
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:EHBP1-BCL11A (FusionGDB2 ID:25525) |
Fusion Gene Summary for EHBP1-BCL11A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: EHBP1-BCL11A | Fusion gene ID: 25525 | Hgene | Tgene | Gene symbol | EHBP1 | BCL11A | Gene ID | 23301 | 53335 |
| Gene name | EH domain binding protein 1 | BAF chromatin remodeling complex subunit BCL11A | |
| Synonyms | HPC12|NACSIN | BCL11A-L|BCL11A-S|BCL11A-XL|BCL11a-M|CTIP1|DILOS|EVI9|HBFQTL5|ZNF856 | |
| Cytomap | 2p15 | 2p16.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | EH domain-binding protein 1NPF calponin-like proteintestis tissue sperm-binding protein Li 50e | B-cell lymphoma/leukemia 11AB cell CLL/lymphoma 11AB-cell CLL/lymphoma 11A (zinc finger protein) isoform 2BCL11A B-cell CLL/lymphoma 11A (zinc finger protein) isoform 1BCL11A, BAF complex componentC2H2-type zinc finger proteinCOUP-TF-interacting pro | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8NDI1 | Q9H165 | |
| Ensembl transtripts involved in fusion gene | ENST00000263991, ENST00000354487, ENST00000405015, ENST00000405289, ENST00000431489, ENST00000496857, | ENST00000335712, ENST00000358510, ENST00000477659, ENST00000356842, ENST00000359629, ENST00000538214, ENST00000537768, | |
| Fusion gene scores | * DoF score | 8 X 12 X 8=768 | 10 X 9 X 3=270 |
| # samples | 20 | 9 | |
| ** MAII score | log2(20/768*10)=-1.94110631094643 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/270*10)=-1.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: EHBP1 [Title/Abstract] AND BCL11A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | EHBP1(63101667)-BCL11A(60679801), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | EHBP1-BCL11A seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. EHBP1-BCL11A seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. EHBP1-BCL11A seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | BCL11A | GO:0000122 | negative regulation of transcription by RNA polymerase II | 19153051 |
 Fusion gene breakpoints across EHBP1 (5'-gene) Fusion gene breakpoints across EHBP1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
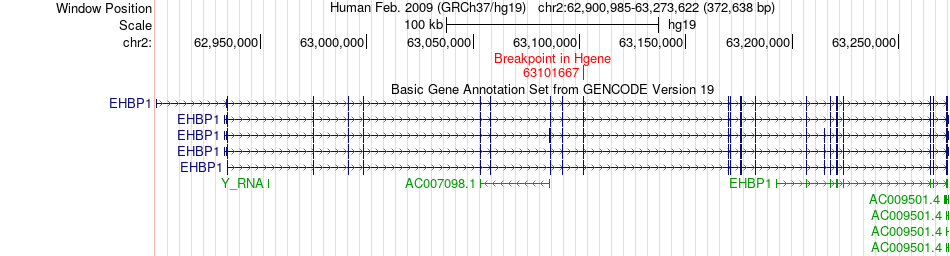 |
 Fusion gene breakpoints across BCL11A (3'-gene) Fusion gene breakpoints across BCL11A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
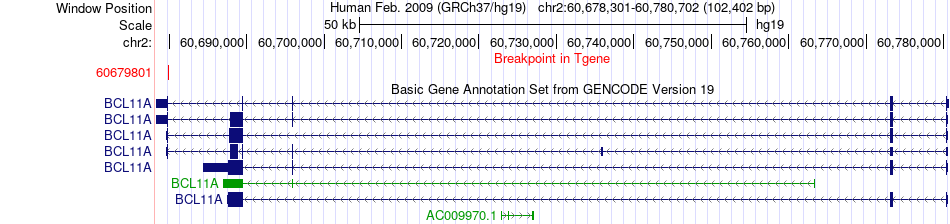 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-D8-A147-01A | EHBP1 | chr2 | 63101667 | - | BCL11A | chr2 | 60679801 | - |
| ChimerDB4 | BRCA | TCGA-D8-A147-01A | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
Top |
Fusion Gene ORF analysis for EHBP1-BCL11A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000263991 | ENST00000335712 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| 5CDS-intron | ENST00000263991 | ENST00000358510 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| 5CDS-intron | ENST00000263991 | ENST00000477659 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| 5CDS-intron | ENST00000354487 | ENST00000335712 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| 5CDS-intron | ENST00000354487 | ENST00000358510 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| 5CDS-intron | ENST00000354487 | ENST00000477659 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| 5CDS-intron | ENST00000405015 | ENST00000335712 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| 5CDS-intron | ENST00000405015 | ENST00000358510 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| 5CDS-intron | ENST00000405015 | ENST00000477659 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| 5CDS-intron | ENST00000405289 | ENST00000335712 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| 5CDS-intron | ENST00000405289 | ENST00000358510 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| 5CDS-intron | ENST00000405289 | ENST00000477659 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| 5CDS-intron | ENST00000431489 | ENST00000335712 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| 5CDS-intron | ENST00000431489 | ENST00000358510 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| 