
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:EHD1-ADRBK2 (FusionGDB2 ID:25546) |
Fusion Gene Summary for EHD1-ADRBK2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: EHD1-ADRBK2 | Fusion gene ID: 25546 | Hgene | Tgene | Gene symbol | EHD1 | ADRBK2 | Gene ID | 10938 | 157 |
| Gene name | EH domain containing 1 | G protein-coupled receptor kinase 3 | |
| Synonyms | H-PAST|HPAST1|PAST|PAST1 | ADRBK2|BARK2 | |
| Cytomap | 11q13.1 | 22q12.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | EH domain-containing protein 1PAST homolog 1testilin | beta-adrenergic receptor kinase 2adrenergic, beta, receptor kinase 2beta-ARK-2 | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | Q9H4M9 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000359393, ENST00000320631, ENST00000488711, | ENST00000324198, | |
| Fusion gene scores | * DoF score | 6 X 7 X 6=252 | 3 X 2 X 3=18 |
| # samples | 8 | 3 | |
| ** MAII score | log2(8/252*10)=-1.65535182861255 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: EHD1 [Title/Abstract] AND ADRBK2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | EHD1(64627396)-ADRBK2(26057543), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | EHD1-ADRBK2 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. EHD1-ADRBK2 seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | ADRBK2 | GO:0031623 | receptor internalization | 20074556 |
 Fusion gene breakpoints across EHD1 (5'-gene) Fusion gene breakpoints across EHD1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
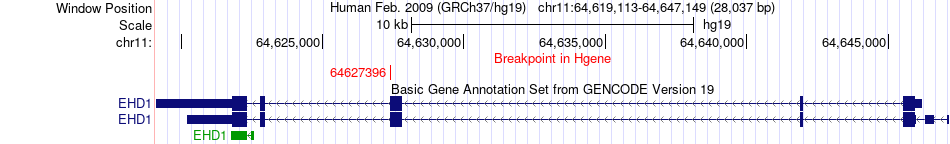 |
 Fusion gene breakpoints across ADRBK2 (3'-gene) Fusion gene breakpoints across ADRBK2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
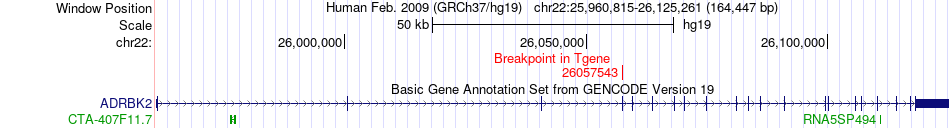 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LGG | TCGA-HT-7686-01A | EHD1 | chr11 | 64627396 | - | ADRBK2 | chr22 | 26057543 | + |
| ChimerDB4 | LGG | TCGA-HT-7686 | EHD1 | chr11 | 64627396 | - | ADRBK2 | chr22 | 26057543 | + |
Top |
Fusion Gene ORF analysis for EHD1-ADRBK2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000359393 | ENST00000324198 | EHD1 | chr11 | 64627396 | - | ADRBK2 | chr22 | 26057543 | + |
| In-frame | ENST00000320631 | ENST00000324198 | EHD1 | chr11 | 64627396 | - | ADRBK2 | chr22 | 26057543 | + |
