
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:EEF1A1-COL3A1 (FusionGDB2 ID:25023) |
Fusion Gene Summary for EEF1A1-COL3A1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: EEF1A1-COL3A1 | Fusion gene ID: 25023 | Hgene | Tgene | Gene symbol | EEF1A1 | COL3A1 | Gene ID | 1915 | 1281 |
| Gene name | eukaryotic translation elongation factor 1 alpha 1 | collagen type III alpha 1 chain | |
| Synonyms | CCS-3|CCS3|EE1A1|EEF-1|EEF1A|EF-Tu|EF1A|GRAF-1EF|LENG7|PTI1|eEF1A-1 | EDS4A|EDSVASC|PMGEDSV | |
| Cytomap | 6q13 | 2q32.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | elongation factor 1-alpha 1CTCL tumor antigenEF-1-alpha-1EF1a-like proteincervical cancer suppressor 3elongation factor 1 alpha subunitelongation factor Tuepididymis secretory sperm binding proteineukaryotic elongation factor 1 A-1eukaryotic tran | collagen alpha-1(III) chainEhlers-Danlos syndrome type IV, autosomal dominantalpha-1 type III collagenalpha1 (III) collagencollagen, fetalcollagen, type III, alpha 1 | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | P68104 | P02461 | |
| Ensembl transtripts involved in fusion gene | ENST00000309268, ENST00000316292, ENST00000331523, ENST00000491404, | ENST00000304636, ENST00000317840, | |
| Fusion gene scores | * DoF score | 24 X 34 X 12=9792 | 34 X 39 X 8=10608 |
| # samples | 38 | 43 | |
| ** MAII score | log2(38/9792*10)=-4.6875322343617 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(43/10608*10)=-4.62467221052313 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: EEF1A1 [Title/Abstract] AND COL3A1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | EEF1A1(74228077)-COL3A1(189872852), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | EEF1A1-COL3A1 seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. EEF1A1-COL3A1 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. EEF1A1-COL3A1 seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. EEF1A1-COL3A1 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | EEF1A1 | GO:0071364 | cellular response to epidermal growth factor stimulus | 9852145 |
| Hgene | EEF1A1 | GO:1900022 | regulation of D-erythro-sphingosine kinase activity | 18263879 |
| Tgene | COL3A1 | GO:0007160 | cell-matrix adhesion | 16912226 |
| Tgene | COL3A1 | GO:0007179 | transforming growth factor beta receptor signaling pathway | 16360482 |
| Tgene | COL3A1 | GO:0009314 | response to radiation | 14736764 |
| Tgene | COL3A1 | GO:0018149 | peptide cross-linking | 16754721 |
| Tgene | COL3A1 | GO:0034097 | response to cytokine | 9076960|16360482 |
| Tgene | COL3A1 | GO:0042060 | wound healing | 1466622 |
 Fusion gene breakpoints across EEF1A1 (5'-gene) Fusion gene breakpoints across EEF1A1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
