
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:EEF1G-ENG (FusionGDB2 ID:25201) |
Fusion Gene Summary for EEF1G-ENG |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: EEF1G-ENG | Fusion gene ID: 25201 | Hgene | Tgene | Gene symbol | EEF1G | ENG | Gene ID | 1937 | 2022 |
| Gene name | eukaryotic translation elongation factor 1 gamma | endoglin | |
| Synonyms | EF1G|GIG35 | END|HHT1|ORW1 | |
| Cytomap | 11q12.3 | 9q34.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | elongation factor 1-gammaEF-1-gammaPRO1608eEF-1B gammapancreatic tumor-related proteintranslation elongation factor eEF-1 gamma chain | endoglinCD105 antigen | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | P26641 | Q8NFI3 | |
| Ensembl transtripts involved in fusion gene | ENST00000329251, ENST00000378019, ENST00000532986, | ENST00000480266, ENST00000344849, ENST00000373203, | |
| Fusion gene scores | * DoF score | 13 X 13 X 9=1521 | 6 X 7 X 4=168 |
| # samples | 21 | 6 | |
| ** MAII score | log2(21/1521*10)=-2.85655892005837 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/168*10)=-1.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: EEF1G [Title/Abstract] AND ENG [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | EEF1G(62334870)-ENG(130605524), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | ENG | GO:0001934 | positive regulation of protein phosphorylation | 12015308 |
| Tgene | ENG | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation | 12015308 |
| Tgene | ENG | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 15702480 |
| Tgene | ENG | GO:0030336 | negative regulation of cell migration | 19736306 |
| Tgene | ENG | GO:0030513 | positive regulation of BMP signaling pathway | 17068149 |
| Tgene | ENG | GO:0031953 | negative regulation of protein autophosphorylation | 12015308 |
 Fusion gene breakpoints across EEF1G (5'-gene) Fusion gene breakpoints across EEF1G (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
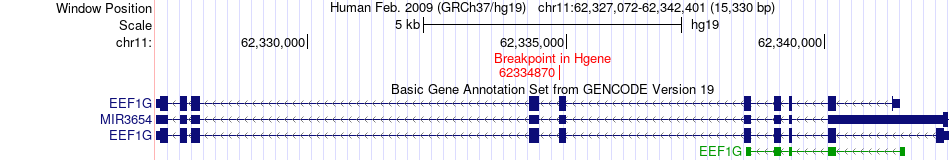 |
 Fusion gene breakpoints across ENG (3'-gene) Fusion gene breakpoints across ENG (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
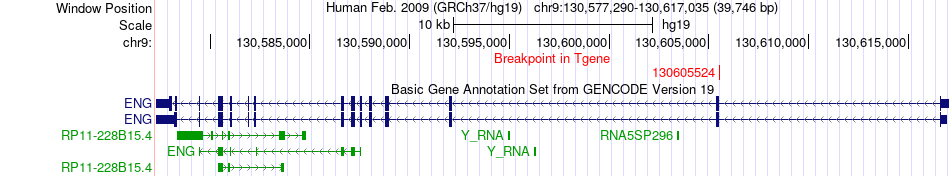 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCEC | TCGA-EO-A1Y5 | EEF1G | chr11 | 62334870 | - | ENG | chr9 | 130605524 | - |
Top |
Fusion Gene ORF analysis for EEF1G-ENG |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000329251 | ENST00000480266 | EEF1G | chr11 | 62334870 | - | ENG | chr9 | 130605524 | - |
| 5CDS-intron | ENST00000378019 | ENST00000480266 | EEF1G | chr11 | 62334870 | - | ENG | chr9 | 130605524 | - |
| In-frame | ENST00000329251 | ENST00000344849 | EEF1G | chr11 | 62334870 | - | ENG | chr9 | 130605524 | - |
| In-frame | ENST00000329251 | ENST00000373203 | EEF1G | chr11 | 62334870 | - | ENG | chr9 | 130605524 | - |
| In-frame | ENST00000378019 | ENST00000344849 | EEF1G | chr11 | 62334870 | - | ENG | chr9 | 130605524 | - |
| In-frame | ENST00000378019 | ENST00000373203 | EEF1G | chr11 | 62334870 | - | ENG | chr9 | 130605524 | - |
| intron-3CDS | ENST00000532986 | ENST00000344849 | EEF1G | chr11 | 62334870 | - | ENG | chr9 | 130605524 | - |
| intron-3CDS | ENST00000532986 | ENST00000373203 | EEF1G | chr11 | 62334870 | - | ENG | chr9 | 130605524 | - |