5CDS-intron | ENST00000431489 | ENST00000477659 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| Frame-shift | ENST00000263991 | ENST00000356842 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| Frame-shift | ENST00000263991 | ENST00000359629 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| Frame-shift | ENST00000263991 | ENST00000538214 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| Frame-shift | ENST00000354487 | ENST00000356842 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| Frame-shift | ENST00000354487 | ENST00000359629 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| Frame-shift | ENST00000354487 | ENST00000538214 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| Frame-shift | ENST00000405015 | ENST00000356842 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| Frame-shift | ENST00000405015 | ENST00000359629 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| Frame-shift | ENST00000405015 | ENST00000538214 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| Frame-shift | ENST00000405289 | ENST00000356842 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| Frame-shift | ENST00000405289 | ENST00000359629 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| Frame-shift | ENST00000405289 | ENST00000538214 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| Frame-shift | ENST00000431489 | ENST00000356842 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| Frame-shift | ENST00000431489 | ENST00000359629 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| Frame-shift | ENST00000431489 | ENST00000538214 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| In-frame | ENST00000263991 | ENST00000537768 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| In-frame | ENST00000354487 | ENST00000537768 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| In-frame | ENST00000405015 | ENST00000537768 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| In-frame | ENST00000405289 | ENST00000537768 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| In-frame | ENST00000431489 | ENST00000537768 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| intron-3CDS | ENST00000496857 | ENST00000356842 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| intron-3CDS | ENST00000496857 | ENST00000359629 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| intron-3CDS | ENST00000496857 | ENST00000537768 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| intron-3CDS | ENST00000496857 | ENST00000538214 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| intron-intron | ENST00000496857 | ENST00000335712 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| intron-intron | ENST00000496857 | ENST00000358510 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
| intron-intron | ENST00000496857 | ENST00000477659 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000405015 | EHBP1 | chr2 | 63101667 | + | ENST00000537768 | BCL11A | chr2 | 60679801 | - | 1880 | 1713 | 519 | 1814 | 431 |
| ENST00000431489 | EHBP1 | chr2 | 63101667 | + | ENST00000537768 | BCL11A | chr2 | 60679801 | - | 1958 | 1791 | 597 | 1892 | 431 |
| ENST00000354487 | EHBP1 | chr2 | 63101667 | + | ENST00000537768 | BCL11A | chr2 | 60679801 | - | 1834 | 1667 | 473 | 1768 | 431 |
| ENST00000263991 | EHBP1 | chr2 | 63101667 | + | ENST00000537768 | BCL11A | chr2 | 60679801 | - | 1939 | 1772 | 473 | 1873 | 466 |
| ENST00000405289 | EHBP1 | chr2 | 63101667 | + | ENST00000537768 | BCL11A | chr2 | 60679801 | - | 1352 | 1185 | 0 | 1286 | 428 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000405015 | ENST00000537768 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - | 0.00142861 | 0.9985714 |
| ENST00000431489 | ENST00000537768 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - | 0.001398618 | 0.9986014 |
| ENST00000354487 | ENST00000537768 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - | 0.001373942 | 0.9986261 |
| ENST00000263991 | ENST00000537768 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - | 0.001268179 | 0.9987318 |