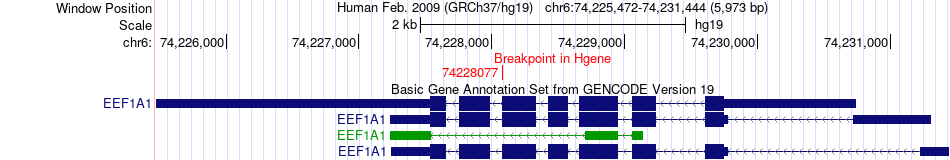 |
 Fusion gene breakpoints across COL3A1 (3'-gene) Fusion gene breakpoints across COL3A1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
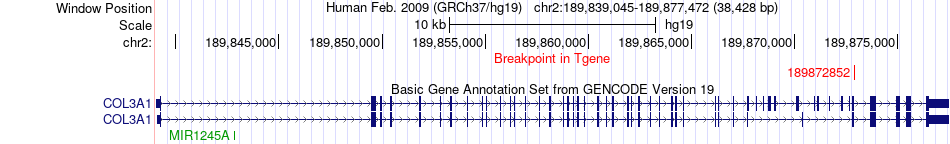 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-E2-A1B1-01A | EEF1A1 | chr6 | 74228077 | - | COL3A1 | chr2 | 189872852 | + |
Top |
Fusion Gene ORF analysis for EEF1A1-COL3A1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000309268 | ENST00000304636 | EEF1A1 | chr6 | 74228077 | - | COL3A1 | chr2 | 189872852 | + |
| Frame-shift | ENST00000316292 | ENST00000317840 | EEF1A1 | chr6 | 74228077 | - | COL3A1 | chr2 | 189872852 | + |
| Frame-shift | ENST00000331523 | ENST00000304636 | EEF1A1 | chr6 | 74228077 | - | COL3A1 | chr2 | 189872852 | + |
| In-frame | ENST00000309268 | ENST00000317840 | EEF1A1 | chr6 | 74228077 | - | COL3A1 | chr2 | 189872852 | + |
| In-frame | ENST00000316292 | ENST00000304636 | EEF1A1 | chr6 | 74228077 | - | COL3A1 | chr2 | 189872852 | + |
| In-frame | ENST00000331523 | ENST00000317840 | EEF1A1 | chr6 | 74228077 | - | COL3A1 | chr2 | 189872852 | + |
| intron-3CDS | ENST00000491404 | ENST00000304636 | EEF1A1 | chr6 | 74228077 | - | COL3A1 | chr2 | 189872852 | + |
| intron-3CDS | ENST00000491404 | ENST00000317840 | EEF1A1 | chr6 | 74228077 | - | COL3A1 | chr2 | 189872852 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000316292 | EEF1A1 | chr6 | 74228077 | - | ENST00000304636 | COL3A1 | chr2 | 189872852 | + | 3885 | 2021 | 992 | 2023 | 343 |
| ENST00000309268 | EEF1A1 | chr6 | 74228077 | - | ENST00000317840 | COL3A1 | chr2 | 189872852 | + | 3512 | 1648 | 619 | 1650 | 343 |
| ENST00000331523 | EEF1A1 | chr6 | 74228077 | - | ENST00000317840 | COL3A1 | chr2 | 189872852 | + | 3138 | 1274 | 245 | 1276 | 343 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000316292 | ENST00000304636 | EEF1A1 | chr6 | 74228077 | - | COL3A1 | chr2 | 189872852 | + | 0.000406032 | 0.99959403 |
| ENST00000309268 | ENST00000317840 | EEF1A1 | chr6 | 74228077 | - | COL3A1 | chr2 | 189872852 | + | 0.00046004 | 0.99954 |
| ENST00000331523 | ENST00000317840 | EEF1A1 | chr6 | 74228077 | - | COL3A1 | chr2 | 189872852 | + | 0.000347327 | 0.9996526 |
Top |
Fusion Genomic Features for EEF1A1-COL3A1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
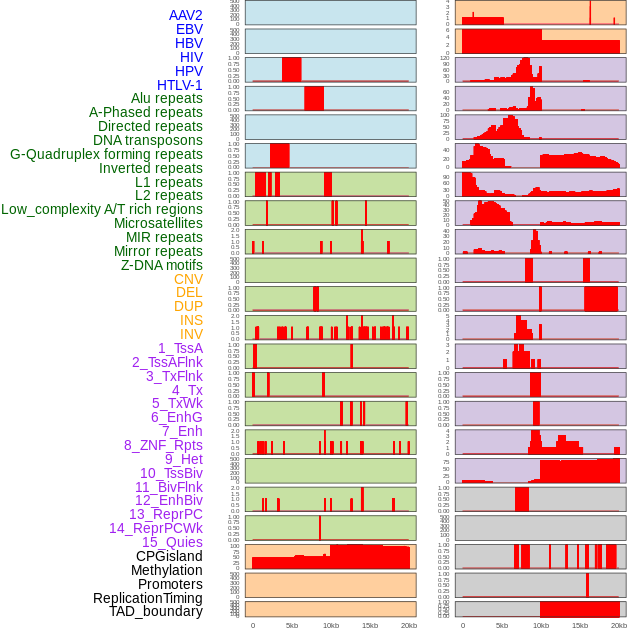 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for EEF1A1-COL3A1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr6:74228077/chr2:189872852) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| EEF1A1 | COL3A1 |