| intron-3CDS | ENST00000488711 | ENST00000324198 | EHD1 | chr11 | 64627396 | - | ADRBK2 | chr22 | 26057543 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000320631 | EHD1 | chr11 | 64627396 | - | ENST00000324198 | ADRBK2 | chr22 | 26057543 | + | 9817 | 1170 | 123 | 2972 | 949 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000320631 | ENST00000324198 | EHD1 | chr11 | 64627396 | - | ADRBK2 | chr22 | 26057543 | + | 0.000148637 | 0.99985135 |
Top |
Fusion Genomic Features for EHD1-ADRBK2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| EHD1 | chr11 | 64627395 | - | ADRBK2 | chr22 | 26057542 | + | 0.00012209 | 0.9998779 |
| EHD1 | chr11 | 64627395 | - | ADRBK2 | chr22 | 26057542 | + | 0.00012209 | 0.9998779 |
| EHD1 | chr11 | 64627395 | - | ADRBK2 | chr22 | 26057542 | + | 0.00012209 | 0.9998779 |
| EHD1 | chr11 | 64627395 | - | ADRBK2 | chr22 | 26057542 | + | 0.00012209 | 0.9998779 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
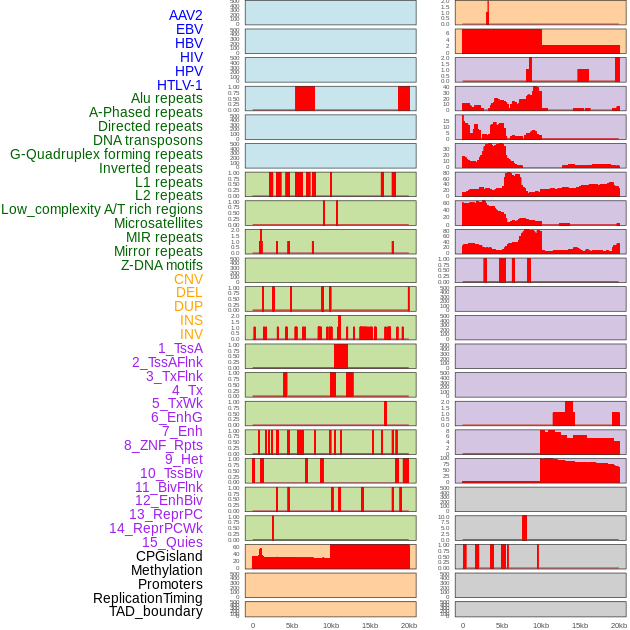 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
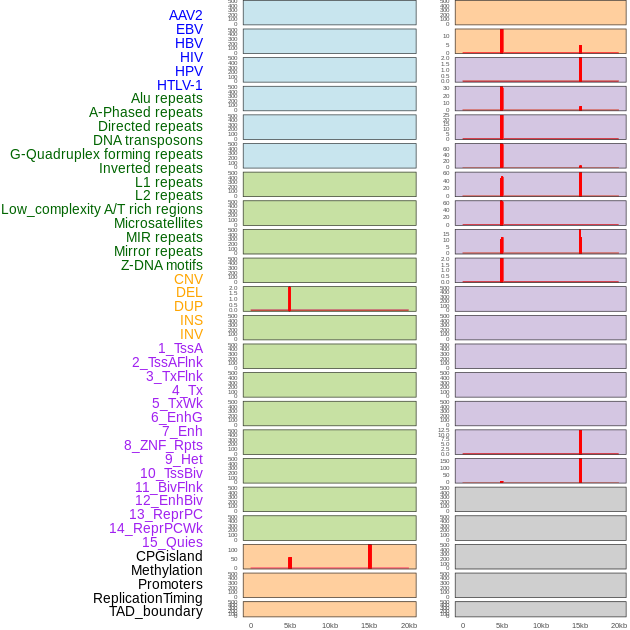 |
Top |
Fusion Protein Features for EHD1-ADRBK2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:64627396/chr22:26057543) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| EHD1 | . |
| FUNCTION: ATP- and membrane-binding protein that controls membrane reorganization/tubulation upon ATP hydrolysis. In vitro causes vesiculation of endocytic membranes (PubMed:24019528). Acts in early endocytic membrane fusion and membrane trafficking of recycling endosomes (PubMed:15020713, PubMed:17233914, PubMed:20801876). Recruited to endosomal membranes upon nerve growth factor stimulation, indirectly regulates neurite outgrowth (By similarity). Plays a role in myoblast fusion (By similarity). Involved in the unidirectional retrograde dendritic transport of endocytosed BACE1 and in efficient sorting of BACE1 to axons implicating a function in neuronal APP processing (By similarity). Plays a role in the formation of the ciliary vesicle (CV), an early step in cilium biogenesis. Proposed to be required for the fusion of distal appendage vesicles (DAVs) to form the CV by recruiting SNARE complex component SNAP29. Is required for recruitment of transition zone proteins CEP290, RPGRIP1L, TMEM67 and B9D2, and of IFT20 following DAV reorganization before Rab8-dependent ciliary membrane extension. Required for the loss of CCP110 form the mother centriole essential for the maturation of the basal body during ciliogenesis (PubMed:25686250). {ECO:0000250|UniProtKB:Q641Z6, ECO:0000250|UniProtKB:Q9WVK4, ECO:0000269|PubMed:15020713, ECO:0000269|PubMed:17233914, ECO:0000269|PubMed:20801876, ECO:0000269|PubMed:24019528, ECO:0000269|PubMed:25686250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000320631 | - | 3 | 5 | 198_227 | 305 | 535.