATTGAATTGTCAAGTCAGCAACAAAAAGAACAGCAGCTGCCTCCACGAGCTGTACAAATTGTTTCCCACCCATTTTTTGGCAGGGTA GTATTGACTGCTGGACCAGCTCAGTTTGGGCTGGATCTGTCTAAACATAAAGAGGCTGGCCTTGGGTGACCCTCTCTGTCCTGGGATTCC TGTACTGCTACTCCCACGTGGGCATTGCCTGGGCCCAGACCTACGCCACGGACTAATGCTGTTGGGCCCAGGCCAGTCCTTGTTGCTGGC CTCCAAGGCAAATAGTGCTTCACCCTGACCTCTCACTCCAGGACAGCCTCTAAGGGATTTGATCTGCTCATCTTCAGTTGAATGCCCTCA CTCCAAGACTGGATGCTGGATCTCATAGAAAATTCACAGCCAGACAATCTTCTAATCTGGAGTCTTTGAGATCTTCTACCCCAACTCATC ATTTTCCTATTGAGGAAACGGGTCCAGGGCAGTCGTGTGTCTTACCCAGCTACACAGGGTGACATTTTGGTCTAGAACCTAGTCTCATGA GCCCTTTGCATTCTTTCCCCTTAAGCAAGAATAGAATGTAGTGGAAATTTATTGATTGAGACACAGAAATCCTGATTAGTGATAGGCCTG TCTTCTGGGCAATATTTCTAAAATATTTGAGTGACATTGATGTTTAAGTGACCTCCTTCATCACATCCTGCAGTATCTCCAGAAGCAGCA CTAGGGTTTGAGTTTCATACTTCAGCCCCTCTGTAGGAATGAGACCAAGCCACAGCTGTCTTTTGAATTACGTAGTCGAAGAAGACGGTA GCAGCCCTGTAGCATTCTAAGGCATCTATACCCAAGGAGTCCTGTGATTTGAGCTTCAGCAGGGTGATCTACATTTGGGTGCTTGTTTCT GAGATTTGCAGAGAAGTATAACATGGTAGTTCCTCTACCTTACAGTTAATCGTTTCTTAATAAAGAAGCAGAATTTAGAAACCACAGGAT AGTGTACCCACAGATGGGTGTTATCAAGGCCAGTCATGAGGATGGTGTCCTGGAGTCTTGTCCACCCTCTCCATACAAGTCTCAAAAGTC ATCCTCCTACTCAGTGATTCACGTTTAGTGGTTTATATTATTAAGGTTTGATTCAAACAGAGCCTTTTCTGTCCTGTAGATAATCTACAT GTTTGTAGAATTATTTTGAATATGTTTGAGGAAAATGTTTAAAATCTAAATATACTCACATAACTTGATTATTCACTCCTCTGAAAAGAT GCTGGATAGGCTACCAAAGTTCCCAAGTGGTAGATAATTCAGAAGACTTGTTTGAATTTGGATTTTTTTTTTTTTTGGAGTGGGGAAGGG TATAAAGGAGGCTTAAAATTTGAATCCATAATATATCTAATTACAGGAGAATTTACAACATCTCAAGTACGTAAATTAAGTTGTCATTGA GTGAAAGGTTCACTTGGACCTAGTGCTGCCTCCTGTTTATTACATAGCATGGCCCTTATGTCTTGAGTTGAGGTTATCATCTCAATGAGG CTTAAGCTCCTAGAGTACAGGACCATTTTGTTGATTGTCTTTCTTCATAGCTTCTCTGCTTGGCAAAGAGATGGGAGGGGGCCAGATACT GACTACCTGGGGTAGGCACATTATGTGTTAAAGCAAGACAGAGGCCAGAGAGGGGCAGGTAGACCTGCATAGCAGCAGCCTCAGCAGCTG TCTTGGTAAAGGAGAGAGAGAGACATGGGGCCAGTAATTCCGGGGTGCTCAGAAGTTTTAGGAGGGAATGAGCCTCAGGGAGGAGTGAGC ACCTAAATGAACGCAGTAAACCTTCATGGACCAACAGTGATTGAGGATTTGTGGGCAGCCAGAGGGAGTCTGACTGAAGTTTACTTGGAA