| ENST00000405289 | ENST00000537768 | EHBP1 | chr2 | 63101667 | + | BCL11A | chr2 | 60679801 | - | 0.002551905 | 0.9974481 |
Top |
Fusion Genomic Features for EHBP1-BCL11A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
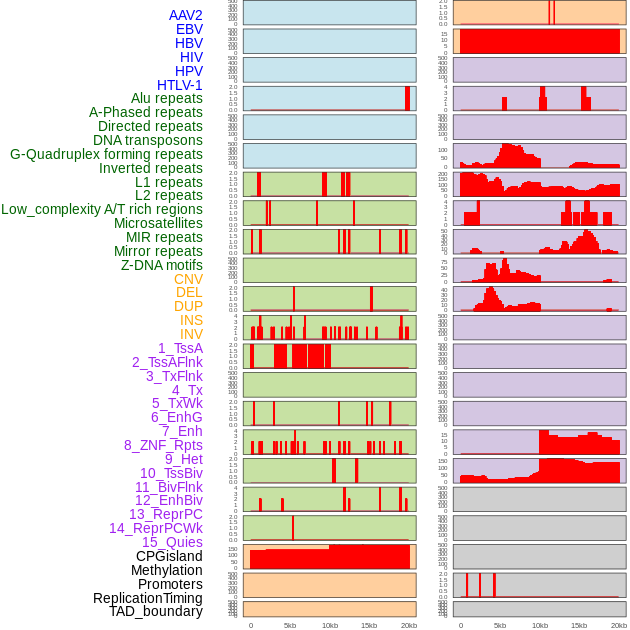 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for EHBP1-BCL11A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:63101667/chr2:60679801) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| EHBP1 | BCL11A |
| FUNCTION: May play a role in actin reorganization. Links clathrin-mediated endocytosis to the actin cytoskeleton. May act as Rab effector protein and play a role in vesicle trafficking (PubMed:14676205, PubMed:27552051). Required for perinuclear sorting and insulin-regulated recycling of SLC2A4/GLUT4 in adipocytes (By similarity). {ECO:0000250|UniProtKB:Q69ZW3, ECO:0000269|PubMed:14676205, ECO:0000305|PubMed:27552051}. | FUNCTION: Transcription factor associated with the BAF SWI/SNF chromatin remodeling complex (By similarity). Repressor of fetal hemoglobin (HbF) level (PubMed:26375765). Involved in brain development (PubMed:27453576). May play a role in hematopoiesis. Essential factor in lymphopoiesis required for B-cell formation in fetal liver. May function as a modulator of the transcriptional repression activity of ARP1 (By similarity). {ECO:0000250|UniProtKB:Q9QYE3, ECO:0000303|PubMed:26375765, ECO:0000303|PubMed:27453576}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000263991 | + | 11 | 25 | 185_210 | 430 | 1232.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000354487 | + | 10 | 24 | 185_210 | 395 | 1197.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405015 | + | 10 | 23 | 185_210 | 395 | 1161.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405289 | + | 9 | 23 | 185_210 | 395 | 1197.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000431489 | + | 10 | 23 | 185_210 | 395 | 1161.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000263991 | + | 11 | 25 | 362_365 | 430 | 1232.0 | Compositional bias | Note=Poly-Glu |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000354487 | + | 10 | 24 | 362_365 | 395 | 1197.0 | Compositional bias | Note=Poly-Glu |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405015 | + | 10 | 23 | 362_365 | 395 | 1161.0 | Compositional bias | Note=Poly-Glu |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405289 | + | 9 | 23 | 362_365 | 395 | 1197.0 | Compositional bias | Note=Poly-Glu |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000431489 | + | 10 | 23 | 362_365 | 395 | 1161.0 | Compositional bias | Note=Poly-Glu |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000263991 | + | 11 | 25 | 8_158 | 430 | 1232.0 | Domain | C2 NT-type |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000354487 | + | 10 | 24 | 8_158 | 395 | 1197.0 | Domain | C2 NT-type |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405015 | + | 10 | 23 | 8_158 | 395 | 1161.0 | Domain | C2 NT-type |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405289 | + | 9 | 23 | 8_158 | 395 | 1197.0 | Domain | C2 NT-type |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000431489 | + | 10 | 23 | 8_158 | 395 | 1161.0 | Domain | C2 NT-type |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000335712 | 0 | 4 | 260_373 | 0 | 836.0 | Compositional bias | Note=Pro-rich | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000335712 | 0 | 4 | 481_509 | 0 | 836.0 | Compositional bias | Note=Glu-rich | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000358510 | 0 | 3 | 260_373 | 0 | 802.0 | Compositional bias | Note=Pro-rich | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000358510 | 0 | 3 | 481_509 | 0 | 802.0 | Compositional bias | Note=Glu-rich | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000359629 | 3 | 5 | 260_373 | 210 | 244.0 | Compositional bias | Note=Pro-rich | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000359629 | 3 | 5 | 481_509 | 210 | 244.0 | Compositional bias | Note=Glu-rich | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000335712 | 0 | 4 | 1_210 | 0 | 836.0 | Region | Required for nuclear body formation and for