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000359393 | - | 5 | 7 | 198_227 | 305 | 535.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000320631 | - | 3 | 5 | 55_286 | 305 | 535.0 | Domain | Dynamin-type G |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000359393 | - | 5 | 7 | 55_286 | 305 | 535.0 | Domain | Dynamin-type G |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000320631 | - | 3 | 5 | 153_156 | 305 | 535.0 | Region | G3 motif |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000320631 | - | 3 | 5 | 219_222 | 305 | 535.0 | Region | G4 motif |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000320631 | - | 3 | 5 | 243_243 | 305 | 535.0 | Region | G5 motif |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000320631 | - | 3 | 5 | 65_72 | 305 | 535.0 | Region | G1 motif |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000320631 | - | 3 | 5 | 91_92 | 305 | 535.0 | Region | G2 motif |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000359393 | - | 5 | 7 | 153_156 | 305 | 535.0 | Region | G3 motif |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000359393 | - | 5 | 7 | 219_222 | 305 | 535.0 | Region | G4 motif |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000359393 | - | 5 | 7 | 243_243 | 305 | 535.0 | Region | G5 motif |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000359393 | - | 5 | 7 | 65_72 | 305 | 535.0 | Region | G1 motif |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000359393 | - | 5 | 7 | 91_92 | 305 | 535.0 | Region | G2 motif |
| Tgene | ADRBK2 | chr11:64627396 | chr22:26057543 | ENST00000324198 | 2 | 21 | 191_453 | 88 | 689.0 | Domain | Protein kinase | |
| Tgene | ADRBK2 | chr11:64627396 | chr22:26057543 | ENST00000324198 | 2 | 21 | 454_521 | 88 | 689.0 | Domain | AGC-kinase C-terminal | |
| Tgene | ADRBK2 | chr11:64627396 | chr22:26057543 | ENST00000324198 | 2 | 21 | 558_652 | 88 | 689.0 | Domain | PH | |