| FUNCTION: This protein promotes the GTP-dependent binding of aminoacyl-tRNA to the A-site of ribosomes during protein biosynthesis. Plays a role in the positive regulation of IFNG transcription in T-helper 1 cells as part of an IFNG promoter-binding complex with TXK and PARP1 (PubMed:17177976). {ECO:0000269|PubMed:17177976}. | FUNCTION: Collagen type III occurs in most soft connective tissues along with type I collagen. Involved in regulation of cortical development. Is the major ligand of ADGRG1 in the developing brain and binding to ADGRG1 inhibits neuronal migration and activates the RhoA pathway by coupling ADGRG1 to GNA13 and possibly GNA12. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000309268 | - | 6 | 8 | 5_242 | 343 | 463.0 | Domain | Note=tr-type G |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000316292 | - | 5 | 7 | 5_242 | 343 | 463.0 | Domain | Note=tr-type G |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000331523 | - | 6 | 8 | 5_242 | 343 | 463.0 | Domain | Note=tr-type G |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000309268 | - | 6 | 8 | 91_95 | 343 | 463.0 | Nucleotide binding | GTP |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000316292 | - | 5 | 7 | 91_95 | 343 | 463.0 | Nucleotide binding | GTP |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000331523 | - | 6 | 8 | 91_95 | 343 | 463.0 | Nucleotide binding | GTP |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000309268 | - | 6 | 8 | 14_21 | 343 | 463.0 | Region | G1 |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000309268 | - | 6 | 8 | 153_156 | 343 | 463.0 | Region | G4 |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000309268 | - | 6 | 8 | 194_196 | 343 | 463.0 | Region | G5 |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000309268 | - | 6 | 8 | 70_74 | 343 | 463.0 | Region | G2 |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000309268 | - | 6 | 8 | 91_94 | 343 | 463.0 | Region | G3 |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000316292 | - | 5 | 7 | 14_21 | 343 | 463.0 | Region | G1 |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000316292 | - | 5 | 7 | 153_156 | 343 | 463.0 | Region | G4 |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000316292 | - | 5 | 7 | 194_196 | 343 | 463.0 | Region | G5 |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000316292 | - | 5 | 7 | 70_74 | 343 | 463.0 | Region | G2 |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000316292 | - | 5 | 7 | 91_94 | 343 | 463.0 | Region | G3 |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000331523 | - | 6 | 8 | 14_21 | 343 | 463.0 | Region | G1 |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000331523 | - | 6 | 8 | 153_156 | 343 | 463.0 | Region | G4 |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000331523 | - | 6 | 8 | 194_196 | 343 | 463.0 | Region | G5 |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000331523 | - | 6 | 8 | 70_74 | 343 | 463.0 | Region | G2 |