AGAAAGGGCTTGCTAAGAAAAAAGGGAGTAAAAATGATGATAGGGAAGTGTCTAATGTATGTGCACATATAAGTAATACAAAAGTTTTGA GCTCTTCCAAGTATACCATTTATATACAAACAAATAGGTTTATTCATTCATTAAACTACTTTGGAAGCGTCAGTGGATATATTTGAAAGT >24964_24964_2_EDEM3-HHAT_EDEM3_chr1_184677287_ENST00000318130_HHAT_chr1_210847630_ENST00000541565_length(amino acids)=750AA_BP= MRAEGGGSAGAGDRLGGFGKQFGAGGRPCYSASRPVSARLAAPWPPASSNVSHRPGVAAARARNRGAMSEAGGRGCGSPVPQRARWRLVA ATAAFCLVSATSVWTAGAEPMSREEKQKLGNQVLEMFDHAYGNYMEHAYPADELMPLTCRGRVRGQEPSRGDVDDALGKFSLTLIDSLDT LVVLNKTKEFEDAVRKVLRDVNLDNDVVVSVFETNIRVLGGLLGGHSLAIMLKEKGEYMQWYNDELLQMAKQLGYKLLPAFNTTSGLPYP RINLKFGIRKPEARTGTETDTCTACAGTLILEFAALSRFTGATIFEEYARKALDFLWEKRQRSSNLVGVTINIHTGDWVRKDSGVGAGID SYYEYLLKAYVLLGDDSFLERFNTHYDAIMRYISQPPLLLDVHIHKPMLNARTWMDALLAFFPGLQVLKGDIRPAIETHEMLYQVIKKHN FLPEAFTTDFRVHWAQHPLRPEFAESTYFLYKATGDPYYLEVGKTLIENLNKYARVPCGFAAMKDVRTGSHEDRMDSFFLAEMFKYLYLL FADKEDIIFDIEDYIFTTEAHLLPLWLSTTNQSISKKNTTSEYTELDDSNFDWTCPNTQILFPNDPLYAQSIREPLKNVVDKSCPRGIIR VEESFRSGAKPPLRARDFMATNPEHLEILKKMGVSLIHLKDGRVQLVQHAIQAASSIDAEDGLRFMQEMIELSSQQQKEQQLPPRAVQIV -------------------------------------------------------------- >24964_24964_3_EDEM3-HHAT_EDEM3_chr1_184677287_ENST00000367512_HHAT_chr1_210847630_ENST00000413764_length(transcript)=3967nt_BP=1958nt CGTTCTGCCTGGTGTCGGCCACCTCCGTGTGGACGGCGGGGGCCGAGCCCATGAGTAGGGAGGAGAAACAGAAGCTTGGGAATCAAGTAC TGGAAATGTTTGATCATGCTTATGGTAACTATATGGAACATGCTTACCCTGCTGATGAACTCATGCCTTTAACCTGTAGAGGTCGAGTTA GAGGCCAAGAGCCAAGTCGCGGTGACGTTGATGATGCCTTGGGAAAATTTTCTCTGACACTGATTGATTCTTTGGACACTCTTGTGGTTT TAAATAAAACTAAAGAATTTGAAGATGCAGTGAGAAAAGTTTTAAGAGATGTTAATTTAGATAACGATGTAGTCGTATCAGTCTTTGAAA CAAACATCAGAGTTCTTGGGGGTCTTTTGGGTGGGCACTCCCTGGCAATCATGCTGAAAGAAAAAGGTGAATATATGCAGTGGTACAATG ATGAACTTCTCCAAATGGCAAAGCAGTTAGGTTACAAACTTTTACCGGCTTTCAACACTACCAGTGGCCTTCCTTATCCAAGAATTAATT TAAAGTTTGGCATCAGAAAACCAGAAGCTCGGACAGGAACTGAGACAGATACCTGTACAGCTTGTGCAGGTACCTTGATCCTTGAATTTG