| Tgene | ADRBK2 | chr11:64627396 | chr22:26057543 | ENST00000324198 | 2 | 21 | 197_205 | 88 | 689.0 | Nucleotide binding | ATP |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000320631 | - | 3 | 5 | 489_500 | 305 | 535.0 | Calcium binding | Ontology_term=ECO:0000269,ECO:0007744,ECO:0007744 |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000359393 | - | 5 | 7 | 489_500 | 305 | 535.0 | Calcium binding | Ontology_term=ECO:0000269,ECO:0007744,ECO:0007744 |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000320631 | - | 3 | 5 | 444_532 | 305 | 535.0 | Domain | EH |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000320631 | - | 3 | 5 | 476_511 | 305 | 535.0 | Domain | EF-hand |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000359393 | - | 5 | 7 | 444_532 | 305 | 535.0 | Domain | EH |
| Hgene | EHD1 | chr11:64627396 | chr22:26057543 | ENST00000359393 | - | 5 | 7 | 476_511 | 305 | 535.0 | Domain | EF-hand |
| Tgene | ADRBK2 | chr11:64627396 | chr22:26057543 | ENST00000324198 | 2 | 21 | 54_175 | 88 | 689.0 | Domain | RGS | |
| Tgene | ADRBK2 | chr11:64627396 | chr22:26057543 | ENST00000324198 | 2 | 21 | 1_190 | 88 | 689.0 | Region | Note=N-terminal |
Top |
Fusion Gene Sequence for EHD1-ADRBK2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >25546_25546_1_EHD1-ADRBK2_EHD1_chr11_64627396_ENST00000320631_ADRBK2_chr22_26057543_ENST00000324198_length(transcript)=9817nt_BP=1170nt CCCTACCGCCCCCAATTCCGCCCTGCCCCCGCCGCGGCGGCGCTAGCCGCCACTGAGGGACCGACCCTATAAAGGCCGCTCCGCGAGGGG TGCGCAGCATTCGGCAGAGGGCGCTTCGACGGGCTGGGCTGTGCGCCTGCGCAGTGTGGGTCGCTCCCGATTCCCTGCCCCGGCCGGCCC CGCCTCGGCTCCGCACCCTCGCCCCGCTCTCAGCCGCCGCTCTGCCCCGCAGCAGCCAGCCCCGTGTCCGGCAGTATGTTCAGCTGGGTC AGCAAGGATGCCCGCCGCAAGAAGGAGCCGGAGCTCTTCCAGACGGTGGCTGAGGGGCTGCGGCAGCTGTACGCGCAGAAGCTGCTACCC CTGGAGGAGCACTACCGCTTCCACGAGTTCCACTCGCCCGCGCTGGAGGACGCTGACTTCGACAACAAGCCTATGGTGCTCCTCGTGGGG CAGTACAGCACGGGCAAGACCACCTTCATCCGACACCTGATCGAGCAGGACTTCCCGGGGATGCGCATCGGGCCCGAGCCCACCACCGAC TCCTTCATCGCCGTCATGCACGGCCCCACTGAGGGCGTGGTGCCGGGCAACGCGCTCGTGGTGGACCCGCGGCGCCCCTTCCGCAAGCTC AACGCGTTTGGCAACGCTTTCCTCAACAGGTTCATGTGTGCCCAGCTGCCCAACCCCGTCCTGGACAGCATCAGCATCATCGACACCCCC GGGATCCTGTCTGGAGAGAAGCAGCGGATCAGCAGAGGCTATGACTTTGCAGCCGTCCTGGAGTGGTTCGCGGAGCGTGTGGACCGCATC ATCCTGCTCTTCGACGCCCACAAGCTGGACATCTCCGATGAGTTCTCGGAAGTGATCAAGGCTCTGAAGAACCATGAGGACAAGATCCGC GTGGTGCTGAACAAGGCAGACCAGATCGAGACGCAGCAGCTGATGCGGGTGTACGGGGCCCTCATGTGGTCCCTGGGCAAGATCATCAAC ACCCCCGAGGTGGTCAGGGTCTACATCGGCTCCTTCTGGTCCCACCCGCTCCTCATCCCCGACAACCGCAAGCTCTTTGAGGCCGAGGAG CAGGACCTCTTCAAGGACATCCAGTCACTGCCCCGAAACGCCGCCCTCAGGAAGCTCAATGACCTGATCAAGCGGGCACGGCTGGCCAAG ATAAAGGAATATGAAAAACTTGATAATGAGGAAGACCGCCTTTGCAGAAGTCGACAAATTTATGATGCCTACATCATGAAGGAACTTCTT TCCTGTTCACATCCTTTCTCAAAGCAAGCTGTAGAACACGTACAAAGTCATTTATCCAAGAAACAAGTGACATCAACTCTTTTTCAGCCA