| Hgene | EEF1A1 | chr6:74228077 | chr2:189872852 | ENST00000331523 | - | 6 | 8 | 91_94 | 343 | 463.0 | Region | G3 |
| Tgene | COL3A1 | chr6:74228077 | chr2:189872852 | ENST00000304636 | 0 | 51 | 1232_1466 | 0 | 1467.0 | Domain | Fibrillar collagen NC1 | |
| Tgene | COL3A1 | chr6:74228077 | chr2:189872852 | ENST00000304636 | 0 | 51 | 30_89 | 0 | 1467.0 | Domain | VWFC | |
| Tgene | COL3A1 | chr6:74228077 | chr2:189872852 | ENST00000304636 | 0 | 51 | 1197_1205 | 0 | 1467.0 | Region | Note=Nonhelical region (C-terminal) | |
| Tgene | COL3A1 | chr6:74228077 | chr2:189872852 | ENST00000304636 | 0 | 51 | 149_167 | 0 | 1467.0 | Region | Note=Nonhelical region (N-terminal) | |
| Tgene | COL3A1 | chr6:74228077 | chr2:189872852 | ENST00000304636 | 0 | 51 | 168_1196 | 0 | 1467.0 | Region | Note=Triple-helical region |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Gene Sequence for EEF1A1-COL3A1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >25023_25023_1_EEF1A1-COL3A1_EEF1A1_chr6_74228077_ENST00000309268_COL3A1_chr2_189872852_ENST00000317840_length(transcript)=3512nt_BP=1648nt CGCAGAGGAAGACGCTCTAGGGATTTGTCCCGGACTAGCGAGATGGCAAGGCTGAGGACGGGAGGCTGATTGAGAGGCGAAGGTACACCC TAATCTCAATACAACCTTTGGAGCTAAGCCAGCAATGGTAGAGGGAAGATTCTGCACGTCCCTTCCAGGCGGCCTCCCCGTCACCACCCC CCCCAACCCGCCCCGACCGGAGCTGAGAGTAATTCATACAAAAGGACTCGCCCCTGCCTTGGGGAATCCCAGGGACCGTCGTTAAACTCC CACTAACGTAGAACCCAGAGATCGCTGCGTTCCCGCCCCCTCACCCGCCCGCTCTCGTCATCACTGAGGTGGAGAAGAGCATGCGTGAGG CTCCGGTGCCCGTCAGTGGGCAGAGCGCACATCGCCCACAGTCCCCGAGAAGTTGGGGGGAGGGGTCGGCAATTGAACCGGTGCCTAGAG AAGGTGGCGCGGGGTAAACTGGGAAAGTGATGTCGTGTACTGGCTCCGCCTTTTTCCCGAGGGTGGGGGAGAACCGTATATAAGTGCAGT AGTCGCCGTGAACGTTCTTTTTCGCAACGGGTTTGCCGCCAGAACACAGGTGTCGTGAAAACTACCCCTAAAAGCCAAAATGGGAAAGGA AAAGACTCATATCAACATTGTCGTCATTGGACACGTAGATTCGGGCAAGTCCACCACTACTGGCCATCTGATCTATAAATGCGGTGGCAT CGACAAAAGAACCATTGAAAAATTTGAGAAGGAGGCTGCTGAGATGGGAAAGGGCTCCTTCAAGTATGCCTGGGTCTTGGATAAACTGAA AGCTGAGCGTGAACGTGGTATCACCATTGATATCTCCTTGTGGAAATTTGAGACCAGCAAGTACTATGTGACTATCATTGATGCCCCAGG ACACAGAGACTTTATCAAAAACATGATTACAGGGACATCTCAGGCTGACTGTGCTGTCCTGATTGTTGCTGCTGGTGTTGGTGAATTTGA AGCTGGTATCTCCAAGAATGGGCAGACCCGAGAGCATGCCCTTCTGGCTTACACACTGGGTGTGAAACAACTAATTGTCGGTGTTAACAA AATGGATTCCACTGAGCCACCCTACAGCCAGAAGAGATATGAGGAAATTGTTAAGGAAGTCAGCACTTACATTAAGAAAATTGGCTACAA CCCCGACACAGTAGCATTTGTGCCAATTTCTGGTTGGAATGGTGACAACATGCTGGAGCCAAGTGCTAACATGCCTTGGTTCAAGGGATG GAAAGTCACCCGTAAGGATGGCAATGCCAGTGGAACCACGCTGCTTGAGGCTCTGGACTGCATCCTACCACCAACTCGTCCAACTGACAA GCCCTTGCGCCTGCCTCTCCAGGATGTCTACAAAATTGGTGGTATTGGTACTGTTCCTGTTGGCCGAGTGGAGACTGGTGTTCTCAAACC CGGTATGGTGGTCACCTTTGCTCCAGTCAACGTTACAACGGAAGTAAAATCTGTCGAAATGCACCATGAAGCTTTGAGTGAAGCTCTTCC