CTGCTTTAAGTCGATTCACAGGAGCAACAATATTTGAGGAATATGCCAGAAAAGCTCTTGATTTTCTCTGGGAAAAAAGACAGCGAAGTA GTAATTTAGTGGGCGTGACTATAAATATTCATACTGGAGATTGGGTACGAAAAGATAGTGGAGTTGGAGCAGGGATTGATTCATATTATG AATATCTGTTGAAAGCCTATGTCTTGCTTGGAGATGACAGTTTTCTGGAAAGATTTAACACACACTATGATGCCATAATGAGGTATATTA GCCAGCCACCTCTTCTACTTGATGTGCATATCCACAAACCAATGCTGAATGCTCGGACTTGGATGGATGCTTTGCTTGCCTTCTTCCCAG GCTTGCAGGTGTTAAAGGGGGATATTAGACCTGCTATTGAAACTCATGAAATGTTATATCAGGTGATTAAAAAACACAATTTTCTACCAG AGGCATTTACCACAGATTTCAGAGTACACTGGGCTCAACATCCTTTAAGGCCAGAATTTGCAGAAAGTACCTACTTCTTATATAAAGCTA CAGGAGATCCTTACTACCTTGAAGTAGGGAAGACACTTATTGAAAATTTAAATAAATATGCTAGAGTGCCTTGCGGATTTGCTGCCATGA AGGATGTTCGTACTGGAAGTCATGAGGACAGAATGGATTCTTTCTTCTTGGCTGAAATGTTTAAATATCTTTACCTGTTATTTGCTGATA AAGAAGACATTATTTTTGACATAGAAGATTACATCTTTACAACAGAAGCTCATCTGTTACCTCTTTGGCTCTCTACTACAAATCAAAGCA TCTCTAAAAAGAACACAACCTCGGAATATACAGAACTGGATGACAGTAACTTCGATTGGACTTGTCCAAATACTCAGATCCTCTTTCCTA ATGACCCATTGTATGCTCAAAGTATTCGTGAGCCCTTGAAAAATGTGGTGGATAAGAGCTGTCCTAGAGGCATCATCAGAGTAGAGGAGA GTTTCAGGAGTGGAGCTAAACCCCCTCTGAGAGCCAGAGATTTCATGGCCACTAACCCTGAGCATTTAGAAATCCTGAAGAAGATGGGGG TGAGTTTGATTCACCTCAAAGATGGGAGAGTCCAGTTGGTCCAACATGCAATCCAAGCTGCTAGTTCAATCGACGCTGAAGATGGGTTGA GGTTCATGCAGGAGATGATTGAATTGTCAAGTCAGCAACAAAAAGAACAGCAGCTGCCTCCACGAGCTGTACAAATTGTTTCCCACCCAT TTTTTGGCAGGGTAGTATTGACTGCTGGACCAGCTCAGTTTGGGCTGGATCTGTCTAAACATAAAGAGGCTGGCCTTGGGTGACCCTCTC TGTCCTGGGATTCCTGTACTGCTACTCCCACGTGGGCATTGCCTGGGCCCAGACCTACGCCACGGACTAATGCTGTTGGGCCCAGGCCAG TCCTTGTTGCTGGCCTCCAAGGCAAATAGTGCTTCACCCTGACCTCTCACTCCAGGACAGCCTCTAAGGGATTTGATCTGCTCATCTTCA GTTGAATGCCCTCACTCCAAGACTGGATGCTGGATCTCATAGAAAATTCACAGCCAGACAATCTTCTAATCTGGAGTCTTTGAGATCTTC TACCCCAACTCATCATTTTCCTATTGAGGAAACGGGTCCAGGGCAGTCGTGTGTCTTACCCAGCTACACAGGGTGACATTTTGGTCTAGA ACCTAGTCTCATGAGCCCTTTGCATTCTTTCCCCTTAAGCAAGAATAGAATGTAGTGGAAATTTATTGATTGAGACACAGAAATCCTGAT