TACATAGAAGAAATTTGTGAAAGCCTTCGAGGTGACATTTTTCAAAAATTTATGGAAAGTGACAAGTTCACTAGATTTTGTCAGTGGAAA AACGTTGAATTAAATATCCATTTGACCATGAATGAGTTCAGTGTGCATAGGATTATTGGACGAGGAGGATTCGGGGAAGTTTATGGTTGC AGGAAAGCAGACACTGGAAAAATGTATGCAATGAAATGCTTAGATAAGAAGAGGATCAAAATGAAACAAGGAGAAACATTAGCCTTAAAT GAAAGAATCATGTTGTCTCTTGTCAGCACAGGAGACTGTCCTTTCATTGTATGTATGACCTATGCCTTCCATACCCCAGATAAACTCTGC TTCATCCTGGATCTGATGAACGGGGGCGATTTGCACTACCACCTTTCACAACACGGTGTGTTCTCTGAGAAGGAGATGCGGTTTTATGCC ACTGAAATCATTCTGGGTCTGGAACACATGCACAATCGGTTTGTTGTCTACAGAGATTTGAAGCCAGCAAATATTCTCTTGGATGAACAT GGACACGCAAGAATATCAGATCTTGGTCTTGCCTGCGATTTTTCCAAAAAGAAGCCTCATGCGAGTGTTGGCACCCATGGGTACATGGCT CCCGAGGTGCTGCAGAAGGGGACGGCCTATGACAGCAGTGCCGACTGGTTCTCCCTGGGCTGCATGCTTTTCAAACTTCTGAGAGGTCAC AGCCCTTTCAGACAACATAAAACCAAAGACAAGCATGAAATTGACCGAATGACACTCACCGTGAATGTGGAACTTCCAGACACCTTCTCT CCTGAACTGAAGTCCCTTTTGGAGGGCTTGCTTCAGCGAGACGTTAGCAAGCGGCTGGGCTGTCACGGAGGCGGCTCACAGGAAGTAAAA GAGCACAGCTTTTTCAAAGGTGTTGACTGGCAGCATGTCTACTTACAAAAGTACCCACCACCCTTGATTCCTCCCCGGGGAGAAGTCAAT GCTGCTGATGCCTTTGATATTGGCTCATTTGATGAAGAGGATACCAAAGGGATTAAGCTACTTGATTGCGACCAAGAACTCTACAAGAAC TTCCCTTTGGTCATCTCTGAACGCTGGCAGCAAGAAGTAACGGAAACAGTTTATGAAGCAGTAAATGCAGACACAGATAAAATCGAGGCC AGGAAGAGAGCTAAAAATAAGCAACTTGGCCACGAAGAAGATTACGCTCTGGGGAAGGACTGTATTATGCACGGGTACATGCTGAAACTG GGAAACCCATTTCTGACTCAGTGGCAGCGTCGCTATTTTTACCTCTTTCCAAATAGACTTGAATGGAGAGGAGAGGGAGAGTCCCGGCAA AATTTACTGACAATGGAACAGATTCTCTCTGTGGAAGAAACTCAAATTAAAGACAAAAAATGCATTTTGTTCAGAATAAAAGGAGGGAAA CAATTTGTCTTGCAATGTGAGAGTGATCCAGAGTTTGTGCAGTGGAAGAAAGAGTTGAACGAAACCTTCAAGGAGGCCCAGCGGCTATTG CGTCGTGCCCCGAAGTTCCTCAACAAACCTCGGTCAGGTACTGTGGAGCTCCCAAAGCCATCCCTCTGTCACAGAAACAGCAACGGCCTC TAGCACCCAGAAACAGGGAGGGTCCTCGAGGAGGACACACCAGGGTCTCAGCCTTTTGGGGTGAACGAGGATGAGGCATCTGATCTATTC GCTACCGGGACTCCTCCAGGCTCCCGAGAGGAGTCGGGACCCTTCGGCTTGGGGTCAGCTCAGCTCCCTGCCTTGTCACATTTGTCTGCA TTAGAAACTACTGAAGAAATAAAAGTTCTTTTTCTTTGCTACACACTTTGGTACCTATGAACCTAGAACTTGAAGTGACTCCTACTTATC ACGTAAATTTTTATGTCTGATATCAAACACATCTTAGACTCCCCAGAATGGAATTTAAAGATGTTCAGTGTTGGGTAACAGATTGCCCTA AGCATTGCCACATATTCTGTCTAGTCACTGCTGATTTTCTATGTCTTTGCTCCATACTGCTGGGGGATGGGAGAGCCACAGTGTGTTTCT TTTGTGCACTTCGCAACTGACTTCTTGTCCTGGGGTTAAAAGTTGAAGATATTTTCTGATGATATTAAAAGTTGAAGATATTTCTGCACT TGGGCCCTCCTCTGGGAGCCGCACCCACATGACTGCCCTGCCTCTGACCAGTCTGTTCCGGGGCCCCCTCAGCCAGGTGGGAATGACGGA CACGTACTATCCAAGTGTATGGGATTAACTAATCATTGAAGGCATTCATCCGTCCATCATTGGAAAGATTTACAGTGATTCTGAAGGACA GGCCGTGGAGTTTTAGGTTTCAGGGGCAAGAGCAGTTTTCAAAAGTCTTTGAGTCCAGTGTGCACGAGTCGACAAGCAGTACCTGGCATG