| intron-intron | ENST00000532986 | ENST00000480266 | EEF1G | chr11 | 62334870 | - | ENG | chr9 | 130605524 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000329251 | EEF1G | chr11 | 62334870 | - | ENST00000373203 | ENG | chr9 | 130605524 | - | 3363 | 783 | 5 | 2692 | 895 |
| ENST00000329251 | EEF1G | chr11 | 62334870 | - | ENST00000344849 | ENG | chr9 | 130605524 | - | 3494 | 783 | 5 | 2593 | 862 |
| ENST00000378019 | EEF1G | chr11 | 62334870 | - | ENST00000373203 | ENG | chr9 | 130605524 | - | 3473 | 893 | 91 | 2802 | 903 |
| ENST00000378019 | EEF1G | chr11 | 62334870 | - | ENST00000344849 | ENG | chr9 | 130605524 | - | 3604 | 893 | 91 | 2703 | 870 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000329251 | ENST00000373203 | EEF1G | chr11 | 62334870 | - | ENG | chr9 | 130605524 | - | 0.010438094 | 0.98956186 |
| ENST00000329251 | ENST00000344849 | EEF1G | chr11 | 62334870 | - | ENG | chr9 | 130605524 | - | 0.010686318 | 0.98931366 |
| ENST00000378019 | ENST00000373203 | EEF1G | chr11 | 62334870 | - | ENG | chr9 | 130605524 | - | 0.005064794 | 0.9949352 |
| ENST00000378019 | ENST00000344849 | EEF1G | chr11 | 62334870 | - | ENG | chr9 | 130605524 | - | 0.005283155 | 0.9947168 |
Top |
Fusion Genomic Features for EEF1G-ENG |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
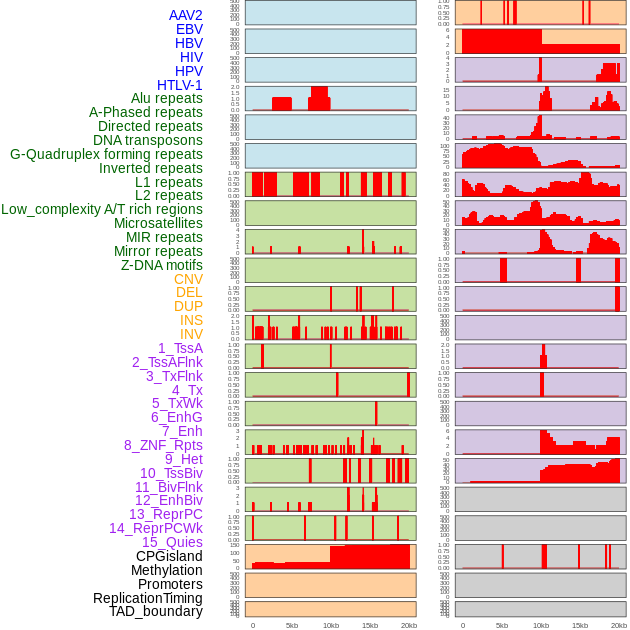 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for EEF1G-ENG |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:62334870/chr9:130605524) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| EEF1G | ENG |
| FUNCTION: Probably plays a role in anchoring the complex to other cellular components. | FUNCTION: Endoglycosidase that releases N-glycans from glycoproteins by cleaving the beta-1,4-glycosidic bond in the N,N'-diacetylchitobiose core. Involved in the processing of free oligosaccharides in the cytosol. {ECO:0000269|PubMed:12114544}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EEF1G | chr11:62334870 | chr9:130605524 | ENST00000329251 | - | 6 | 10 | 2_87 | 217 | 438.0 | Domain | Note=GST N-terminal |
| Hgene | EEF1G | chr11:62334870 | chr9:130605524 | ENST00000329251 | - | 6 | 10 | 88_216 | 217 | 438.0 | Domain | Note=GST C-terminal |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000344849 | 0 | 14 | 336_576 | 22 | 626.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000373203 | 0 | 15 | 336_576 | 22 | 659.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000344849 | 0 | 14 | 363_533 | 22 | 626.0 | Domain | ZP | |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000373203 | 0 | 15 | 363_533 | 22 | 659.0 | Domain | ZP | |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000344849 | 0 | 14 | 399_401 | 22 | 626.0 | Motif | Cell attachment site | |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000373203 | 0 | 15 | 399_401 | 22 | 659.0 | Motif | Cell attachment site | |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000344849 | 0 | 14 | 200_330 | 22 | 626.0 | Region | OR1%2C C-terminal part | |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000344849 | 0 | 14 | 26_46 | 22 | 626.0 | Region | OR1%2C N-terminal part | |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000344849 | 0 | 14 | 47_199 | 22 | 626.0 | Region | OR2 | |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000373203 | 0 | 15 | 200_330 | 22 | 659.0 | Region | OR1%2C C-terminal part | |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000373203 | 0 | 15 | 26_46 | 22 | 659.0 | Region | OR1%2C N-terminal part | |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000373203 | 0 | 15 | 47_199 | 22 | 659.0 | Region | OR2 | |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000344849 | 0 | 14 | 26_586 | 22 | 626.0 | Topological domain | Extracellular | |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000344849 | 0 | 14 | 612_658 | 22 | 626.0 | Topological domain | Cytoplasmic | |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000373203 | 0 | 15 | 26_586 | 22 | 659.0 | Topological domain | Extracellular | |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000373203 | 0 | 15 | 612_658 | 22 | 659.0 | Topological domain | Cytoplasmic | |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000344849 | 0 | 14 | 587_611 | 22 | 626.0 | Transmembrane | Helical | |
| Tgene | ENG | chr11:62334870 | chr9:130605524 | ENST00000373203 | 0 | 15 | 587_611 | 22 | 659.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EEF1G | chr11:62334870 | chr9:130605524 | ENST00000329251 | - | 6 | 10 | 276_437 | 217 | 438.0 | Domain | EF-1-gamma C-terminal |
Top |
Fusion Gene Sequence for EEF1G-ENG |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >25201_25201_1_EEF1G-ENG_EEF1G_chr11_62334870_ENST00000329251_ENG_chr9_130605524_ENST00000344849_length(transcript)=3494nt_BP=783nt GTCTTCTGTTTCGTCCTCGCTTTCCGGCTGCTGTTTCTCCACGGCTCTCCTCTTTCCCCCTCCCTTCTCTCCCGGGCGGCTTACTTTGCG GCAGCGCCGAGAACCCCACCCCCTTTCTTTGCGGAATCACCATGGCGGCTGGGACCCTGTACACGTATCCTGAAAACTGGAGGGCCTTCA AGGCTCTCATCGCTGCTCAGTACAGCGGGGCTCAGGTCCGCGTGCTCTCCGCACCACCCCACTTCCATTTTGGCCAAACCAACCGCACCC CTGAATTTCTCCGCAAATTTCCTGCCGGCAAGGTCCCAGCATTTGAGGGTGATGATGGATTCTGTGTGTTTGAGAGCAACGCCATTGCCT ACTATGTGAGCAATGAGGAGCTGCGGGGAAGTACTCCAGAGGCAGCAGCCCAGGTGGTGCAGTGGGTGAGCTTTGCTGATTCCGATATAG TGCCCCCAGCCAGTACCTGGGTGTTCCCCACCTTGGGCATCATGCACCACAACAAACAGGCCACTGAGAATGCAAAGGAGGAAGTGAGGC GAATTCTGGGGCTGCTGGATGCTTACTTGAAGACGAGGACTTTTCTGGTGGGCGAACGAGTGACATTGGCTGACATCACAGTTGTCTGCA CCCTGTTGTGGCTCTATAAGCAGGTTCTAGAGCCTTCTTTCCGCCAGGCCTTTCCCAATACCAACCGCTGGTTCCTCACCTGCATTAACC AGCCCCAGTTCCGGGCTGTCTTGGGCGAAGTGAAACTGTGTGAGAAGATGGCCCAGTTTGATGGTCTTGCAGAAACAGTCCATTGTGACC TTCAGCCTGTGGGCCCCGAGAGGGGCGAGGTGACATATACCACTAGCCAGGTCTCGAAGGGCTGCGTGGCTCAGGCCCCCAATGCCATCC TTGAAGTCCATGTCCTCTTCCTGGAGTTCCCAACGGGCCCGTCACAGCTGGAGCTGACTCTCCAGGCATCCAAGCAAAATGGCACCTGGC CCCGAGAGGTGCTTCTGGTCCTCAGTGTAAACAGCAGTGTCTTCCTGCATCTCCAGGCCCTGGGAATCCCACTGCACTTGGCCTACAATT CCAGCCTGGTCACCTTCCAAGAGCCCCCGGGGGTCAACACCACAGAGCTGCCATCCTTCCCCAAGACCCAGATCCTTGAGTGGGCAGCTG AGAGGGGCCCCATCACCTCTGCTGCTGAGCTGAATGACCCCCAGAGCATCCTCCTCCGACTGGGCCAAGCCCAGGGGTCACTGTCCTTCT GCATGCTGGAAGCCAGCCAGGACATGGGCCGCACGCTCGAGTGGCGGCCGCGTACTCCAGCCTTGGTCCGGGGCTGCCACTTGGAAGGCG TGGCCGGCCACAAGGAGGCGCACATCCTGAGGGTCCTGCCGGGCCACTCGGCCGGGCCCCGGACGGTGACGGTGAAGGTGGAACTGAGCT GCGCACCCGGGGATCTCGATGCCGTCCTCATCCTGCAGGGTCCCCCCTACGTGTCCTGGCTCATCGACGCCAACCACAACATGCAGATCT GGACCACTGGAGAATACTCCTTCAAGATCTTTCCAGAGAAAAACATTCGTGGCTTCAAGCTCCCAGACACACCTCAAGGCCTCCTGGGGG AGGCCCGGATGCTCAATGCCAGCATTGTGGCATCCTTCGTGGAGCTACCGCTGGCCAGCATTGTCTCACTTCATGCCTCCAGCTGCGGTG GTAGGCTGCAGACCTCACCCGCACCGATCCAGACCACTCCTCCCAAGGACACTTGTAGCCCGGAGCTGCTCATGTCCTTGATCCAGACAA AGTGTGCCGACGACGCCATGACCCTGGTACTAAAGAAAGAGCTTGTTGCGCATTTGAAGTGCACCATCACGGGCCTGACCTTCTGGGACC