TAGTGATAGGCCTGTCTTCTGGGCAATATTTCTAAAATATTTGAGTGACATTGATGTTTAAGTGACCTCCTTCATCACATCCTGCAGTAT CTCCAGAAGCAGCACTAGGGTTTGAGTTTCATACTTCAGCCCCTCTGTAGGAATGAGACCAAGCCACAGCTGTCTTTTGAATTACGTAGT CGAAGAAGACGGTAGCAGCCCTGTAGCATTCTAAGGCATCTATACCCAAGGAGTCCTGTGATTTGAGCTTCAGCAGGGTGATCTACATTT GGGTGCTTGTTTCTGAGATTTGCAGAGAAGTATAACATGGTAGTTCCTCTACCTTACAGTTAATCGTTTCTTAATAAAGAAGCAGAATTT AGAAACCACAGGATAGTGTACCCACAGATGGGTGTTATCAAGGCCAGTCATGAGGATGGTGTCCTGGAGTCTTGTCCACCCTCTCCATAC AAGTCTCAAAAGTCATCCTCCTACTCAGTGATTCACGTTTAGTGGTTTATATTATTAAGGTTTGATTCAAACAGAGCCTTTTCTGTCCTG TAGATAATCTACATGTTTGTAGAATTATTTTGAATATGTTTGAGGAAAATGTTTAAAATCTAAATATACTCACATAACTTGATTATTCAC TCCTCTGAAAAGATGCTGGATAGGCTACCAAAGTTCCCAAGTGGTAGATAATTCAGAAGACTTGTTTGAATTTGGATTTTTTTTTTTTTT GGAGTGGGGAAGGGTATAAAGGAGGCTTAAAATTTGAATCCATAATATATCTAATTACAGGAGAATTTACAACATCTCAAGTACGTAAAT TAAGTTGTCATTGAGTGAAAGGTTCACTTGGACCTAGTGCTGCCTCCTGTTTATTACATAGCATGGCCCTTATGTCTTGAGTTGAGGTTA TCATCTCAATGAGGCTTAAGCTCCTAGAGTACAGGACCATTTTGTTGATTGTCTTTCTTCATAGCTTCTCTGCTTGGCAAAGAGATGGGA GGGGGCCAGATACTGACTACCTGGGGTAGGCACATTATGTGTTAAAGCAAGACAGAGGCCAGAGAGGGGCAGGTAGACCTGCATAGCAGC AGCCTCAGCAGCTGTCTTGGTAAAGGAGAGAGAGAGACATGGGGCCAGTAATTCCGGGGTGCTCAGAAGTTTTAGGAGGGAATGAGCCTC AGGGAGGAGTGAGCACCTAAATGAACGCAGTAAACCTTCATGGACCAACAGTGATTGAGGATTTGTGGGCAGCCAGAGGGAGTCTGACTG AAGTTTACTTGGAAAGAAAGGGCTTGCTAAGAAAAAAGGGAGTAAAAATGATGATAGGGAAGTGTCTAATGTATGTGCACATATAAGTAA TACAAAAGTTTTGAGCTCTTCCAAGTATACCATTTATATACAAACAAATAGGTTTATTCATTCATTAAACTACTTTGGAAGCGTCAGTGG ATATATTTGAAAGTGGTAATCCTGAATCTCTTTTAAACTATTATATGATTCATAATGGTTCTCAGGAATTAATAAATGATTACTGTGTTT >24964_24964_3_EDEM3-HHAT_EDEM3_chr1_184677287_ENST00000367512_HHAT_chr1_210847630_ENST00000413764_length(amino acids)=654AA_BP= MVSATSVWTAGAEPMSREEKQKLGNQVLEMFDHAYGNYMEHAYPADELMPLTCRGRVRGQEPSRGDVDDALGKFSLTLIDSLDTLVVLNK TKEFEDAVRKVLRDVNLDNDVVVSVFETNIRVLGGLLGGHSLAIMLKEKGEYMQWYNDELLQMAKQLGYKLLPAFNTTSGLPYPRINLKF GIRKPEARTGTETDTCTACAGTLILEFAALSRFTGATIFEEYARKALDFLWEKRQRSSNLVGVTINIHTGDWVRKDSGVGAGIDSYYEYL