CAGGAGCACTCATGGGTGAGTCCGTCTCAGGTCTCGACAATTAGCAGTTGTGTGACAGTCATTCTGGTTCCTTCTGCCTGACCCTGGGAG ACATATCAGTAATGGATGTACAAAAGCAGGTCTGTTTTATGTCTTAGTATAATTTCAGATGAATTGTATTGAAAAAATGCTGAGGAATGA ATGTGTCAAAATGGGTTAACTGTGTATATTGACTTTCATGTCGTCATGCATCTGTCATGAATGAATGATACTTTGCACTGGGCTGTACGA CAGTGAGGACCTTAGGGCATGAAGCCTTTTTCCTGGTCCCAGCAGCATCTGCCCTGTGAAGTTTGTTTTCTCCCACTGCCTCCAGGCCCC ACTGATACCCCCAAATAGATGCTGGGTTATGAGAACCAGCGAAATCCCCCATGTCATCAGTCTTAAAAAAAAAATTTTACAAATCCACGT ATTTGTCCCATTCTTGGAGTAGTTTTAGTGTATGTCTTTACATTAACTACTAACAGTATAAATAACTTGACATCGTAATTGTCTGCATCC TGTCCTTGATATTTTTAGCAGTTCCAAATCTTTGTTTTTGTATTTGTTTGCTGTGTTCATGGGCAAAGTAAGTACTTTTTAATGCAGTTA TTTTGAGAGTTTGGAAGATAATTACCAAAAGGGTCCATTATTTCATAAGAGTTACTTTGCAAAAAAAAAAATGTGGGTTTTTTTTTTTGT CTATCTCAACTACTAGTTGGGGTTTAAATTAACATACATTTTCTACTATCTGTTATTTCCAGTGTGGGAGGAGGGATGTACTACTTACAT GCATTCTCCTTATTTAAAAAGGAAGAATAGTATTCAAATTCTGTTGAAACACACACACACACACACACACACACACACACACACTCCAGA AGCAGAAAAGCCATTGTTCTTAAAGAGTGAATGTCTTCCCAGCCCTGGTTAATTATAGCTGTGACTGATGCCGTTCCCGTCTGCATCTCA AGCTCATAGGTTCTCAGCATGTGCAGTTGAGGATGCGCTGGGCCTCATGCCTGTTCTAGATCTCCAGGATAAAGGGCCTGCTGTTGACTC CACCAGGGTCTGGGCTTAGCGTCTAATATCTCGTACCTAGGGCGTGAGCTGCACAAACGTGTTCAGAAAGATTATTCAACTTTCCCATAC TTGTTCTAAAATTGAGCTGATCCGCATCTCTTTCAAAAACTAGAATTTCTGCTCTAAGAATAGAACATAAGGCTCCACTCCCTTTTAGAA AAGATATATGAATTGGAAAATGCTCTGAAAGTCCTTTTGCTTCAAACAAAAGTGTAAACTTTTACACTTCCCCAACTCACATTTGATTTG TAATGATATGGTTGAGAAGTACATCTAGATGTCATTTATTAAAAGTGCTTTGTAAGACTAGATTGAGCTGTTTCTGAGGGCGGTCACCAG TTGTGTTGGGGTCTGGTTTGAGTGCCTTCTGCCAAAATGTTGTGATGGAGGTGTTTCTGCGACCAGACACAGGATACCGCTGTGTCTGCA CCCGGTTGCCTGCATGGCCAGAGGAAAAGTCAGTTGGATTAAACATCATGGTATACTTGGCTGTTGTTTTTTTTTAATTTTTTAATTTTT TGGGATAGGGCCTCGCTCTGTCACCCAGGCTGGAGAACAGTGGGATGATCATGGCTCACTGCAGCCTTGAATTCCTAGGTTCAAGCAATC CTCCCACGTCAGCCTCCTGAGTAGCTAGGACTACAGGTGCATGCCACCTTTCCTGGCTAATTTATTTTTTGGGTAGAGATGGGGTCTTGA ACTCTTAGGCTCAAGTGATCCTCCTTCCTTGGCCTCCCAAAATGCTGGAATTAGAGATGTAAGCCACCATGCCCAGCCATAGTACTTGGA TGTTTTAGAAGGTTTTCCAAGTATTACATAATTCCTAGATGTTCACCCTTATTACACTCCAACTATTAAAAAGGTCAAAATTCAGCCTAT TTTTTTTCATTATTTTAGATTCCTGTGGTTGGGATATTTTAACATTGATGAGAAAAATAATTGAGGTTGATATTTTTACAAAATCATGCG GTAATAAGTCTTGATTTCATGATTCAAAAGAATCAATAAAGCCTAAAAATAATAGATTACTTTAAGCTGCTATGTAAGATATATATGGAA TAAATTAAAAACCTTTGTGAATTCAGGTTTATTATTTTTAACCTAAAACATTCTCTTTGGTTCATTCATCCCCTCATGTCATGGGGGCTC ATTGGTTTTCCTTCTTTGTCATATTTAAGTATGATTTTTCAACAAAACTTCTAGAAGTCAGCTTATTATGTCACCATTCATGCAAAGTGC TCATGCCTCTGATTGGTCCATTCACTGACGTGACAATTTCAGGTCCTATGTTTAAAAAGAAGGGGCTGGCCGGGCACGATGGCTCACGCC