SUMO1 recruitment | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000358510 | 0 | 3 | 1_210 | 0 | 802.0 | Region | Required for nuclear body formation and for SUMO1 recruitment | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000335712 | 0 | 4 | 170_193 | 0 | 836.0 | Zinc finger | C2H2-type 1 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000335712 | 0 | 4 | 377_399 | 0 | 836.0 | Zinc finger | C2H2-type 2 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000335712 | 0 | 4 | 405_429 | 0 | 836.0 | Zinc finger | C2H2-type 3 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000335712 | 0 | 4 | 742_764 | 0 | 836.0 | Zinc finger | C2H2-type 4 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000335712 | 0 | 4 | 770_792 | 0 | 836.0 | Zinc finger | C2H2-type 5 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000335712 | 0 | 4 | 800_823 | 0 | 836.0 | Zinc finger | C2H2-type 6 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000356842 | 3 | 5 | 742_764 | 743 | 774.0 | Zinc finger | C2H2-type 4 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000356842 | 3 | 5 | 770_792 | 743 | 774.0 | Zinc finger | C2H2-type 5 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000356842 | 3 | 5 | 800_823 | 743 | 774.0 | Zinc finger | C2H2-type 6 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000358510 | 0 | 3 | 170_193 | 0 | 802.0 | Zinc finger | C2H2-type 1 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000358510 | 0 | 3 | 377_399 | 0 | 802.0 | Zinc finger | C2H2-type 2 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000358510 | 0 | 3 | 405_429 | 0 | 802.0 | Zinc finger | C2H2-type 3 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000358510 | 0 | 3 | 742_764 | 0 | 802.0 | Zinc finger | C2H2-type 4 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000358510 | 0 | 3 | 770_792 | 0 | 802.0 | Zinc finger | C2H2-type 5 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000358510 | 0 | 3 | 800_823 | 0 | 802.0 | Zinc finger | C2H2-type 6 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000359629 | 3 | 5 | 377_399 | 210 | 244.0 | Zinc finger | C2H2-type 2 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000359629 | 3 | 5 | 405_429 | 210 | 244.0 | Zinc finger | C2H2-type 3 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000359629 | 3 | 5 | 742_764 | 210 | 244.0 | Zinc finger | C2H2-type 4 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000359629 | 3 | 5 | 770_792 | 210 | 244.0 | Zinc finger | C2H2-type 5 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000359629 | 3 | 5 | 800_823 | 210 | 244.0 | Zinc finger | C2H2-type 6 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000263991 | + | 11 | 25 | 1076_1100 | 430 | 1232.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000263991 | + | 11 | 25 | 1136_1230 | 430 | 1232.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000263991 | + | 11 | 25 | 808_879 | 430 | 1232.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000354487 | + | 10 | 24 | 1076_1100 | 395 | 1197.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000354487 | + | 10 | 24 | 1136_1230 | 395 | 1197.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000354487 | + | 10 | 24 | 808_879 | 395 | 1197.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405015 | + | 10 | 23 | 1076_1100 | 395 | 1161.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405015 | + | 10 | 23 | 1136_1230 | 395 | 1161.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405015 | + | 10 | 23 | 808_879 | 395 | 1161.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405289 | + | 9 | 23 | 1076_1100 | 395 | 1197.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405289 | + | 9 | 23 | 1136_1230 | 395 | 1197.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405289 | + | 9 | 23 | 808_879 | 395 | 1197.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000431489 | + | 10 | 23 | 1076_1100 | 395 | 1161.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000431489 | + | 10 | 23 | 1136_1230 | 395 | 1161.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000431489 | + | 10 | 23 | 808_879 | 395 | 1161.