LKAYVLLGDDSFLERFNTHYDAIMRYISQPPLLLDVHIHKPMLNARTWMDALLAFFPGLQVLKGDIRPAIETHEMLYQVIKKHNFLPEAF TTDFRVHWAQHPLRPEFAESTYFLYKATGDPYYLEVGKTLIENLNKYARVPCGFAAMKDVRTGSHEDRMDSFFLAEMFKYLYLLFADKED IIFDIEDYIFTTEAHLLPLWLSTTNQSISKKNTTSEYTELDDSNFDWTCPNTQILFPNDPLYAQSIREPLKNVVDKSCPRGIIRVEESFR SGAKPPLRARDFMATNPEHLEILKKMGVSLIHLKDGRVQLVQHAIQAASSIDAEDGLRFMQEMIELSSQQQKEQQLPPRAVQIVSHPFFG -------------------------------------------------------------- >24964_24964_4_EDEM3-HHAT_EDEM3_chr1_184677287_ENST00000367512_HHAT_chr1_210847630_ENST00000541565_length(transcript)=3967nt_BP=1958nt CGTTCTGCCTGGTGTCGGCCACCTCCGTGTGGACGGCGGGGGCCGAGCCCATGAGTAGGGAGGAGAAACAGAAGCTTGGGAATCAAGTAC TGGAAATGTTTGATCATGCTTATGGTAACTATATGGAACATGCTTACCCTGCTGATGAACTCATGCCTTTAACCTGTAGAGGTCGAGTTA GAGGCCAAGAGCCAAGTCGCGGTGACGTTGATGATGCCTTGGGAAAATTTTCTCTGACACTGATTGATTCTTTGGACACTCTTGTGGTTT TAAATAAAACTAAAGAATTTGAAGATGCAGTGAGAAAAGTTTTAAGAGATGTTAATTTAGATAACGATGTAGTCGTATCAGTCTTTGAAA CAAACATCAGAGTTCTTGGGGGTCTTTTGGGTGGGCACTCCCTGGCAATCATGCTGAAAGAAAAAGGTGAATATATGCAGTGGTACAATG ATGAACTTCTCCAAATGGCAAAGCAGTTAGGTTACAAACTTTTACCGGCTTTCAACACTACCAGTGGCCTTCCTTATCCAAGAATTAATT TAAAGTTTGGCATCAGAAAACCAGAAGCTCGGACAGGAACTGAGACAGATACCTGTACAGCTTGTGCAGGTACCTTGATCCTTGAATTTG CTGCTTTAAGTCGATTCACAGGAGCAACAATATTTGAGGAATATGCCAGAAAAGCTCTTGATTTTCTCTGGGAAAAAAGACAGCGAAGTA GTAATTTAGTGGGCGTGACTATAAATATTCATACTGGAGATTGGGTACGAAAAGATAGTGGAGTTGGAGCAGGGATTGATTCATATTATG AATATCTGTTGAAAGCCTATGTCTTGCTTGGAGATGACAGTTTTCTGGAAAGATTTAACACACACTATGATGCCATAATGAGGTATATTA GCCAGCCACCTCTTCTACTTGATGTGCATATCCACAAACCAATGCTGAATGCTCGGACTTGGATGGATGCTTTGCTTGCCTTCTTCCCAG GCTTGCAGGTGTTAAAGGGGGATATTAGACCTGCTATTGAAACTCATGAAATGTTATATCAGGTGATTAAAAAACACAATTTTCTACCAG AGGCATTTACCACAGATTTCAGAGTACACTGGGCTCAACATCCTTTAAGGCCAGAATTTGCAGAAAGTACCTACTTCTTATATAAAGCTA CAGGAGATCCTTACTACCTTGAAGTAGGGAAGACACTTATTGAAAATTTAAATAAATATGCTAGAGTGCCTTGCGGATTTGCTGCCATGA