CCAGCTGTGAGGCAGAGGACAGGGGTGACAAGTTTGTCTTGCGCAGTGCTTACTCCAGCTGTGGCATGCAGGTGTCAGCAAGTATGATCA GCAATGAGGCGGTGGTCAATATCCTGTCGAGCTCATCACCACAGCGGAAAAAGGTGCACTGCCTCAACATGGACAGCCTCTCTTTCCAGC TGGGCCTCTACCTCAGCCCACACTTCCTCCAGGCCTCCAACACCATCGAGCCGGGGCAGCAGAGCTTTGTGCAGGTCAGAGTGTCCCCAT CCGTCTCCGAGTTCCTGCTCCAGTTAGACAGCTGCCACCTGGACTTGGGGCCTGAGGGAGGCACCGTGGAACTCATCCAGGGCCGGGCGG CCAAGGGCAACTGTGTGAGCCTGCTGTCCCCAAGCCCCGAGGGTGACCCGCGCTTCAGCTTCCTCCTCCACTTCTACACAGTACCCATAC CCAAAACCGGCACCCTCAGCTGCACGGTAGCCCTGCGTCCCAAGACCGGGTCTCAAGACCAGGAAGTCCATAGGACTGTCTTCATGCGCT TGAACATCATCAGCCCTGACCTGTCTGGTTGCACAAGCAAAGGCCTCGTCCTGCCCGCCGTGCTGGGCATCACCTTTGGTGCCTTCCTCA TCGGGGCCCTGCTCACTGCTGCACTCTGGTACATCTACTCGCACACGCGTGAGTACCCCAGGCCCCCACAGTGAGCATGCCGGGCCCCTC CATCCACCCGGGGGAGCCCAGTGAAGCCTCTGAGGGATTGAGGGGCCCTGGCCAGGACCCTGACCTCCGCCCCTGCCCCCGCTCCCGCTC CCAGGTTCCCCCAGCAAGCGGGAGCCCGTGGTGGCGGTGGCTGCCCCGGCCTCCTCGGAGAGCAGCAGCACCAACCACAGCATCGGGAGC ACCCAGAGCACCCCCTGCTCCACCAGCAGCATGGCATAGCCCCGGCCCCCCGCGCTCGCCCAGCAGGAGAGACTGAGCAGCCGCCAGCTG GGAGCACTGGTGTGAACTCACCCTGGGAGCCAGTCCTCCACTCGACCCAGAATGGAGCCTGCTCTCCGCGCCTACCCTTCCCGCCTCCCT CTCAGAGGCCTGCTGCCAGTGCAGCCACTGGCTTGGAACACCTTGGGGTCCCTCCACCCCACAGAACCTTCAACCCAGTGGGTCTGGGAT ATGGCTGCCCAGGAGACAGACCACTTGCCACGCTGTTGTAAAAACCCAAGTCCCTGTCATTTGAACCTGGATCCAGCACTGGTGAACTGA GCTGGGCAGGAAGGGAGAACTTGAAACAGATTCAGGCCAGCCCAGCCAGGCCAACAGCACCTCCCCGCTGGGAAGAGAAGAGGGCCCAGC CCAGAGCCACCTGGATCTATCCCTGCGGCCTCCACACCTGAACTTGCCTAACTAACTGGCAGGGGAGACAGGAGCCTAGCGGAGCCCAGC CTGGGAGCCCAGAGGGTGGCAAGAACAGTGGGCGTTGGGAGCCTAGCTCCTGCCACATGGAGCCCCCTCTGCCGGTCGGGCAGCCAGCAG >25201_25201_1_EEF1G-ENG_EEF1G_chr11_62334870_ENST00000329251_ENG_chr9_130605524_ENST00000344849_length(amino acids)=862AA_BP=259 MFRPRFPAAVSPRLSSFPLPSLPGGLLCGSAENPTPFLCGITMAAGTLYTYPENWRAFKALIAAQYSGAQVRVLSAPPHFHFGQTNRTPE FLRKFPAGKVPAFEGDDGFCVFESNAIAYYVSNEELRGSTPEAAAQVVQWVSFADSDIVPPASTWVFPTLGIMHHNKQATENAKEEVRRI LGLLDAYLKTRTFLVGERVTLADITVVCTLLWLYKQVLEPSFRQAFPNTNRWFLTCINQPQFRAVLGEVKLCEKMAQFDGLAETVHCDLQ PVGPERGEVTYTTSQVSKGCVAQAPNAILEVHVLFLEFPTGPSQLELTLQASKQNGTWPREVLLVLSVNSSVFLHLQALGIPLHLAYNSS LVTFQEPPGVNTTELPSFPKTQILEWAAERGPITSAAELNDPQSILLRLGQAQGSLSFCMLEASQDMGRTLEWRPRTPALVRGCHLEGVA GHKEAHILRVLPGHSAGPRTVTVKVELSCAPGDLDAVLILQGPPYVSWLIDANHNMQIWTTGEYSFKIFPEKNIRGFKLPDTPQGLLGEA RMLNASIVASFVELPLASIVSLHASSCGGRLQTSPAPIQTTPPKDTCSPELLMSLIQTKCADDAMTLVLKKELVAHLKCTITGLTFWDPS CEAEDRGDKFVLRSAYSSCGMQVSASMISNEAVVNILSSSSPQRKKVHCLNMDSLSFQLGLYLSPHFLQASNTIEPGQQSFVQVRVSPSV SEFLLQLDSCHLDLGPEGGTVELIQGRAAKGNCVSLLSPSPEGDPRFSFLLHFYTVPIPKTGTLSCTVALRPKTGSQDQEVHRTVFMRLN -------------------------------------------------------------- >25201_25201_2_EEF1G-ENG_EEF1G_chr11_62334870_ENST00000329251_ENG_chr9_130605524_ENST00000373203_length(transcript)=3363nt_BP=783nt GTCTTCTGTTTCGTCCTCGCTTTCCGGCTGCTGTTTCTCCACGGCTCTCCTCTTTCCCCCTCCCTTCTCTCCCGGGCGGCTTACTTTGCG GCAGCGCCGAGAACCCCACCCCCTTTCTTTGCGGAATCACCATGGCGGCTGGGACCCTGTACACGTATCCTGAAAACTGGAGGGCCTTCA AGGCTCTCATCGCTGCTCAGTACAGCGGGGCTCAGGTCCGCGTGCTCTCCGCACCACCCCACTTCCATTTTGGCCAAACCAACCGCACCC CTGAATTTCTCCGCAAATTTCCTGCCGGCAAGGTCCCAGCATTTGAGGGTGATGATGGATTCTGTGTGTTTGAGAGCAACGCCATTGCCT ACTATGTGAGCAATGAGGAGCTGCGGGGAAGTACTCCAGAGGCAGCAGCCCAGGTGGTGCAGTGGGTGAGCTTTGCTGATTCCGATATAG TGCCCCCAGCCAGTACCTGGGTGTTCCCCACCTTGGGCATCATGCACCACAACAAACAGGCCACTGAGAATGCAAAGGAGGAAGTGAGGC GAATTCTGGGGCTGCTGGATGCTTACTTGAAGACGAGGACTTTTCTGGTGGGCGAACGAGTGACATTGGCTGACATCACAGTTGTCTGCA CCCTGTTGTGGCTCTATAAGCAGGTTCTAGAGCCTTCTTTCCGCCAGGCCTTTCCCAATACCAACCGCTGGTTCCTCACCTGCATTAACC