TGGGGACAATGTGGGCTTCAATGTCAAGAATGTGTCTGTCAAGGATGTTCGTCGTGGCAACGTTGCTGGTGACAGCAAAAATGACCCACC AATGGAAGCAGCTGGCTTCACTGCTCAGTGAAAGAGGATCTGAGGGCTCCCCAGGCCACCCAGGGCAACCAGGCCCTCCTGGACCTCCTG GTGCCCCTGGTCCTTGCTGTGGTGGTGTTGGAGCCGCTGCCATTGCTGGGATTGGAGGTGAAAAAGCTGGCGGTTTTGCCCCGTATTATG GAGATGAACCAATGGATTTCAAAATCAACACCGATGAGATTATGACTTCACTCAAGTCTGTTAATGGACAAATAGAAAGCCTCATTAGTC CTGATGGTTCTCGTAAAAACCCCGCTAGAAACTGCAGAGACCTGAAATTCTGCCATCCTGAACTCAAGAGTGGAGAATACTGGGTTGACC CTAACCAAGGATGCAAATTGGATGCTATCAAGGTATTCTGTAATATGGAAACTGGGGAAACATGCATAAGTGCCAATCCTTTGAATGTTC CACGGAAACACTGGTGGACAGATTCTAGTGCTGAGAAGAAACACGTTTGGTTTGGAGAGTCCATGGATGGTGGTTTTCAGTTTAGCTACG GCAATCCTGAACTTCCTGAAGATGTCCTTGATGTGCATCTGGCATTCCTTCGACTTCTCTCCAGCCGAGCTTCCCAGAACATCACATATC ACTGCAAAAATAGCATTGCATACATGGATCAGGCCAGTGGAAATGTAAAGAAGGCCCTGAAGCTGATGGGGTCAAATGAAGGTGAATTCA AGGCTGAAGGAAATAGCAAATTCACCTACACAGTTCTGGAGGATGGTTGCACGAAACACACTGGGGAATGGAGCAAAACAGTCTTTGAAT ATCGAACACGCAAGGCTGTGAGACTACCTATTGTAGATATTGCACCCTATGACATTGGTGGTCCTGATCAAGAATTTGGTGTGGACGTTG GCCCTGTTTGCTTTTTATAAACCAAACTCTATCTGAAATCCCAACAAAAAAAATTTAACTCCATATGTGTTCCTCTTGTTCTAATCTTGT CAACCAGTGCAAGTGACCGACAAAATTCCAGTTATTTATTTCCAAAATGTTTGGAAACAGTATAATTTGACAAAGAAAAATGATACTTCT CTTTTTTTGCTGTTCCACCAAATACAATTCAAATGCTTTTTGTTTTATTTTTTTACCAATTCCAATTTCAAAATGTCTCAATGGTGCTAT AATAAATAAACTTCAACACTCTTTATGATAACAACACTGTGTTATATTCTTTGAATCCTAGCCCATCTGCAGAGCAATGACTGTGCTCAC CAGTAAAAGATAACCTTTCTTTCTGAAATAGTCAAATACGAAATTAGAAAAGCCCTCCCTATTTTAACTACCTCAACTGGTCAGAAACAC AGATTGTATTCTATGAGTCCCAGAAGATGAAAAAAATTTTATACGTTGATAAAACTTATAAATTTCATTGATTAATCTCCTGGAAGATTG GTTTAAAAAGAAAAGTGTAATGCAAGAATTTAAAGAAATATTTTTAAAGCCACAATTATTTTAATATTGGATATCAACTGCTTGTAAAGG TGCTCCTCTTTTTTCTTGTCATTGCTGGTCAAGATTACTAATATTTGGGAAGGCTTTAAAGACGCATGTTATGGTGCTAATGTACTTTCA CTTTTAAACTCTAGATCAGAATTGTTGACTTGCATTCAGAACATAAATGCACAAAATCTGTACATGTCTCCCATCAGAAAGATTCATTGG CATGCCACAGGGGATTCTCCTCCTTCATCCTGTAAAGGTCAACAATAAAAACCAAATTATGGGGCTGCTTTTGTCACACTAGCATAGAGA ATGTGTTGAAATTTAACTTTGTAAGCTTGTATGTGGTTGTTGATCTTTTTTTTCCTTACAGACACCCATAATAAAATATCATATTAAAAT >25023_25023_1_EEF1A1-COL3A1_EEF1A1_chr6_74228077_ENST00000309268_COL3A1_chr2_189872852_ENST00000317840_length(amino acids)=343AA_BP= MGKEKTHINIVVIGHVDSGKSTTTGHLIYKCGGIDKRTIEKFEKEAAEMGKGSFKYAWVLDKLKAERERGITIDISLWKFETSKYYVTII DAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAGISKNGQTREHALLAYTLGVKQLIVGVNKMDSTEPPYSQKRYEEIVKEVSTYIKK IGYNPDTVAFVPISGWNGDNMLEPSANMPWFKGWKVTRKDGNASGTTLLEALDCILPPTRPTDKPLRLPLQDVYKIGGIGTVPVGRVETG -------------------------------------------------------------- >25023_25023_2_EEF1A1-COL3A1_EEF1A1_chr6_74228077_ENST00000316292_COL3A1_chr2_189872852_ENST00000304636_length(transcript)=3885nt_BP=2021nt GTTTGCCGCCAGAACACAGGTAAGTGCCGTGTGTGGTTCCCGCGGGCCTGGCCTCTTTACGGGTTATGGCCCTTGCGTGCCTTGAATTAC