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000263991 | + | 11 | 25 | 1110_1113 | 430 | 1232.0 | Compositional bias | Note=Poly-Glu |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000354487 | + | 10 | 24 | 1110_1113 | 395 | 1197.0 | Compositional bias | Note=Poly-Glu |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405015 | + | 10 | 23 | 1110_1113 | 395 | 1161.0 | Compositional bias | Note=Poly-Glu |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405289 | + | 9 | 23 | 1110_1113 | 395 | 1197.0 | Compositional bias | Note=Poly-Glu |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000431489 | + | 10 | 23 | 1110_1113 | 395 | 1161.0 | Compositional bias | Note=Poly-Glu |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000263991 | + | 11 | 25 | 1056_1212 | 430 | 1232.0 | Domain | bMERB |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000263991 | + | 11 | 25 | 443_548 | 430 | 1232.0 | Domain | Calponin-homology (CH) |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000354487 | + | 10 | 24 | 1056_1212 | 395 | 1197.0 | Domain | bMERB |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000354487 | + | 10 | 24 | 443_548 | 395 | 1197.0 | Domain | Calponin-homology (CH) |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405015 | + | 10 | 23 | 1056_1212 | 395 | 1161.0 | Domain | bMERB |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405015 | + | 10 | 23 | 443_548 | 395 | 1161.0 | Domain | Calponin-homology (CH) |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405289 | + | 9 | 23 | 1056_1212 | 395 | 1197.0 | Domain | bMERB |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405289 | + | 9 | 23 | 443_548 | 395 | 1197.0 | Domain | Calponin-homology (CH) |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000431489 | + | 10 | 23 | 1056_1212 | 395 | 1161.0 | Domain | bMERB |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000431489 | + | 10 | 23 | 443_548 | 395 | 1161.0 | Domain | Calponin-homology (CH) |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000263991 | + | 11 | 25 | 1228_1231 | 430 | 1232.0 | Motif | CAAX motif |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000354487 | + | 10 | 24 | 1228_1231 | 395 | 1197.0 | Motif | CAAX motif |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405015 | + | 10 | 23 | 1228_1231 | 395 | 1161.0 | Motif | CAAX motif |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000405289 | + | 9 | 23 | 1228_1231 | 395 | 1197.0 | Motif | CAAX motif |
| Hgene | EHBP1 | chr2:63101667 | chr2:60679801 | ENST00000431489 | + | 10 | 23 | 1228_1231 | 395 | 1161.0 | Motif | CAAX motif |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000356842 | 3 | 5 | 260_373 | 743 | 774.0 | Compositional bias | Note=Pro-rich | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000356842 | 3 | 5 | 481_509 | 743 | 774.0 | Compositional bias | Note=Glu-rich | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000356842 | 3 | 5 | 1_210 | 743 | 774.0 | Region | Required for nuclear body formation and for SUMO1 recruitment | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000359629 | 3 | 5 | 1_210 | 210 | 244.0 | Region | Required for nuclear body formation and for SUMO1 recruitment | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000356842 | 3 | 5 | 170_193 | 743 | 774.0 | Zinc finger | C2H2-type 1 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000356842 | 3 | 5 | 377_399 | 743 | 774.0 | Zinc finger | C2H2-type 2 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000356842 | 3 | 5 | 405_429 | 743 | 774.0 | Zinc finger | C2H2-type 3 | |
| Tgene | BCL11A | chr2:63101667 | chr2:60679801 | ENST00000359629 | 3 | 5 | 170_193 | 210 | 244.0 | Zinc finger | C2H2-type 1 |
Top |