AGCCCCAGTTCCGGGCTGTCTTGGGCGAAGTGAAACTGTGTGAGAAGATGGCCCAGTTTGATGGTCTTGCAGAAACAGTCCATTGTGACC TTCAGCCTGTGGGCCCCGAGAGGGGCGAGGTGACATATACCACTAGCCAGGTCTCGAAGGGCTGCGTGGCTCAGGCCCCCAATGCCATCC TTGAAGTCCATGTCCTCTTCCTGGAGTTCCCAACGGGCCCGTCACAGCTGGAGCTGACTCTCCAGGCATCCAAGCAAAATGGCACCTGGC CCCGAGAGGTGCTTCTGGTCCTCAGTGTAAACAGCAGTGTCTTCCTGCATCTCCAGGCCCTGGGAATCCCACTGCACTTGGCCTACAATT CCAGCCTGGTCACCTTCCAAGAGCCCCCGGGGGTCAACACCACAGAGCTGCCATCCTTCCCCAAGACCCAGATCCTTGAGTGGGCAGCTG AGAGGGGCCCCATCACCTCTGCTGCTGAGCTGAATGACCCCCAGAGCATCCTCCTCCGACTGGGCCAAGCCCAGGGGTCACTGTCCTTCT GCATGCTGGAAGCCAGCCAGGACATGGGCCGCACGCTCGAGTGGCGGCCGCGTACTCCAGCCTTGGTCCGGGGCTGCCACTTGGAAGGCG TGGCCGGCCACAAGGAGGCGCACATCCTGAGGGTCCTGCCGGGCCACTCGGCCGGGCCCCGGACGGTGACGGTGAAGGTGGAACTGAGCT GCGCACCCGGGGATCTCGATGCCGTCCTCATCCTGCAGGGTCCCCCCTACGTGTCCTGGCTCATCGACGCCAACCACAACATGCAGATCT GGACCACTGGAGAATACTCCTTCAAGATCTTTCCAGAGAAAAACATTCGTGGCTTCAAGCTCCCAGACACACCTCAAGGCCTCCTGGGGG AGGCCCGGATGCTCAATGCCAGCATTGTGGCATCCTTCGTGGAGCTACCGCTGGCCAGCATTGTCTCACTTCATGCCTCCAGCTGCGGTG GTAGGCTGCAGACCTCACCCGCACCGATCCAGACCACTCCTCCCAAGGACACTTGTAGCCCGGAGCTGCTCATGTCCTTGATCCAGACAA AGTGTGCCGACGACGCCATGACCCTGGTACTAAAGAAAGAGCTTGTTGCGCATTTGAAGTGCACCATCACGGGCCTGACCTTCTGGGACC CCAGCTGTGAGGCAGAGGACAGGGGTGACAAGTTTGTCTTGCGCAGTGCTTACTCCAGCTGTGGCATGCAGGTGTCAGCAAGTATGATCA GCAATGAGGCGGTGGTCAATATCCTGTCGAGCTCATCACCACAGCGGAAAAAGGTGCACTGCCTCAACATGGACAGCCTCTCTTTCCAGC TGGGCCTCTACCTCAGCCCACACTTCCTCCAGGCCTCCAACACCATCGAGCCGGGGCAGCAGAGCTTTGTGCAGGTCAGAGTGTCCCCAT CCGTCTCCGAGTTCCTGCTCCAGTTAGACAGCTGCCACCTGGACTTGGGGCCTGAGGGAGGCACCGTGGAACTCATCCAGGGCCGGGCGG CCAAGGGCAACTGTGTGAGCCTGCTGTCCCCAAGCCCCGAGGGTGACCCGCGCTTCAGCTTCCTCCTCCACTTCTACACAGTACCCATAC CCAAAACCGGCACCCTCAGCTGCACGGTAGCCCTGCGTCCCAAGACCGGGTCTCAAGACCAGGAAGTCCATAGGACTGTCTTCATGCGCT TGAACATCATCAGCCCTGACCTGTCTGGTTGCACAAGCAAAGGCCTCGTCCTGCCCGCCGTGCTGGGCATCACCTTTGGTGCCTTCCTCA TCGGGGCCCTGCTCACTGCTGCACTCTGGTACATCTACTCGCACACGCGTTCCCCCAGCAAGCGGGAGCCCGTGGTGGCGGTGGCTGCCC CGGCCTCCTCGGAGAGCAGCAGCACCAACCACAGCATCGGGAGCACCCAGAGCACCCCCTGCTCCACCAGCAGCATGGCATAGCCCCGGC CCCCCGCGCTCGCCCAGCAGGAGAGACTGAGCAGCCGCCAGCTGGGAGCACTGGTGTGAACTCACCCTGGGAGCCAGTCCTCCACTCGAC CCAGAATGGAGCCTGCTCTCCGCGCCTACCCTTCCCGCCTCCCTCTCAGAGGCCTGCTGCCAGTGCAGCCACTGGCTTGGAACACCTTGG GGTCCCTCCACCCCACAGAACCTTCAACCCAGTGGGTCTGGGATATGGCTGCCCAGGAGACAGACCACTTGCCACGCTGTTGTAAAAACC CAAGTCCCTGTCATTTGAACCTGGATCCAGCACTGGTGAACTGAGCTGGGCAGGAAGGGAGAACTTGAAACAGATTCAGGCCAGCCCAGC CAGGCCAACAGCACCTCCCCGCTGGGAAGAGAAGAGGGCCCAGCCCAGAGCCACCTGGATCTATCCCTGCGGCCTCCACACCTGAACTTG CCTAACTAACTGGCAGGGGAGACAGGAGCCTAGCGGAGCCCAGCCTGGGAGCCCAGAGGGTGGCAAGAACAGTGGGCGTTGGGAGCCTAG CTCCTGCCACATGGAGCCCCCTCTGCCGGTCGGGCAGCCAGCAGAGGGGGAGTAGCCAAGCTGCTTGTCCTGGGCCTGCCCCTGTGTATT >25201_25201_2_EEF1G-ENG_EEF1G_chr11_62334870_ENST00000329251_ENG_chr9_130605524_ENST00000373203_length(amino acids)=895AA_BP=259 MFRPRFPAAVSPRLSSFPLPSLPGGLLCGSAENPTPFLCGITMAAGTLYTYPENWRAFKALIAAQYSGAQVRVLSAPPHFHFGQTNRTPE FLRKFPAGKVPAFEGDDGFCVFESNAIAYYVSNEELRGSTPEAAAQVVQWVSFADSDIVPPASTWVFPTLGIMHHNKQATENAKEEVRRI LGLLDAYLKTRTFLVGERVTLADITVVCTLLWLYKQVLEPSFRQAFPNTNRWFLTCINQPQFRAVLGEVKLCEKMAQFDGLAETVHCDLQ PVGPERGEVTYTTSQVSKGCVAQAPNAILEVHVLFLEFPTGPSQLELTLQASKQNGTWPREVLLVLSVNSSVFLHLQALGIPLHLAYNSS LVTFQEPPGVNTTELPSFPKTQILEWAAERGPITSAAELNDPQSILLRLGQAQGSLSFCMLEASQDMGRTLEWRPRTPALVRGCHLEGVA GHKEAHILRVLPGHSAGPRTVTVKVELSCAPGDLDAVLILQGPPYVSWLIDANHNMQIWTTGEYSFKIFPEKNIRGFKLPDTPQGLLGEA