TTCCACGCCCCTGGCTGCAGTACGTGATTCTTGATCCCGAGCTTCGGGTTGGAAGTGGGTGGGAGAGTTCGAGGCCTTGCGCTTAAGGAG CCCCTTCGCCTCGTGCTTGAGTTGAGGCCTGGCTTGGGCGCTGGGGCCGCCGCGTGCGAATCTGGTGGCACCTTCGCGCCTGTCTCGCTG CTTTCGATAAGTCTCTAGCCATTTAAAATTTTTGATGACCTGCTGCGACGCTTTTTTTCTGGCAAGATAGTCTTGTAAATGCGGGCCAAG ATCTGCACACTGGTATTTCGGTTTTTGGGGCCGCGGGCGGCGACGGGGCCCGTGCGTCCCAGCGCACATGTTCGGCGAGGCGGGGCCTGC GAGCGCGGCCACCGAGAATCGGACGGGGGTAGTCTCAAGCTGGCCGGCCTGCTCTGGTGCCTGGCCTCGCGCCGCCGTGTATCGCCCCGC CCTGGGCGGCAAGGCTGGCCCGGTCGGCACCAGTTGCGTGAGCGGAAAGATGGCCGCTTCCCGGCCCTGCTGCAGGGAGCTCAAAATGGA GGACGCGGCGCTCGGGAGAGCGGGCGGGTGAGTCACCCACACAAAGGAAAAGGGCCTTTCCGTCCTCAGCCGTCGCTTCATGTGACTCCA CGGAGTACCGGGCGCCGTCCAGGCACCTCGATTAGTTCTCGAGCTTTTGGAGTACGTCGTCTTTAGGTTGGGGGGAGGGGTTTTATGCGA TGGAGTTTCCCCACACTGAGTGGGTGGAGACTGAAGTTAGGCCAGCTTGGCACTTGATGTAATTCTCCTTGGAATTTGCCCTTTTTGAGT TTGGATCTTGGTTCATTCTCAAGCCTCAGACAGTGGTTCAAAGTTTTTTTCTTCCATTTCAGGTGTCGTGAAAACTACCCCTAAAAGCCA AAATGGGAAAGGAAAAGACTCATATCAACATTGTCGTCATTGGACACGTAGATTCGGGCAAGTCCACCACTACTGGCCATCTGATCTATA AATGCGGTGGCATCGACAAAAGAACCATTGAAAAATTTGAGAAGGAGGCTGCTGAGATGGGAAAGGGCTCCTTCAAGTATGCCTGGGTCT TGGATAAACTGAAAGCTGAGCGTGAACGTGGTATCACCATTGATATCTCCTTGTGGAAATTTGAGACCAGCAAGTACTATGTGACTATCA TTGATGCCCCAGGACACAGAGACTTTATCAAAAACATGATTACAGGGACATCTCAGGCTGACTGTGCTGTCCTGATTGTTGCTGCTGGTG TTGGTGAATTTGAAGCTGGTATCTCCAAGAATGGGCAGACCCGAGAGCATGCCCTTCTGGCTTACACACTGGGTGTGAAACAACTAATTG TCGGTGTTAACAAAATGGATTCCACTGAGCCACCCTACAGCCAGAAGAGATATGAGGAAATTGTTAAGGAAGTCAGCACTTACATTAAGA AAATTGGCTACAACCCCGACACAGTAGCATTTGTGCCAATTTCTGGTTGGAATGGTGACAACATGCTGGAGCCAAGTGCTAACATGCCTT GGTTCAAGGGATGGAAAGTCACCCGTAAGGATGGCAATGCCAGTGGAACCACGCTGCTTGAGGCTCTGGACTGCATCCTACCACCAACTC GTCCAACTGACAAGCCCTTGCGCCTGCCTCTCCAGGATGTCTACAAAATTGGTGGTATTGGTACTGTTCCTGTTGGCCGAGTGGAGACTG GTGTTCTCAAACCCGGTATGGTGGTCACCTTTGCTCCAGTCAACGTTACAACGGAAGTAAAATCTGTCGAAATGCACCATGAAGCTTTGA GTGAAGCTCTTCCTGGGGACAATGTGGGCTTCAATGTCAAGAATGTGTCTGTCAAGGATGTTCGTCGTGGCAACGTTGCTGGTGACAGCA AAAATGACCCACCAATGGAAGCAGCTGGCTTCACTGCTCAGTGAAAGAGGATCTGAGGGCTCCCCAGGCCACCCAGGGCAACCAGGCCCT CCTGGACCTCCTGGTGCCCCTGGTCCTTGCTGTGGTGGTGTTGGAGCCGCTGCCATTGCTGGGATTGGAGGTGAAAAAGCTGGCGGTTTT GCCCCGTATTATGGAGATGAACCAATGGATTTCAAAATCAACACCGATGAGATTATGACTTCACTCAAGTCTGTTAATGGACAAATAGAA AGCCTCATTAGTCCTGATGGTTCTCGTAAAAACCCCGCTAGAAACTGCAGAGACCTGAAATTCTGCCATCCTGAACTCAAGAGTGGAGAA TACTGGGTTGACCCTAACCAAGGATGCAAATTGGATGCTATCAAGGTATTCTGTAATATGGAAACTGGGGAAACATGCATAAGTGCCAAT CCTTTGAATGTTCCACGGAAACACTGGTGGACAGATTCTAGTGCTGAGAAGAAACACGTTTGGTTTGGAGAGTCCATGGATGGTGGTTTT CAGTTTAGCTACGGCAATCCTGAACTTCCTGAAGATGTCCTTGATGTGCATCTGGCATTCCTTCGACTTCTCTCCAGCCGAGCTTCCCAG AACATCACATATCACTGCAAAAATAGCATTGCATACATGGATCAGGCCAGTGGAAATGTAAAGAAGGCCCTGAAGCTGATGGGGTCAAAT GAAGGTGAATTCAAGGCTGAAGGAAATAGCAAATTCACCTACACAGTTCTGGAGGATGGTTGCACGAAACACACTGGGGAATGGAGCAAA