Fusion Gene Sequence for EHBP1-BCL11A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >25525_25525_1_EHBP1-BCL11A_EHBP1_chr2_63101667_ENST00000263991_BCL11A_chr2_60679801_ENST00000537768_length(transcript)=1939nt_BP=1772nt GGGCGCCGTCCGCCTGAGGGGCTGCAGATCCCGGCTACCGCGAGCGCCGTGGCTGGCTTGGCTGTTCCATGTGGCTCCGGCGGGGATGGA GGACTCGGTGGCGGCGGCGGCTGCTGCTGCGGCGGCGACGGAGGAGCGGCTATGAGAAGGGAACGGGGCCAGACTCAGCCCGACAGCGGG ATTAGAGCCCTCCAGAATACCCATCATATAGCCCCTGAGGTGGCATGGTGATGTCTCCATGAGGGAACCCCTTCCCACTCATCCTGTCAC GTATATCATAGTGTTCTTGACTGGGCCATTCATCTAAGATGGGATTTACCCTGTGAAACAGGGAGAAGACTTATGGACCCCAAGCATCAT TTCGAGTTGTAGTTGAGTTTTTAAAAGACATACATGCAAAGTTCCTTTGCTTTGGACCCTCTGCATTATTAAAGCTGCTGTATTGCTAAC CCAGAACTGCTCCAGTGTCTTGACTGATCATCATGGCTTCAGTTTGGAAGAGACTGCAGCGTGTGGGAAAACATGCATCCAAGTTCCAGT TTGTGGCCTCCTACCAGGAGCTCATGGTTGAGTGTACGAAGAAATGGCAACCAGATAAACTGGTGGTAGTTTGGACCAGAAGAAGCCGAA GGAAGTCTTCTAAGGCACATAGCTGGCAACCTGGAATAAAAAATCCCTATCGTGGTGTTGTTGTGTGGCCTGTTCCTGAAAACATTGAAA TCACTGTAACACTTTTTAAGGATCCTCATGCGGAAGAATTTGAAGACAAAGAGTGGACATTTGTCATAGAAAATGAATCCCCTTCTGGTC GAAGGAAAGCTCTTGCTACTAGCAGCATCAATATGAAACAGTATGCAAGCCCTATGCCAACTCAGACTGATGTCAAGTTAAAATTCAAGC CATTATCTAAAAAAGTTGTATCTGCCGCTCTTCAGTTTTCATTATCTTGCATTTTTCTGAGGGAAGGAAAAGCCACAGATGAAGACATGC AAAGTTTGGCTAGTTTGATGAGTATGAAGCAGGCTGACATTGGCAATTTAGATGACTTCGAAGAAGATAATGAAGATGATGATGAGAACA GAGTGAACCAAGAAGAAAAGGCAGCTAAAATTACAGAAATTGTAAACCAGTTGAATGCTCTGAGCAGCTTAGATGAAGATCAAGATGACT GCATAAAGCAAGCAAATATGCGTTCAGCTAAATCAGCCAGTTCCTCTGAAGAGCTTATCAACAAACTTAACTTTTTGGATGAAGCAGAAA AGGACTTGGCCACCGTGAATTCAAATCCATTTGATGATCCTGATGCTGCAGAATTAAATCCATTTGGAGATCCTGACTCAGAAGAACCTA TCACTGAAACAGCTTCACCTAGAAAAACAGAAGACTCTTTTTATAATAACAGCTATAATCCCTTTAAAGAGGTGCAGACTCCACAGTATT TGAACCCATTCGATGAGCCAGAAGCATTTGTGACCATAAAGGATTCTCCTCCCCAGTCTACAAAAAGAAAAAATATAAGACCTGTGGATA TGAGCAAGTACCTCTATGCTGATAGTTCTAAAACTGAAGAAGAAGAATTGGATGAATCAAATCCTTTTTATGAACCTAAATCAACTCCTC CTCCAAATAATTTGGTAAATCCTGTTCAAGAACTAGAAACTGAAAGGCGAGTGAAAAGAAAGGCCCCGGCTCCACCAGTCCTCTCACCAA AAACAGGAGTATTAAATGAAAACACAGTTTCTGCAGGAAAAGATCTCTCTACTTCTCCTAAGGTTCTTCACACACCCCCATTCGGCGTAG TACCCAGAGAGCTCAAGATGTGTGGCAGTTTTCGGATGGAAGCTCGAGAGCCCTTAAGTTCTGAGAAAATTTGAAGCCCCCAGGGGTGGG >25525_25525_1_EHBP1-BCL11A_EHBP1_chr2_63101667_ENST00000263991_BCL11A_chr2_60679801_ENST00000537768_length(amino acids)=466AA_BP=429 MIIMASVWKRLQRVGKHASKFQFVASYQELMVECTKKWQPDKLVVVWTRRSRRKSSKAHSWQPGIKNPYRGVVVWPVPENIEITVTLFKD PHAEEFEDKEWTFVIENESPSGRRKALATSSINMKQYASPMPTQTDVKLKFKPLSKKVVSAALQFSLSCIFLREGKATDEDMQSLASLMS MKQADIGNLDDFEEDNEDDDENRVNQEEKAAKITEIVNQLNALSSLDEDQDDCIKQANMRSAKSASSSEELINKLNFLDEAEKDLATVNS NPFDDPDAAELNPFGDPDSEEPITETASPRKTEDSFYNNSYNPFKEVQTPQYLNPFDEPEAFVTIKDSPPQSTKRKNIRPVDMSKYLYAD SSKTEEEELDESNPFYEPKSTPPPNNLVNPVQELETERRVKRKAPAPPVLSPKTGVLNENTVSAGKDLSTSPKVLHTPPFGVVPRELKMC -------------------------------------------------------------- >25525_25525_2_EHBP1-BCL11A_EHBP1_chr2_63101667_ENST00000354487_BCL11A_chr2_60679801_ENST00000537768_length(transcript)=1834nt_BP=1667nt GGGCGCCGTCCGCCTGAGGGGCTGCAGATCCCGGCTACCGCGAGCGCCGTGGCTGGCTTGGCTGTTCCATGTGGCTCCGGCGGGGATGGA GGACTCGGTGGCGGCGGCGGCTGCTGCTGCGGCGGCGACGGAGGAGCGGCTATGAGAAGGGAACGGGGCCAGACTCAGCCCGACAGCGGG ATTAGAGCCCTCCAGAATACCCATCATATAGCCCCTGAGGTGGCATGGTGATGTCTCCATGAGGGAACCCCTTCCCACTCATCCTGTCAC GTATATCATAGTGTTCTTGACTGGGCCATTCATCTAAGATGGGATTTACCCTGTGAAACAGGGAGAAGACTTATGGACCCCAAGCATCAT TTCGAGTTGTAGTTGAGTTTTTAAAAGACATACATGCAAAGTTCCTTTGCTTTGGACCCTCTGCATTATTAAAGCTGCTGTATTGCTAAC CCAGAACTGCTCCAGTGTCTTGACTGATCATCATGGCTTCAGTTTGGAAGAGACTGCAGCGTGTGGGAAAACATGCATCCAAGTTCCAGT TTGTGGCCTCCTACCAGGAGCTCATGGTTGAGTGTACGAAGAAATGGCAACCAGATAAACTGGTGGTAGTTTGGACCAGAAGAAGCCGAA GGAAGTCTTCTAAGGCACATAGCTGGCAACCTGGAATAAAAAATCCCTATCGTGGTGTTGTTGTGTGGCCTGTTCCTGAAAACATTGAAA TCACTGTAACACTTTTTAAGGATCCTCATGCGGAAGAATTTGAAGACAAAGAGTGGACATTTGTCATAGAAAATGAATCCCCTTCTGGTC GAAGGAAAGCTCTTGCTACTAGCAGCATCAATATGAAACAGTATGCAAGCCCTATGCCAACTCAGACTGATGTCAAGTTAAAATTCAAGC CATTATCTAAAAAAGTTGTATCTGCCGCTCTTCAGTTTTCATTATCTTGCATTTTTCTGAGGGAAGGAAAAGCCACAGATGAAGACATGC AAAGTTTGGCTAGTTTGATGAGTATGAAGCAGGCTGACATTGGCAATTTAGATGACTTCGAAGAAGATAATGAAGATGATGATGAGAACA GAGTGAACCAAGAAGAAAAGGCAGCTAAAATTACAGAGCTTATCAACAAACTTAACTTTTTGGATGAAGCAGAAAAGGACTTGGCCACCG TGAATTCAAATCCATTTGATGATCCTGATGCTGCAGAATTAAATCCATTTGGAGATCCTGACTCAGAAGAACCTATCACTGAAACAGCTT CACCTAGAAAAACAGAAGACTCTTTTTATAATAACAGCTATAATCCCTTTAAAGAGGTGCAGACTCCACAGTATTTGAACCCATTCGATG AGCCAGAAGCATTTGTGACCATAAAGGATTCTCCTCCCCAGTCTACAAAAAGAAAAAATATAAGACCTGTGGATATGAGCAAGTACCTCT ATGCTGATAGTTCTAAAACTGAAGAAGAAGAATTGGATGAATCAAATCCTTTTTATGAACCTAAATCAACTCCTCCTCCAAATAATTTGG