RMLNASIVASFVELPLASIVSLHASSCGGRLQTSPAPIQTTPPKDTCSPELLMSLIQTKCADDAMTLVLKKELVAHLKCTITGLTFWDPS CEAEDRGDKFVLRSAYSSCGMQVSASMISNEAVVNILSSSSPQRKKVHCLNMDSLSFQLGLYLSPHFLQASNTIEPGQQSFVQVRVSPSV SEFLLQLDSCHLDLGPEGGTVELIQGRAAKGNCVSLLSPSPEGDPRFSFLLHFYTVPIPKTGTLSCTVALRPKTGSQDQEVHRTVFMRLN -------------------------------------------------------------- >25201_25201_3_EEF1G-ENG_EEF1G_chr11_62334870_ENST00000378019_ENG_chr9_130605524_ENST00000344849_length(transcript)=3604nt_BP=893nt ACCAGCTTCTCCTTCCTACAGTTCGCACAGCTCGAGGGGAGTGCGGGGGCAGCACCGCGCGTGGCACCTGGCACCCGCGCAGAGTTCCTG AATGGCAGAGCGGTGGGTGGCTCCAGCAGTACTGAGAAGAGCCAGGTTTGCCTCCACATTCTTCCTGTCGCCTCAGATTTATGCGCACAA GGACGGAGATTTGCGTTCAGCGTTCTTCATTCTCTCCTTTAAACGAGGAGAGTTCATCCCGTTCTTAAACTGGACCCTGTACACGTATCC TGAAAACTGGAGGGCCTTCAAGGCTCTCATCGCTGCTCAGTACAGCGGGGCTCAGGTCCGCGTGCTCTCCGCACCACCCCACTTCCATTT TGGCCAAACCAACCGCACCCCTGAATTTCTCCGCAAATTTCCTGCCGGCAAGGTCCCAGCATTTGAGGGTGATGATGGATTCTGTGTGTT TGAGAGCAACGCCATTGCCTACTATGTGAGCAATGAGGAGCTGCGGGGAAGTACTCCAGAGGCAGCAGCCCAGGTGGTGCAGTGGGTGAG CTTTGCTGATTCCGATATAGTGCCCCCAGCCAGTACCTGGGTGTTCCCCACCTTGGGCATCATGCACCACAACAAACAGGCCACTGAGAA TGCAAAGGAGGAAGTGAGGCGAATTCTGGGGCTGCTGGATGCTTACTTGAAGACGAGGACTTTTCTGGTGGGCGAACGAGTGACATTGGC TGACATCACAGTTGTCTGCACCCTGTTGTGGCTCTATAAGCAGGTTCTAGAGCCTTCTTTCCGCCAGGCCTTTCCCAATACCAACCGCTG GTTCCTCACCTGCATTAACCAGCCCCAGTTCCGGGCTGTCTTGGGCGAAGTGAAACTGTGTGAGAAGATGGCCCAGTTTGATGGTCTTGC AGAAACAGTCCATTGTGACCTTCAGCCTGTGGGCCCCGAGAGGGGCGAGGTGACATATACCACTAGCCAGGTCTCGAAGGGCTGCGTGGC TCAGGCCCCCAATGCCATCCTTGAAGTCCATGTCCTCTTCCTGGAGTTCCCAACGGGCCCGTCACAGCTGGAGCTGACTCTCCAGGCATC CAAGCAAAATGGCACCTGGCCCCGAGAGGTGCTTCTGGTCCTCAGTGTAAACAGCAGTGTCTTCCTGCATCTCCAGGCCCTGGGAATCCC ACTGCACTTGGCCTACAATTCCAGCCTGGTCACCTTCCAAGAGCCCCCGGGGGTCAACACCACAGAGCTGCCATCCTTCCCCAAGACCCA GATCCTTGAGTGGGCAGCTGAGAGGGGCCCCATCACCTCTGCTGCTGAGCTGAATGACCCCCAGAGCATCCTCCTCCGACTGGGCCAAGC CCAGGGGTCACTGTCCTTCTGCATGCTGGAAGCCAGCCAGGACATGGGCCGCACGCTCGAGTGGCGGCCGCGTACTCCAGCCTTGGTCCG GGGCTGCCACTTGGAAGGCGTGGCCGGCCACAAGGAGGCGCACATCCTGAGGGTCCTGCCGGGCCACTCGGCCGGGCCCCGGACGGTGAC GGTGAAGGTGGAACTGAGCTGCGCACCCGGGGATCTCGATGCCGTCCTCATCCTGCAGGGTCCCCCCTACGTGTCCTGGCTCATCGACGC CAACCACAACATGCAGATCTGGACCACTGGAGAATACTCCTTCAAGATCTTTCCAGAGAAAAACATTCGTGGCTTCAAGCTCCCAGACAC ACCTCAAGGCCTCCTGGGGGAGGCCCGGATGCTCAATGCCAGCATTGTGGCATCCTTCGTGGAGCTACCGCTGGCCAGCATTGTCTCACT TCATGCCTCCAGCTGCGGTGGTAGGCTGCAGACCTCACCCGCACCGATCCAGACCACTCCTCCCAAGGACACTTGTAGCCCGGAGCTGCT CATGTCCTTGATCCAGACAAAGTGTGCCGACGACGCCATGACCCTGGTACTAAAGAAAGAGCTTGTTGCGCATTTGAAGTGCACCATCAC GGGCCTGACCTTCTGGGACCCCAGCTGTGAGGCAGAGGACAGGGGTGACAAGTTTGTCTTGCGCAGTGCTTACTCCAGCTGTGGCATGCA GGTGTCAGCAAGTATGATCAGCAATGAGGCGGTGGTCAATATCCTGTCGAGCTCATCACCACAGCGGAAAAAGGTGCACTGCCTCAACAT GGACAGCCTCTCTTTCCAGCTGGGCCTCTACCTCAGCCCACACTTCCTCCAGGCCTCCAACACCATCGAGCCGGGGCAGCAGAGCTTTGT GCAGGTCAGAGTGTCCCCATCCGTCTCCGAGTTCCTGCTCCAGTTAGACAGCTGCCACCTGGACTTGGGGCCTGAGGGAGGCACCGTGGA ACTCATCCAGGGCCGGGCGGCCAAGGGCAACTGTGTGAGCCTGCTGTCCCCAAGCCCCGAGGGTGACCCGCGCTTCAGCTTCCTCCTCCA CTTCTACACAGTACCCATACCCAAAACCGGCACCCTCAGCTGCACGGTAGCCCTGCGTCCCAAGACCGGGTCTCAAGACCAGGAAGTCCA TAGGACTGTCTTCATGCGCTTGAACATCATCAGCCCTGACCTGTCTGGTTGCACAAGCAAAGGCCTCGTCCTGCCCGCCGTGCTGGGCAT CACCTTTGGTGCCTTCCTCATCGGGGCCCTGCTCACTGCTGCACTCTGGTACATCTACTCGCACACGCGTGAGTACCCCAGGCCCCCACA GTGAGCATGCCGGGCCCCTCCATCCACCCGGGGGAGCCCAGTGAAGCCTCTGAGGGATTGAGGGGCCCTGGCCAGGACCCTGACCTCCGC