ACAGTCTTTGAATATCGAACACGCAAGGCTGTGAGACTACCTATTGTAGATATTGCACCCTATGACATTGGTGGTCCTGATCAAGAATTT GGTGTGGACGTTGGCCCTGTTTGCTTTTTATAAACCAAACTCTATCTGAAATCCCAACAAAAAAAATTTAACTCCATATGTGTTCCTCTT GTTCTAATCTTGTCAACCAGTGCAAGTGACCGACAAAATTCCAGTTATTTATTTCCAAAATGTTTGGAAACAGTATAATTTGACAAAGAA AAATGATACTTCTCTTTTTTTGCTGTTCCACCAAATACAATTCAAATGCTTTTTGTTTTATTTTTTTACCAATTCCAATTTCAAAATGTC TCAATGGTGCTATAATAAATAAACTTCAACACTCTTTATGATAACAACACTGTGTTATATTCTTTGAATCCTAGCCCATCTGCAGAGCAA TGACTGTGCTCACCAGTAAAAGATAACCTTTCTTTCTGAAATAGTCAAATACGAAATTAGAAAAGCCCTCCCTATTTTAACTACCTCAAC TGGTCAGAAACACAGATTGTATTCTATGAGTCCCAGAAGATGAAAAAAATTTTATACGTTGATAAAACTTATAAATTTCATTGATTAATC TCCTGGAAGATTGGTTTAAAAAGAAAAGTGTAATGCAAGAATTTAAAGAAATATTTTTAAAGCCACAATTATTTTAATATTGGATATCAA CTGCTTGTAAAGGTGCTCCTCTTTTTTCTTGTCATTGCTGGTCAAGATTACTAATATTTGGGAAGGCTTTAAAGACGCATGTTATGGTGC TAATGTACTTTCACTTTTAAACTCTAGATCAGAATTGTTGACTTGCATTCAGAACATAAATGCACAAAATCTGTACATGTCTCCCATCAG AAAGATTCATTGGCATGCCACAGGGGATTCTCCTCCTTCATCCTGTAAAGGTCAACAATAAAAACCAAATTATGGGGCTGCTTTTGTCAC ACTAGCATAGAGAATGTGTTGAAATTTAACTTTGTAAGCTTGTATGTGGTTGTTGATCTTTTTTTTCCTTACAGACACCCATAATAAAAT >25023_25023_2_EEF1A1-COL3A1_EEF1A1_chr6_74228077_ENST00000316292_COL3A1_chr2_189872852_ENST00000304636_length(amino acids)=343AA_BP= MGKEKTHINIVVIGHVDSGKSTTTGHLIYKCGGIDKRTIEKFEKEAAEMGKGSFKYAWVLDKLKAERERGITIDISLWKFETSKYYVTII DAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAGISKNGQTREHALLAYTLGVKQLIVGVNKMDSTEPPYSQKRYEEIVKEVSTYIKK IGYNPDTVAFVPISGWNGDNMLEPSANMPWFKGWKVTRKDGNASGTTLLEALDCILPPTRPTDKPLRLPLQDVYKIGGIGTVPVGRVETG -------------------------------------------------------------- >25023_25023_3_EEF1A1-COL3A1_EEF1A1_chr6_74228077_ENST00000331523_COL3A1_chr2_189872852_ENST00000317840_length(transcript)=3138nt_BP=1274nt CGTTAAGGAGGCCCCCAGTCCCGACCCTTCGCCCCAAGCCCCTCGGGGTCCCCGGGCCTGGTACTCCTTGCCACACGGGAGGGGCGCGGA AGCCGGGGCGGAGGAGGAGCCAACCCCGGGCTGGGCTGAGACCCGCAGAGGAAGACGCTCTAGGGATTTGTCCCGGACTAGCGAGATGGC AAGGCTGAGGACGGGAGGCTGATTGAGAGGCGAAGGTGTCGTGAAAACTACCCCTAAAAGCCAAAATGGGAAAGGAAAAGACTCATATCA ACATTGTCGTCATTGGACACGTAGATTCGGGCAAGTCCACCACTACTGGCCATCTGATCTATAAATGCGGTGGCATCGACAAAAGAACCA TTGAAAAATTTGAGAAGGAGGCTGCTGAGATGGGAAAGGGCTCCTTCAAGTATGCCTGGGTCTTGGATAAACTGAAAGCTGAGCGTGAAC GTGGTATCACCATTGATATCTCCTTGTGGAAATTTGAGACCAGCAAGTACTATGTGACTATCATTGATGCCCCAGGACACAGAGACTTTA TCAAAAACATGATTACAGGGACATCTCAGGCTGACTGTGCTGTCCTGATTGTTGCTGCTGGTGTTGGTGAATTTGAAGCTGGTATCTCCA AGAATGGGCAGACCCGAGAGCATGCCCTTCTGGCTTACACACTGGGTGTGAAACAACTAATTGTCGGTGTTAACAAAATGGATTCCACTG AGCCACCCTACAGCCAGAAGAGATATGAGGAAATTGTTAAGGAAGTCAGCACTTACATTAAGAAAATTGGCTACAACCCCGACACAGTAG CATTTGTGCCAATTTCTGGTTGGAATGGTGACAACATGCTGGAGCCAAGTGCTAACATGCCTTGGTTCAAGGGATGGAAAGTCACCCGTA AGGATGGCAATGCCAGTGGAACCACGCTGCTTGAGGCTCTGGACTGCATCCTACCACCAACTCGTCCAACTGACAAGCCCTTGCGCCTGC