TATAATCCCAGCACTTTGGGAGGCCGAGAGGGGCGGTTCACGAGGTCAGGAGATTGAGACCATCCTGGTTAGCAGAGTGAAACCCCGTCT CTACTAAAAATACAAATAAAAATTAGCCGGGCGTGGTGGCGGGCGCCTGTAGTCCCAGCTACTTGGGAGGCTGAGGCAGGAGAATGGCAT GAACCCGGGAGGCAGAGCTTGCAGTGAGCCGAGATTGCGCCACTGCACTCCAGCCTGGGCGACAGAGCGAGACTCTGTCTCAAAAAAAAA AAGGAGGGGGGCTAAATATCCAGTGAGATGCACTGAGGAAAGGAAGCATTTTGCTGAAGACAGCAGCAGCAACAAACAATGGTCTGTTTG TTGCAAACAAGATGTAGCTTGATTTCTGGTCTGACATATGCCATATACAGATATTAGAAACGACTGTTTGAAGGCCACACTGGTCATCTA CAAAGTAATGTTTACCAATTGACGACAGGGATTTAACTAGATTAAAAAGATCAAAGTGTGGTTTTTCTCTGCTTTTTAAAATTTCACTCG GAATTTGTAGCTGGGCCAATTCAACACATTTTACTTTTCAGTGGAATTGATTTTTCTAATGTTTCAGAATTTTAACATATCAAGAAGAAA ACAACGTTCTCAAAGTCTGGCCTCTTTAGCATGATGTAAACCTATAGAAATGCTTTGAAATGTGCTGGTGTAAGATAAGAGTTATCTTGT ATGATTTAATCATATGCAGTGTTGTCTCAGTTACGTTCAGGGAAATGTTTCTGTGTCATTCAGAGATGCTTGATGAATTAACACCTCCCA CCCTGAGTGAGGGGTTGACTTGTTGGGAGATGATTTGGGCTTCACTGGGATCTGTGACAGGTGGGGGCTGGGCTGGGTGTCACAAAGAGA ATAGTGGTAGAAATCGGGCGAAGGAAGAAAGAAGTTACTGGTAAAAATCATTACACCATAAAGCACCAAGGAAATAACTGAGTTAAAATA GGTGAAGTTTCTTTTTTCCCCCCTGTAACAGGAGAGTTTTCCTTATGATAATTATTCTGAGACTTGGTCACTTTGTTTTTGAATGTGGAG CTGCTGAACTCATTCAGAAGCCATTTGCTGCCTATCAGGACTTTCTGAAGAAGTTCTTTTGCCTCTGCCTACCCTCTGGCACCCTCCCAT GGAGGCACAGGGGACCCAGAGCTAAAGCATTACCAGGCCATCTCCAAAACACCCCGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTG TGTGTGTGTGCACTTTGCAGCCCCCGAGGTGGAGAGGCAGTGTCTGGATCACTGTGAATGCATTGCCCCATTGGTCAGTTGGGGACACTG TTACAAATCCACTGAAGTCCTGGTAAAACTGTCAAGAGTAACAGGCCTCTTCTGTTCTACCCTGCTCACTTCCACGGTGAGTTACCAGCC TGGGCAACACAGCAAGACCCCATCTCTACAAAAAAAATTTTTTTAAGTAATTAACCGTTTAAATTTTTTCCTAAAGATTTAACATGATTT TTCCCTCCTATGTAAAGTTTACTGGAGAGACTTGAATTACTTAAATTCATGTTAATATGATTTTTTTTTAATCCAGGTCACATTTTAACA AAGTTTATTATGAAACAAATGAAATTTGAACTCTAAAATGGTACTCCTTGGCTTCCTCAAGTCACAATGAACTTTATATTTTCTTTGTCC TTAAGGACTAAGATAGTTGTTTTATTTCAGCCGAATCACAGAGATAACCACTCCTGCAGGCCCCCACAGCTGGCCCAAAGGGGCTGTCTT TCTGACCTGGCTGTGTTAGCACTGATTGAGAAACGCAGGCTCCCAAATTTTAAATTGCCTTTATTAAAAACACAAACTACAGAAAATGGG TTAAGAGTATACGCATTTCATCAAACACATATAGGGGAAAAAATCCTTCAATTTAGAGTTAAATAACTCAGCTTTGTATAGTAGAGTTAG CGCTCCAGTATCTAACAATCTCAGAATCATCTCTGAAAACTGGTAACTATGCTTCCATTTTTAATTTTGTCCTAAATATCAGATGTCTTT GATGTAAGGGTAGGGAATGGAGAAATATTTTCAATTGTGTATTTGTATTACAAAGAACTTGAAATTTACTTTCTTAGTTGATTATATTAA ATGATGTATATATTATATGTGGTTTATAAGCTCAACACTGGCCATTTTTTTAGTTTTATTGTTAAATGGTATTTTTCTATGTTTAATTAT AATAGATCTGGCTTTTTCTGGATAGCATAAAGATCACTGAACTATATATATATAAGAAACAAGAGTTCTATTTTAGCACAAAGGCATTTT ATATTATTTATTGAATCCATAAGTTTGTTTTCGTCAAAAACATTCCATATTATTTCTGCTCCTTTTTATTTGTATAGTTTGTTATTTAAA