CCCTGCCCCCGCTCCCGCTCCCAGGTTCCCCCAGCAAGCGGGAGCCCGTGGTGGCGGTGGCTGCCCCGGCCTCCTCGGAGAGCAGCAGCA CCAACCACAGCATCGGGAGCACCCAGAGCACCCCCTGCTCCACCAGCAGCATGGCATAGCCCCGGCCCCCCGCGCTCGCCCAGCAGGAGA GACTGAGCAGCCGCCAGCTGGGAGCACTGGTGTGAACTCACCCTGGGAGCCAGTCCTCCACTCGACCCAGAATGGAGCCTGCTCTCCGCG CCTACCCTTCCCGCCTCCCTCTCAGAGGCCTGCTGCCAGTGCAGCCACTGGCTTGGAACACCTTGGGGTCCCTCCACCCCACAGAACCTT CAACCCAGTGGGTCTGGGATATGGCTGCCCAGGAGACAGACCACTTGCCACGCTGTTGTAAAAACCCAAGTCCCTGTCATTTGAACCTGG ATCCAGCACTGGTGAACTGAGCTGGGCAGGAAGGGAGAACTTGAAACAGATTCAGGCCAGCCCAGCCAGGCCAACAGCACCTCCCCGCTG GGAAGAGAAGAGGGCCCAGCCCAGAGCCACCTGGATCTATCCCTGCGGCCTCCACACCTGAACTTGCCTAACTAACTGGCAGGGGAGACA GGAGCCTAGCGGAGCCCAGCCTGGGAGCCCAGAGGGTGGCAAGAACAGTGGGCGTTGGGAGCCTAGCTCCTGCCACATGGAGCCCCCTCT GCCGGTCGGGCAGCCAGCAGAGGGGGAGTAGCCAAGCTGCTTGTCCTGGGCCTGCCCCTGTGTATTCACCACCAATAAATCAGACCATGA >25201_25201_3_EEF1G-ENG_EEF1G_chr11_62334870_ENST00000378019_ENG_chr9_130605524_ENST00000344849_length(amino acids)=870AA_BP=267 MAERWVAPAVLRRARFASTFFLSPQIYAHKDGDLRSAFFILSFKRGEFIPFLNWTLYTYPENWRAFKALIAAQYSGAQVRVLSAPPHFHF GQTNRTPEFLRKFPAGKVPAFEGDDGFCVFESNAIAYYVSNEELRGSTPEAAAQVVQWVSFADSDIVPPASTWVFPTLGIMHHNKQATEN AKEEVRRILGLLDAYLKTRTFLVGERVTLADITVVCTLLWLYKQVLEPSFRQAFPNTNRWFLTCINQPQFRAVLGEVKLCEKMAQFDGLA ETVHCDLQPVGPERGEVTYTTSQVSKGCVAQAPNAILEVHVLFLEFPTGPSQLELTLQASKQNGTWPREVLLVLSVNSSVFLHLQALGIP LHLAYNSSLVTFQEPPGVNTTELPSFPKTQILEWAAERGPITSAAELNDPQSILLRLGQAQGSLSFCMLEASQDMGRTLEWRPRTPALVR GCHLEGVAGHKEAHILRVLPGHSAGPRTVTVKVELSCAPGDLDAVLILQGPPYVSWLIDANHNMQIWTTGEYSFKIFPEKNIRGFKLPDT PQGLLGEARMLNASIVASFVELPLASIVSLHASSCGGRLQTSPAPIQTTPPKDTCSPELLMSLIQTKCADDAMTLVLKKELVAHLKCTIT GLTFWDPSCEAEDRGDKFVLRSAYSSCGMQVSASMISNEAVVNILSSSSPQRKKVHCLNMDSLSFQLGLYLSPHFLQASNTIEPGQQSFV QVRVSPSVSEFLLQLDSCHLDLGPEGGTVELIQGRAAKGNCVSLLSPSPEGDPRFSFLLHFYTVPIPKTGTLSCTVALRPKTGSQDQEVH -------------------------------------------------------------- >25201_25201_4_EEF1G-ENG_EEF1G_chr11_62334870_ENST00000378019_ENG_chr9_130605524_ENST00000373203_length(transcript)=3473nt_BP=893nt ACCAGCTTCTCCTTCCTACAGTTCGCACAGCTCGAGGGGAGTGCGGGGGCAGCACCGCGCGTGGCACCTGGCACCCGCGCAGAGTTCCTG AATGGCAGAGCGGTGGGTGGCTCCAGCAGTACTGAGAAGAGCCAGGTTTGCCTCCACATTCTTCCTGTCGCCTCAGATTTATGCGCACAA GGACGGAGATTTGCGTTCAGCGTTCTTCATTCTCTCCTTTAAACGAGGAGAGTTCATCCCGTTCTTAAACTGGACCCTGTACACGTATCC TGAAAACTGGAGGGCCTTCAAGGCTCTCATCGCTGCTCAGTACAGCGGGGCTCAGGTCCGCGTGCTCTCCGCACCACCCCACTTCCATTT TGGCCAAACCAACCGCACCCCTGAATTTCTCCGCAAATTTCCTGCCGGCAAGGTCCCAGCATTTGAGGGTGATGATGGATTCTGTGTGTT TGAGAGCAACGCCATTGCCTACTATGTGAGCAATGAGGAGCTGCGGGGAAGTACTCCAGAGGCAGCAGCCCAGGTGGTGCAGTGGGTGAG CTTTGCTGATTCCGATATAGTGCCCCCAGCCAGTACCTGGGTGTTCCCCACCTTGGGCATCATGCACCACAACAAACAGGCCACTGAGAA TGCAAAGGAGGAAGTGAGGCGAATTCTGGGGCTGCTGGATGCTTACTTGAAGACGAGGACTTTTCTGGTGGGCGAACGAGTGACATTGGC TGACATCACAGTTGTCTGCACCCTGTTGTGGCTCTATAAGCAGGTTCTAGAGCCTTCTTTCCGCCAGGCCTTTCCCAATACCAACCGCTG GTTCCTCACCTGCATTAACCAGCCCCAGTTCCGGGCTGTCTTGGGCGAAGTGAAACTGTGTGAGAAGATGGCCCAGTTTGATGGTCTTGC AGAAACAGTCCATTGTGACCTTCAGCCTGTGGGCCCCGAGAGGGGCGAGGTGACATATACCACTAGCCAGGTCTCGAAGGGCTGCGTGGC TCAGGCCCCCAATGCCATCCTTGAAGTCCATGTCCTCTTCCTGGAGTTCCCAACGGGCCCGTCACAGCTGGAGCTGACTCTCCAGGCATC CAAGCAAAATGGCACCTGGCCCCGAGAGGTGCTTCTGGTCCTCAGTGTAAACAGCAGTGTCTTCCTGCATCTCCAGGCCCTGGGAATCCC ACTGCACTTGGCCTACAATTCCAGCCTGGTCACCTTCCAAGAGCCCCCGGGGGTCAACACCACAGAGCTGCCATCCTTCCCCAAGACCCA GATCCTTGAGTGGGCAGCTGAGAGGGGCCCCATCACCTCTGCTGCTGAGCTGAATGACCCCCAGAGCATCCTCCTCCGACTGGGCCAAGC CCAGGGGTCACTGTCCTTCTGCATGCTGGAAGCCAGCCAGGACATGGGCCGCACGCTCGAGTGGCGGCCGCGTACTCCAGCCTTGGTCCG