AGGATGTTCGTACTGGAAGTCATGAGGACAGAATGGATTCTTTCTTCTTGGCTGAAATGTTTAAATATCTTTACCTGTTATTTGCTGATA AAGAAGACATTATTTTTGACATAGAAGATTACATCTTTACAACAGAAGCTCATCTGTTACCTCTTTGGCTCTCTACTACAAATCAAAGCA TCTCTAAAAAGAACACAACCTCGGAATATACAGAACTGGATGACAGTAACTTCGATTGGACTTGTCCAAATACTCAGATCCTCTTTCCTA ATGACCCATTGTATGCTCAAAGTATTCGTGAGCCCTTGAAAAATGTGGTGGATAAGAGCTGTCCTAGAGGCATCATCAGAGTAGAGGAGA GTTTCAGGAGTGGAGCTAAACCCCCTCTGAGAGCCAGAGATTTCATGGCCACTAACCCTGAGCATTTAGAAATCCTGAAGAAGATGGGGG TGAGTTTGATTCACCTCAAAGATGGGAGAGTCCAGTTGGTCCAACATGCAATCCAAGCTGCTAGTTCAATCGACGCTGAAGATGGGTTGA GGTTCATGCAGGAGATGATTGAATTGTCAAGTCAGCAACAAAAAGAACAGCAGCTGCCTCCACGAGCTGTACAAATTGTTTCCCACCCAT TTTTTGGCAGGGTAGTATTGACTGCTGGACCAGCTCAGTTTGGGCTGGATCTGTCTAAACATAAAGAGGCTGGCCTTGGGTGACCCTCTC TGTCCTGGGATTCCTGTACTGCTACTCCCACGTGGGCATTGCCTGGGCCCAGACCTACGCCACGGACTAATGCTGTTGGGCCCAGGCCAG TCCTTGTTGCTGGCCTCCAAGGCAAATAGTGCTTCACCCTGACCTCTCACTCCAGGACAGCCTCTAAGGGATTTGATCTGCTCATCTTCA GTTGAATGCCCTCACTCCAAGACTGGATGCTGGATCTCATAGAAAATTCACAGCCAGACAATCTTCTAATCTGGAGTCTTTGAGATCTTC TACCCCAACTCATCATTTTCCTATTGAGGAAACGGGTCCAGGGCAGTCGTGTGTCTTACCCAGCTACACAGGGTGACATTTTGGTCTAGA ACCTAGTCTCATGAGCCCTTTGCATTCTTTCCCCTTAAGCAAGAATAGAATGTAGTGGAAATTTATTGATTGAGACACAGAAATCCTGAT TAGTGATAGGCCTGTCTTCTGGGCAATATTTCTAAAATATTTGAGTGACATTGATGTTTAAGTGACCTCCTTCATCACATCCTGCAGTAT CTCCAGAAGCAGCACTAGGGTTTGAGTTTCATACTTCAGCCCCTCTGTAGGAATGAGACCAAGCCACAGCTGTCTTTTGAATTACGTAGT CGAAGAAGACGGTAGCAGCCCTGTAGCATTCTAAGGCATCTATACCCAAGGAGTCCTGTGATTTGAGCTTCAGCAGGGTGATCTACATTT GGGTGCTTGTTTCTGAGATTTGCAGAGAAGTATAACATGGTAGTTCCTCTACCTTACAGTTAATCGTTTCTTAATAAAGAAGCAGAATTT AGAAACCACAGGATAGTGTACCCACAGATGGGTGTTATCAAGGCCAGTCATGAGGATGGTGTCCTGGAGTCTTGTCCACCCTCTCCATAC AAGTCTCAAAAGTCATCCTCCTACTCAGTGATTCACGTTTAGTGGTTTATATTATTAAGGTTTGATTCAAACAGAGCCTTTTCTGTCCTG TAGATAATCTACATGTTTGTAGAATTATTTTGAATATGTTTGAGGAAAATGTTTAAAATCTAAATATACTCACATAACTTGATTATTCAC TCCTCTGAAAAGATGCTGGATAGGCTACCAAAGTTCCCAAGTGGTAGATAATTCAGAAGACTTGTTTGAATTTGGATTTTTTTTTTTTTT