TAAATCCTGTTCAAGAACTAGAAACTGAAAGGCGAGTGAAAAGAAAGGCCCCGGCTCCACCAGTCCTCTCACCAAAAACAGGAGTATTAA ATGAAAACACAGTTTCTGCAGGAAAAGATCTCTCTACTTCTCCTAAGGTTCTTCACACACCCCCATTCGGCGTAGTACCCAGAGAGCTCA AGATGTGTGGCAGTTTTCGGATGGAAGCTCGAGAGCCCTTAAGTTCTGAGAAAATTTGAAGCCCCCAGGGGTGGGGTGGACGCGTGCCGC >25525_25525_2_EHBP1-BCL11A_EHBP1_chr2_63101667_ENST00000354487_BCL11A_chr2_60679801_ENST00000537768_length(amino acids)=431AA_BP=394 MIIMASVWKRLQRVGKHASKFQFVASYQELMVECTKKWQPDKLVVVWTRRSRRKSSKAHSWQPGIKNPYRGVVVWPVPENIEITVTLFKD PHAEEFEDKEWTFVIENESPSGRRKALATSSINMKQYASPMPTQTDVKLKFKPLSKKVVSAALQFSLSCIFLREGKATDEDMQSLASLMS MKQADIGNLDDFEEDNEDDDENRVNQEEKAAKITELINKLNFLDEAEKDLATVNSNPFDDPDAAELNPFGDPDSEEPITETASPRKTEDS FYNNSYNPFKEVQTPQYLNPFDEPEAFVTIKDSPPQSTKRKNIRPVDMSKYLYADSSKTEEEELDESNPFYEPKSTPPPNNLVNPVQELE -------------------------------------------------------------- >25525_25525_3_EHBP1-BCL11A_EHBP1_chr2_63101667_ENST00000405015_BCL11A_chr2_60679801_ENST00000537768_length(transcript)=1880nt_BP=1713nt AACTGGGAGGCTCCCCAGTTTGTGGGTAACATCTGACTCTCCCTCCCCAAAGAGAGACTCCAGCGAGAGACTGGGGATTTGGAGGTCCAA GTCATGGCAGAACTGGGTTGCTTGCTAGGACACAAGGTTAGCATGCCCCTGTAGCAGCAATGTGAGAATCAGAAAAGGTGACTGTGCATC TCAAGAAGCCGAATCAAAAAGCCAAGCCAACCACCTAACCAGTGATTTTCAAGCCCTCCAGAATACCCATCATATAGCCCCTGAGGTGGC ATGGTGATGTCTCCATGAGGGAACCCCTTCCCACTCATCCTGTCACGTATATCATAGTGTTCTTGACTGGGCCATTCATCTAAGATGGGA TTTACCCTGTGAAACAGGGAGAAGACTTATGGACCCCAAGCATCATTTCGAGTTGTAGTTGAGTTTTTAAAAGACATACATGCAAAGTTC CTTTGCTTTGGACCCTCTGCATTATTAAAGCTGCTGTATTGCTAACCCAGAACTGCTCCAGTGTCTTGACTGATCATCATGGCTTCAGTT TGGAAGAGACTGCAGCGTGTGGGAAAACATGCATCCAAGTTCCAGTTTGTGGCCTCCTACCAGGAGCTCATGGTTGAGTGTACGAAGAAA TGGCAACCAGATAAACTGGTGGTAGTTTGGACCAGAAGAAGCCGAAGGAAGTCTTCTAAGGCACATAGCTGGCAACCTGGAATAAAAAAT CCCTATCGTGGTGTTGTTGTGTGGCCTGTTCCTGAAAACATTGAAATCACTGTAACACTTTTTAAGGATCCTCATGCGGAAGAATTTGAA GACAAAGAGTGGACATTTGTCATAGAAAATGAATCCCCTTCTGGTCGAAGGAAAGCTCTTGCTACTAGCAGCATCAATATGAAACAGTAT GCAAGCCCTATGCCAACTCAGACTGATGTCAAGTTAAAATTCAAGCCATTATCTAAAAAAGTTGTATCTGCCGCTCTTCAGTTTTCATTA TCTTGCATTTTTCTGAGGGAAGGAAAAGCCACAGATGAAGACATGCAAAGTTTGGCTAGTTTGATGAGTATGAAGCAGGCTGACATTGGC AATTTAGATGACTTCGAAGAAGATAATGAAGATGATGATGAGAACAGAGTGAACCAAGAAGAAAAGGCAGCTAAAATTACAGAGCTTATC AACAAACTTAACTTTTTGGATGAAGCAGAAAAGGACTTGGCCACCGTGAATTCAAATCCATTTGATGATCCTGATGCTGCAGAATTAAAT CCATTTGGAGATCCTGACTCAGAAGAACCTATCACTGAAACAGCTTCACCTAGAAAAACAGAAGACTCTTTTTATAATAACAGCTATAAT CCCTTTAAAGAGGTGCAGACTCCACAGTATTTGAACCCATTCGATGAGCCAGAAGCATTTGTGACCATAAAGGATTCTCCTCCCCAGTCT ACAAAAAGAAAAAATATAAGACCTGTGGATATGAGCAAGTACCTCTATGCTGATAGTTCTAAAACTGAAGAAGAAGAATTGGATGAATCA AATCCTTTTTATGAACCTAAATCAACTCCTCCTCCAAATAATTTGGTAAATCCTGTTCAAGAACTAGAAACTGAAAGGCGAGTGAAAAGA AAGGCCCCGGCTCCACCAGTCCTCTCACCAAAAACAGGAGTATTAAATGAAAACACAGTTTCTGCAGGAAAAGATCTCTCTACTTCTCCT AAGGTTCTTCACACACCCCCATTCGGCGTAGTACCCAGAGAGCTCAAGATGTGTGGCAGTTTTCGGATGGAAGCTCGAGAGCCCTTAAGT >25525_25525_3_EHBP1-BCL11A_EHBP1_chr2_63101667_ENST00000405015_BCL11A_chr2_60679801_ENST00000537768_length(amino acids)=431AA_BP=394 MIIMASVWKRLQRVGKHASKFQFVASYQELMVECTKKWQPDKLVVVWTRRSRRKSSKAHSWQPGIKNPYRGVVVWPVPENIEITVTLFKD PHAEEFEDKEWTFVIENESPSGRRKALATSSINMKQYASPMPTQTDVKLKFKPLSKKVVSAALQFSLSCIFLREGKATDEDMQSLASLMS MKQADIGNLDDFEEDNEDDDENRVNQEEKAAKITELINKLNFLDEAEKDLATVNSNPFDDPDAAELNPFGDPDSEEPITETASPRKTEDS FYNNSYNPFKEVQTPQYLNPFDEPEAFVTIKDSPPQSTKRKNIRPVDMSKYLYADSSKTEEEELDESNPFYEPKSTPPPNNLVNPVQELE -------------------------------------------------------------- >25525_25525_4_EHBP1-BCL11A_EHBP1_chr2_63101667_ENST00000405289_BCL11A_chr2_60679801_ENST00000537768_length(transcript)=1352nt_BP=1185nt ATGGCTTCAGTTTGGAAGAGACTGCAGCGTGTGGGAAAACATGCATCCAAGTTCCAGTTTGTGGCCTCCTACCAGGAGCTCATGGTTGAG TGTACGAAGAAATGGCAACCAGATAAACTGGTGGTAGTTTGGACCAGAAGAAGCCGAAGGAAGTCTTCTAAGGCACATAGCTGGCAACCT GGAATAAAAAATCCCTATCGTGGTGTTGTTGTGTGGCCTGTTCCTGAAAACATTGAAATCACTGTAACACTTTTTAAGGATCCTCATGCG GAAGAATTTGAAGACAAAGAGTGGACATTTGTCATAGAAAATGAATCCCCTTCTGGTCGAAGGAAAGCTCTTGCTACTAGCAGCATCAAT ATGAAACAGTATGCAAGCCCTATGCCAACTCAGACTGATGTCAAGTTAAAATTCAAGCCATTATCTAAAAAAGTTGTATCTGCCGCTCTT CAGTTTTCATTATCTTGCATTTTTCTGAGGGAAGGAAAAGCCACAGATGAAGACATGCAAAGTTTGGCTAGTTTGATGAGTATGAAGCAG GCTGACATTGGCAATTTAGATGACTTCGAAGAAGATAATGAAGATGATGATGAGAACAGAGTGAACCAAGAAGAAAAGGCAGCTAAAATT