GGGCTGCCACTTGGAAGGCGTGGCCGGCCACAAGGAGGCGCACATCCTGAGGGTCCTGCCGGGCCACTCGGCCGGGCCCCGGACGGTGAC GGTGAAGGTGGAACTGAGCTGCGCACCCGGGGATCTCGATGCCGTCCTCATCCTGCAGGGTCCCCCCTACGTGTCCTGGCTCATCGACGC CAACCACAACATGCAGATCTGGACCACTGGAGAATACTCCTTCAAGATCTTTCCAGAGAAAAACATTCGTGGCTTCAAGCTCCCAGACAC ACCTCAAGGCCTCCTGGGGGAGGCCCGGATGCTCAATGCCAGCATTGTGGCATCCTTCGTGGAGCTACCGCTGGCCAGCATTGTCTCACT TCATGCCTCCAGCTGCGGTGGTAGGCTGCAGACCTCACCCGCACCGATCCAGACCACTCCTCCCAAGGACACTTGTAGCCCGGAGCTGCT CATGTCCTTGATCCAGACAAAGTGTGCCGACGACGCCATGACCCTGGTACTAAAGAAAGAGCTTGTTGCGCATTTGAAGTGCACCATCAC GGGCCTGACCTTCTGGGACCCCAGCTGTGAGGCAGAGGACAGGGGTGACAAGTTTGTCTTGCGCAGTGCTTACTCCAGCTGTGGCATGCA GGTGTCAGCAAGTATGATCAGCAATGAGGCGGTGGTCAATATCCTGTCGAGCTCATCACCACAGCGGAAAAAGGTGCACTGCCTCAACAT GGACAGCCTCTCTTTCCAGCTGGGCCTCTACCTCAGCCCACACTTCCTCCAGGCCTCCAACACCATCGAGCCGGGGCAGCAGAGCTTTGT GCAGGTCAGAGTGTCCCCATCCGTCTCCGAGTTCCTGCTCCAGTTAGACAGCTGCCACCTGGACTTGGGGCCTGAGGGAGGCACCGTGGA ACTCATCCAGGGCCGGGCGGCCAAGGGCAACTGTGTGAGCCTGCTGTCCCCAAGCCCCGAGGGTGACCCGCGCTTCAGCTTCCTCCTCCA CTTCTACACAGTACCCATACCCAAAACCGGCACCCTCAGCTGCACGGTAGCCCTGCGTCCCAAGACCGGGTCTCAAGACCAGGAAGTCCA TAGGACTGTCTTCATGCGCTTGAACATCATCAGCCCTGACCTGTCTGGTTGCACAAGCAAAGGCCTCGTCCTGCCCGCCGTGCTGGGCAT CACCTTTGGTGCCTTCCTCATCGGGGCCCTGCTCACTGCTGCACTCTGGTACATCTACTCGCACACGCGTTCCCCCAGCAAGCGGGAGCC CGTGGTGGCGGTGGCTGCCCCGGCCTCCTCGGAGAGCAGCAGCACCAACCACAGCATCGGGAGCACCCAGAGCACCCCCTGCTCCACCAG CAGCATGGCATAGCCCCGGCCCCCCGCGCTCGCCCAGCAGGAGAGACTGAGCAGCCGCCAGCTGGGAGCACTGGTGTGAACTCACCCTGG GAGCCAGTCCTCCACTCGACCCAGAATGGAGCCTGCTCTCCGCGCCTACCCTTCCCGCCTCCCTCTCAGAGGCCTGCTGCCAGTGCAGCC ACTGGCTTGGAACACCTTGGGGTCCCTCCACCCCACAGAACCTTCAACCCAGTGGGTCTGGGATATGGCTGCCCAGGAGACAGACCACTT GCCACGCTGTTGTAAAAACCCAAGTCCCTGTCATTTGAACCTGGATCCAGCACTGGTGAACTGAGCTGGGCAGGAAGGGAGAACTTGAAA CAGATTCAGGCCAGCCCAGCCAGGCCAACAGCACCTCCCCGCTGGGAAGAGAAGAGGGCCCAGCCCAGAGCCACCTGGATCTATCCCTGC GGCCTCCACACCTGAACTTGCCTAACTAACTGGCAGGGGAGACAGGAGCCTAGCGGAGCCCAGCCTGGGAGCCCAGAGGGTGGCAAGAAC AGTGGGCGTTGGGAGCCTAGCTCCTGCCACATGGAGCCCCCTCTGCCGGTCGGGCAGCCAGCAGAGGGGGAGTAGCCAAGCTGCTTGTCC >25201_25201_4_EEF1G-ENG_EEF1G_chr11_62334870_ENST00000378019_ENG_chr9_130605524_ENST00000373203_length(amino acids)=903AA_BP=267 MAERWVAPAVLRRARFASTFFLSPQIYAHKDGDLRSAFFILSFKRGEFIPFLNWTLYTYPENWRAFKALIAAQYSGAQVRVLSAPPHFHF GQTNRTPEFLRKFPAGKVPAFEGDDGFCVFESNAIAYYVSNEELRGSTPEAAAQVVQWVSFADSDIVPPASTWVFPTLGIMHHNKQATEN AKEEVRRILGLLDAYLKTRTFLVGERVTLADITVVCTLLWLYKQVLEPSFRQAFPNTNRWFLTCINQPQFRAVLGEVKLCEKMAQFDGLA ETVHCDLQPVGPERGEVTYTTSQVSKGCVAQAPNAILEVHVLFLEFPTGPSQLELTLQASKQNGTWPREVLLVLSVNSSVFLHLQALGIP LHLAYNSSLVTFQEPPGVNTTELPSFPKTQILEWAAERGPITSAAELNDPQSILLRLGQAQGSLSFCMLEASQDMGRTLEWRPRTPALVR GCHLEGVAGHKEAHILRVLPGHSAGPRTVTVKVELSCAPGDLDAVLILQGPPYVSWLIDANHNMQIWTTGEYSFKIFPEKNIRGFKLPDT PQGLLGEARMLNASIVASFVELPLASIVSLHASSCGGRLQTSPAPIQTTPPKDTCSPELLMSLIQTKCADDAMTLVLKKELVAHLKCTIT GLTFWDPSCEAEDRGDKFVLRSAYSSCGMQVSASMISNEAVVNILSSSSPQRKKVHCLNMDSLSFQLGLYLSPHFLQASNTIEPGQQSFV QVRVSPSVSEFLLQLDSCHLDLGPEGGTVELIQGRAAKGNCVSLLSPSPEGDPRFSFLLHFYTVPIPKTGTLSCTVALRPKTGSQDQEVH RTVFMRLNIISPDLSGCTSKGLVLPAVLGITFGAFLIGALLTAALWYIYSHTRSPSKREPVVAVAAPASSESSSTNHSIGSTQSTPCSTS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for EEF1G-ENG |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for EEF1G-ENG |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for EEF1G-ENG |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