GAAATGGCAGTCCTTCCTGTTCTTAATACAATAAAATTGAAATAATGCACCTAGTAATGTGGCCGACATCTCTTCTCACCACCATGGACT GTTTTCAACAACAGTTGATCTTCTGGTCTGTGCTGAGAGGCGCATGCATGTCTTTCGTCACGTCGGGCAGCACACCTGCTGTGAAATACT GCTTTCATCTACCTCTTCAGAAGGCTTCTTGCTTGTTGACAAGTACCGCAAAGGCTTTATTCTGGACTGGCTATCTCATAAAAGGATTTC TGTAAGACTTTGCAGTGTCATTCCCTCAGAACCTAGGTTTGTTTCTAAAGCCACGGTATTGTCCAGGAGCCCCTGTGTGTGGGGCAGGTA GCTATCCCTCCCATGTCATTAGTAATCCTTTAGGATTTAAGGTACAACTGGACAGCATCATTCCTTCCCCTTATTGTGCCAAATCCCCAC CATCAGCCTTGCCATTGCCTTAAGATTTGATTATTGCACCCAATTACCTAACCACTAAACAGAAAGGCCACCTTCACTCTTTGAAAAAGG CAAGCTGTGCTTAGAAACACTGCTTTTAAGAGTAGCACATTTGAGTGTGACTTTTTCCCCCCTTCACTATTTCAAAATGGTTTTGAAATG GGGTCTTAAAGGTAAGCGCCCTCATACATGACTGAAACTTTGTGAGAGGTCTTATATTTGAATGGACCCTTAATGATTTATGTGAAATAG AATGAAGTCCTGTCTCTGTGAGAGAACGTGCCTCCTCACTCATTTGTCTCTGTCTGTTTTCATAGCCATCAATATAGTAACATATTTACT ATATTCTTGAATACCCTTGAAGAAAGAAATCCGTTTTCTATTGTGCATTGCTATACGAAGTGAAGCCAGTAAACTAGATACTGTAAATCT AGATATTGTACCTAGACAAAATATCATTGGTTCTATCTCTTTTTGTATCTGTTGTGCCAGGGAAGGTTTATAATCCCTTCTCAGTATACA CTCACTAGTGCACGTCTGAAATAGTATCCCACGGGAGATGCTGCTCCACGTCTGAGGTCACCTGCCCTGTGTGGGGCACACCACCGTCAG CACCACCGTTTTTACAGTTACTTTGGAGCTGCTAGACTGGTTTTCTGTGTTGGTAAATTGCCTATATAAATCTGAATAAAAAGGATCTGT >25546_25546_1_EHD1-ADRBK2_EHD1_chr11_64627396_ENST00000320631_ADRBK2_chr22_26057543_ENST00000324198_length(amino acids)=949AA_BP=349 MGCAPAQCGSLPIPCPGRPRLGSAPSPRSQPPLCPAAASPVSGSMFSWVSKDARRKKEPELFQTVAEGLRQLYAQKLLPLEEHYRFHEFH SPALEDADFDNKPMVLLVGQYSTGKTTFIRHLIEQDFPGMRIGPEPTTDSFIAVMHGPTEGVVPGNALVVDPRRPFRKLNAFGNAFLNRF MCAQLPNPVLDSISIIDTPGILSGEKQRISRGYDFAAVLEWFAERVDRIILLFDAHKLDISDEFSEVIKALKNHEDKIRVVLNKADQIET QQLMRVYGALMWSLGKIINTPEVVRVYIGSFWSHPLLIPDNRKLFEAEEQDLFKDIQSLPRNAALRKLNDLIKRARLAKIKEYEKLDNEE DRLCRSRQIYDAYIMKELLSCSHPFSKQAVEHVQSHLSKKQVTSTLFQPYIEEICESLRGDIFQKFMESDKFTRFCQWKNVELNIHLTMN EFSVHRIIGRGGFGEVYGCRKADTGKMYAMKCLDKKRIKMKQGETLALNERIMLSLVSTGDCPFIVCMTYAFHTPDKLCFILDLMNGGDL HYHLSQHGVFSEKEMRFYATEIILGLEHMHNRFVVYRDLKPANILLDEHGHARISDLGLACDFSKKKPHASVGTHGYMAPEVLQKGTAYD SSADWFSLGCMLFKLLRGHSPFRQHKTKDKHEIDRMTLTVNVELPDTFSPELKSLLEGLLQRDVSKRLGCHGGGSQEVKEHSFFKGVDWQ HVYLQKYPPPLIPPRGEVNAADAFDIGSFDEEDTKGIKLLDCDQELYKNFPLVISERWQQEVTETVYEAVNADTDKIEARKRAKNKQLGH EEDYALGKDCIMHGYMLKLGNPFLTQWQRRYFYLFPNRLEWRGEGESRQNLLTMEQILSVEETQIKDKKCILFRIKGGKQFVLQCESDPE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for EHD1-ADRBK2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for EHD1-ADRBK2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for EHD1-ADRBK2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