ACAGAGCTTATCAACAAACTTAACTTTTTGGATGAAGCAGAAAAGGACTTGGCCACCGTGAATTCAAATCCATTTGATGATCCTGATGCT GCAGAATTAAATCCATTTGGAGATCCTGACTCAGAAGAACCTATCACTGAAACAGCTTCACCTAGAAAAACAGAAGACTCTTTTTATAAT AACAGCTATAATCCCTTTAAAGAGGTGCAGACTCCACAGTATTTGAACCCATTCGATGAGCCAGAAGCATTTGTGACCATAAAGGATTCT CCTCCCCAGTCTACAAAAAGAAAAAATATAAGACCTGTGGATATGAGCAAGTACCTCTATGCTGATAGTTCTAAAACTGAAGAAGAAGAA TTGGATGAATCAAATCCTTTTTATGAACCTAAATCAACTCCTCCTCCAAATAATTTGGTAAATCCTGTTCAAGAACTAGAAACTGAAAGG CGAGTGAAAAGAAAGGCCCCGGCTCCACCAGTCCTCTCACCAAAAACAGGAGTATTAAATGAAAACACAGTTTCTGCAGGAAAAGATCTC TCTACTTCTCCTAAGGTTCTTCACACACCCCCATTCGGCGTAGTACCCAGAGAGCTCAAGATGTGTGGCAGTTTTCGGATGGAAGCTCGA GAGCCCTTAAGTTCTGAGAAAATTTGAAGCCCCCAGGGGTGGGGTGGACGCGTGCCGCCCAGTCGACGTCAGCGTGGTCTGTCATCCTGC >25525_25525_4_EHBP1-BCL11A_EHBP1_chr2_63101667_ENST00000405289_BCL11A_chr2_60679801_ENST00000537768_length(amino acids)=428AA_BP=391 MASVWKRLQRVGKHASKFQFVASYQELMVECTKKWQPDKLVVVWTRRSRRKSSKAHSWQPGIKNPYRGVVVWPVPENIEITVTLFKDPHA EEFEDKEWTFVIENESPSGRRKALATSSINMKQYASPMPTQTDVKLKFKPLSKKVVSAALQFSLSCIFLREGKATDEDMQSLASLMSMKQ ADIGNLDDFEEDNEDDDENRVNQEEKAAKITELINKLNFLDEAEKDLATVNSNPFDDPDAAELNPFGDPDSEEPITETASPRKTEDSFYN NSYNPFKEVQTPQYLNPFDEPEAFVTIKDSPPQSTKRKNIRPVDMSKYLYADSSKTEEEELDESNPFYEPKSTPPPNNLVNPVQELETER -------------------------------------------------------------- >25525_25525_5_EHBP1-BCL11A_EHBP1_chr2_63101667_ENST00000431489_BCL11A_chr2_60679801_ENST00000537768_length(transcript)=1958nt_BP=1791nt GATGATGGTGGCGGAGGAGAAAGGCGGGGGAGGGGGTGCTGGGCGCTGAGCGGTGGCTCTGGCAGGGCTTGGTAGGGTGGGGGACGGTCG CGGGGCGATCTGTCAGGGAGGGGGTGGGCGTGAGGGGCGCCGTCCGCCTGAGGGGCTGCAGATCCCGGCTACCGCGAGCGCCGTGGCTGG CTTGGCTGTTCCATGTGGCTCCGGCGGGGATGGAGGACTCGGTGGCGGCGGCGGCTGCTGCTGCGGCGGCGACGGAGGAGCGGCTATGAG AAGGGAACGGGGCCAGACTCAGCCCGACAGCGGGATTAGAGCCCTCCAGAATACCCATCATATAGCCCCTGAGGTGGCATGGTGATGTCT CCATGAGGGAACCCCTTCCCACTCATCCTGTCACGTATATCATAGTGTTCTTGACTGGGCCATTCATCTAAGATGGGATTTACCCTGTGA AACAGGGAGAAGACTTATGGACCCCAAGCATCATTTCGAGTTGTAGTTGAGTTTTTAAAAGACATACATGCAAAGTTCCTTTGCTTTGGA CCCTCTGCATTATTAAAGCTGCTGTATTGCTAACCCAGAACTGCTCCAGTGTCTTGACTGATCATCATGGCTTCAGTTTGGAAGAGACTG CAGCGTGTGGGAAAACATGCATCCAAGTTCCAGTTTGTGGCCTCCTACCAGGAGCTCATGGTTGAGTGTACGAAGAAATGGCAACCAGAT AAACTGGTGGTAGTTTGGACCAGAAGAAGCCGAAGGAAGTCTTCTAAGGCACATAGCTGGCAACCTGGAATAAAAAATCCCTATCGTGGT GTTGTTGTGTGGCCTGTTCCTGAAAACATTGAAATCACTGTAACACTTTTTAAGGATCCTCATGCGGAAGAATTTGAAGACAAAGAGTGG ACATTTGTCATAGAAAATGAATCCCCTTCTGGTCGAAGGAAAGCTCTTGCTACTAGCAGCATCAATATGAAACAGTATGCAAGCCCTATG CCAACTCAGACTGATGTCAAGTTAAAATTCAAGCCATTATCTAAAAAAGTTGTATCTGCCGCTCTTCAGTTTTCATTATCTTGCATTTTT CTGAGGGAAGGAAAAGCCACAGATGAAGACATGCAAAGTTTGGCTAGTTTGATGAGTATGAAGCAGGCTGACATTGGCAATTTAGATGAC TTCGAAGAAGATAATGAAGATGATGATGAGAACAGAGTGAACCAAGAAGAAAAGGCAGCTAAAATTACAGAGCTTATCAACAAACTTAAC TTTTTGGATGAAGCAGAAAAGGACTTGGCCACCGTGAATTCAAATCCATTTGATGATCCTGATGCTGCAGAATTAAATCCATTTGGAGAT CCTGACTCAGAAGAACCTATCACTGAAACAGCTTCACCTAGAAAAACAGAAGACTCTTTTTATAATAACAGCTATAATCCCTTTAAAGAG GTGCAGACTCCACAGTATTTGAACCCATTCGATGAGCCAGAAGCATTTGTGACCATAAAGGATTCTCCTCCCCAGTCTACAAAAAGAAAA AATATAAGACCTGTGGATATGAGCAAGTACCTCTATGCTGATAGTTCTAAAACTGAAGAAGAAGAATTGGATGAATCAAATCCTTTTTAT GAACCTAAATCAACTCCTCCTCCAAATAATTTGGTAAATCCTGTTCAAGAACTAGAAACTGAAAGGCGAGTGAAAAGAAAGGCCCCGGCT CCACCAGTCCTCTCACCAAAAACAGGAGTATTAAATGAAAACACAGTTTCTGCAGGAAAAGATCTCTCTACTTCTCCTAAGGTTCTTCAC ACACCCCCATTCGGCGTAGTACCCAGAGAGCTCAAGATGTGTGGCAGTTTTCGGATGGAAGCTCGAGAGCCCTTAAGTTCTGAGAAAATT >25525_25525_5_EHBP1-BCL11A_EHBP1_chr2_63101667_ENST00000431489_BCL11A_chr2_60679801_ENST00000537768_length(amino acids)=431AA_BP=394 MIIMASVWKRLQRVGKHASKFQFVASYQELMVECTKKWQPDKLVVVWTRRSRRKSSKAHSWQPGIKNPYRGVVVWPVPENIEITVTLFKD PHAEEFEDKEWTFVIENESPSGRRKALATSSINMKQYASPMPTQTDVKLKFKPLSKKVVSAALQFSLSCIFLREGKATDEDMQSLASLMS MKQADIGNLDDFEEDNEDDDENRVNQEEKAAKITELINKLNFLDEAEKDLATVNSNPFDDPDAAELNPFGDPDSEEPITETASPRKTEDS FYNNSYNPFKEVQTPQYLNPFDEPEAFVTIKDSPPQSTKRKNIRPVDMSKYLYADSSKTEEEELDESNPFYEPKSTPPPNNLVNPVQELE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for EHBP1-BCL11A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for EHBP1-BCL11A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for EHBP1-BCL11A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