GGAGTGGGGAAGGGTATAAAGGAGGCTTAAAATTTGAATCCATAATATATCTAATTACAGGAGAATTTACAACATCTCAAGTACGTAAAT TAAGTTGTCATTGAGTGAAAGGTTCACTTGGACCTAGTGCTGCCTCCTGTTTATTACATAGCATGGCCCTTATGTCTTGAGTTGAGGTTA TCATCTCAATGAGGCTTAAGCTCCTAGAGTACAGGACCATTTTGTTGATTGTCTTTCTTCATAGCTTCTCTGCTTGGCAAAGAGATGGGA GGGGGCCAGATACTGACTACCTGGGGTAGGCACATTATGTGTTAAAGCAAGACAGAGGCCAGAGAGGGGCAGGTAGACCTGCATAGCAGC AGCCTCAGCAGCTGTCTTGGTAAAGGAGAGAGAGAGACATGGGGCCAGTAATTCCGGGGTGCTCAGAAGTTTTAGGAGGGAATGAGCCTC AGGGAGGAGTGAGCACCTAAATGAACGCAGTAAACCTTCATGGACCAACAGTGATTGAGGATTTGTGGGCAGCCAGAGGGAGTCTGACTG AAGTTTACTTGGAAAGAAAGGGCTTGCTAAGAAAAAAGGGAGTAAAAATGATGATAGGGAAGTGTCTAATGTATGTGCACATATAAGTAA TACAAAAGTTTTGAGCTCTTCCAAGTATACCATTTATATACAAACAAATAGGTTTATTCATTCATTAAACTACTTTGGAAGCGTCAGTGG ATATATTTGAAAGTGGTAATCCTGAATCTCTTTTAAACTATTATATGATTCATAATGGTTCTCAGGAATTAATAAATGATTACTGTGTTT >24964_24964_4_EDEM3-HHAT_EDEM3_chr1_184677287_ENST00000367512_HHAT_chr1_210847630_ENST00000541565_length(amino acids)=654AA_BP= MVSATSVWTAGAEPMSREEKQKLGNQVLEMFDHAYGNYMEHAYPADELMPLTCRGRVRGQEPSRGDVDDALGKFSLTLIDSLDTLVVLNK TKEFEDAVRKVLRDVNLDNDVVVSVFETNIRVLGGLLGGHSLAIMLKEKGEYMQWYNDELLQMAKQLGYKLLPAFNTTSGLPYPRINLKF GIRKPEARTGTETDTCTACAGTLILEFAALSRFTGATIFEEYARKALDFLWEKRQRSSNLVGVTINIHTGDWVRKDSGVGAGIDSYYEYL LKAYVLLGDDSFLERFNTHYDAIMRYISQPPLLLDVHIHKPMLNARTWMDALLAFFPGLQVLKGDIRPAIETHEMLYQVIKKHNFLPEAF TTDFRVHWAQHPLRPEFAESTYFLYKATGDPYYLEVGKTLIENLNKYARVPCGFAAMKDVRTGSHEDRMDSFFLAEMFKYLYLLFADKED IIFDIEDYIFTTEAHLLPLWLSTTNQSISKKNTTSEYTELDDSNFDWTCPNTQILFPNDPLYAQSIREPLKNVVDKSCPRGIIRVEESFR SGAKPPLRARDFMATNPEHLEILKKMGVSLIHLKDGRVQLVQHAIQAASSIDAEDGLRFMQEMIELSSQQQKEQQLPPRAVQIVSHPFFG -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for EDEM3-HHAT |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for EDEM3-HHAT |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for EDEM3-HHAT |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