CTCTCCAGGATGTCTACAAAATTGGTGGTATTGGTACTGTTCCTGTTGGCCGAGTGGAGACTGGTGTTCTCAAACCCGGTATGGTGGTCA CCTTTGCTCCAGTCAACGTTACAACGGAAGTAAAATCTGTCGAAATGCACCATGAAGCTTTGAGTGAAGCTCTTCCTGGGGACAATGTGG GCTTCAATGTCAAGAATGTGTCTGTCAAGGATGTTCGTCGTGGCAACGTTGCTGGTGACAGCAAAAATGACCCACCAATGGAAGCAGCTG GCTTCACTGCTCAGTGAAAGAGGATCTGAGGGCTCCCCAGGCCACCCAGGGCAACCAGGCCCTCCTGGACCTCCTGGTGCCCCTGGTCCT TGCTGTGGTGGTGTTGGAGCCGCTGCCATTGCTGGGATTGGAGGTGAAAAAGCTGGCGGTTTTGCCCCGTATTATGGAGATGAACCAATG GATTTCAAAATCAACACCGATGAGATTATGACTTCACTCAAGTCTGTTAATGGACAAATAGAAAGCCTCATTAGTCCTGATGGTTCTCGT AAAAACCCCGCTAGAAACTGCAGAGACCTGAAATTCTGCCATCCTGAACTCAAGAGTGGAGAATACTGGGTTGACCCTAACCAAGGATGC AAATTGGATGCTATCAAGGTATTCTGTAATATGGAAACTGGGGAAACATGCATAAGTGCCAATCCTTTGAATGTTCCACGGAAACACTGG TGGACAGATTCTAGTGCTGAGAAGAAACACGTTTGGTTTGGAGAGTCCATGGATGGTGGTTTTCAGTTTAGCTACGGCAATCCTGAACTT CCTGAAGATGTCCTTGATGTGCATCTGGCATTCCTTCGACTTCTCTCCAGCCGAGCTTCCCAGAACATCACATATCACTGCAAAAATAGC ATTGCATACATGGATCAGGCCAGTGGAAATGTAAAGAAGGCCCTGAAGCTGATGGGGTCAAATGAAGGTGAATTCAAGGCTGAAGGAAAT AGCAAATTCACCTACACAGTTCTGGAGGATGGTTGCACGAAACACACTGGGGAATGGAGCAAAACAGTCTTTGAATATCGAACACGCAAG GCTGTGAGACTACCTATTGTAGATATTGCACCCTATGACATTGGTGGTCCTGATCAAGAATTTGGTGTGGACGTTGGCCCTGTTTGCTTT TTATAAACCAAACTCTATCTGAAATCCCAACAAAAAAAATTTAACTCCATATGTGTTCCTCTTGTTCTAATCTTGTCAACCAGTGCAAGT GACCGACAAAATTCCAGTTATTTATTTCCAAAATGTTTGGAAACAGTATAATTTGACAAAGAAAAATGATACTTCTCTTTTTTTGCTGTT CCACCAAATACAATTCAAATGCTTTTTGTTTTATTTTTTTACCAATTCCAATTTCAAAATGTCTCAATGGTGCTATAATAAATAAACTTC AACACTCTTTATGATAACAACACTGTGTTATATTCTTTGAATCCTAGCCCATCTGCAGAGCAATGACTGTGCTCACCAGTAAAAGATAAC CTTTCTTTCTGAAATAGTCAAATACGAAATTAGAAAAGCCCTCCCTATTTTAACTACCTCAACTGGTCAGAAACACAGATTGTATTCTAT GAGTCCCAGAAGATGAAAAAAATTTTATACGTTGATAAAACTTATAAATTTCATTGATTAATCTCCTGGAAGATTGGTTTAAAAAGAAAA GTGTAATGCAAGAATTTAAAGAAATATTTTTAAAGCCACAATTATTTTAATATTGGATATCAACTGCTTGTAAAGGTGCTCCTCTTTTTT CTTGTCATTGCTGGTCAAGATTACTAATATTTGGGAAGGCTTTAAAGACGCATGTTATGGTGCTAATGTACTTTCACTTTTAAACTCTAG ATCAGAATTGTTGACTTGCATTCAGAACATAAATGCACAAAATCTGTACATGTCTCCCATCAGAAAGATTCATTGGCATGCCACAGGGGA TTCTCCTCCTTCATCCTGTAAAGGTCAACAATAAAAACCAAATTATGGGGCTGCTTTTGTCACACTAGCATAGAGAATGTGTTGAAATTT >25023_25023_3_EEF1A1-COL3A1_EEF1A1_chr6_74228077_ENST00000331523_COL3A1_chr2_189872852_ENST00000317840_length(amino acids)=343AA_BP= MGKEKTHINIVVIGHVDSGKSTTTGHLIYKCGGIDKRTIEKFEKEAAEMGKGSFKYAWVLDKLKAERERGITIDISLWKFETSKYYVTII DAPGHRDFIKNMITGTSQADCAVLIVAAGVGEFEAGISKNGQTREHALLAYTLGVKQLIVGVNKMDSTEPPYSQKRYEEIVKEVSTYIKK IGYNPDTVAFVPISGWNGDNMLEPSANMPWFKGWKVTRKDGNASGTTLLEALDCILPPTRPTDKPLRLPLQDVYKIGGIGTVPVGRVETG -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for EEF1A1-COL3A1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for EEF1A1-COL3A1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for EEF1A1-COL3A1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
