
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:EIF3B-GDI2 (FusionGDB2 ID:25782) |
Fusion Gene Summary for EIF3B-GDI2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: EIF3B-GDI2 | Fusion gene ID: 25782 | Hgene | Tgene | Gene symbol | EIF3B | GDI2 | Gene ID | 8662 | 2665 |
| Gene name | eukaryotic translation initiation factor 3 subunit B | GDP dissociation inhibitor 2 | |
| Synonyms | EIF3-ETA|EIF3-P110|EIF3-P116|EIF3S9|PRT1 | HEL-S-46e|RABGDIB | |
| Cytomap | 7p22.3 | 10p15.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | eukaryotic translation initiation factor 3 subunit Beukaryotic translation initiation factor 3 subunit 9eukaryotic translation initiation factor 3, subunit 9 (eta, 116kD)eukaryotic translation initiation factor 3, subunit 9 eta, 116kDaprt1 homolog | rab GDP dissociation inhibitor betaGDI-2epididymis secretory sperm binding protein Li 46eguanosine diphosphate dissociation inhibitor 2rab GDI betarab GDP-dissociation inhibitor, beta | |
| Modification date | 20200322 | 20200313 | |
| UniProtAcc | P55884 | P50395 | |
| Ensembl transtripts involved in fusion gene | ENST00000360876, ENST00000397011, | ENST00000380132, ENST00000380181, ENST00000479928, ENST00000380191, | |
| Fusion gene scores | * DoF score | 11 X 15 X 5=825 | 19 X 10 X 8=1520 |
| # samples | 16 | 20 | |
| ** MAII score | log2(16/825*10)=-2.36632221424582 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(20/1520*10)=-2.92599941855622 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: EIF3B [Title/Abstract] AND GDI2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | EIF3B(2418877)-GDI2(5828013), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | EIF3B | GO:0006413 | translational initiation | 8995410|17581632 |
| Hgene | EIF3B | GO:0006446 | regulation of translational initiation | 8995410 |
| Hgene | EIF3B | GO:0075522 | IRES-dependent viral translational initiation | 9573242 |
| Hgene | EIF3B | GO:0075525 | viral translational termination-reinitiation | 21347434 |
 Fusion gene breakpoints across EIF3B (5'-gene) Fusion gene breakpoints across EIF3B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
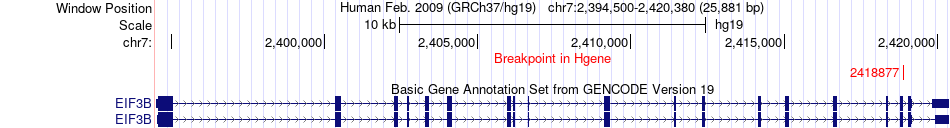 |
 Fusion gene breakpoints across GDI2 (3'-gene) Fusion gene breakpoints across GDI2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
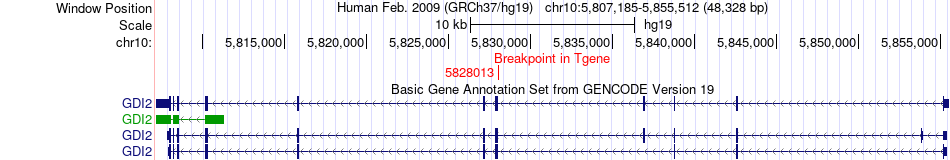 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-SG-A849-01A | EIF3B | chr7 | 2418877 | + | GDI2 | chr10 | 5828013 | - |
Top |
Fusion Gene ORF analysis for EIF3B-GDI2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000360876 | ENST00000380132 | EIF3B | chr7 | 2418877 | + | GDI2 | chr10 | 5828013 | - |
| 5CDS-intron | ENST00000360876 | ENST00000380181 | EIF3B | chr7 | 2418877 | + | GDI2 | chr10 | 5828013 | - |
| 5CDS-intron | ENST00000360876 | ENST00000479928 | EIF3B | chr7 | 2418877 | + | GDI2 | chr10 | 5828013 | - |
| 5CDS-intron | ENST00000397011 | ENST00000380132 | EIF3B | chr7 | 2418877 | + | GDI2 | chr10 | 5828013 | - |
| 5CDS-intron | ENST00000397011 | ENST00000380181 | EIF3B | chr7 | 2418877 | + | GDI2 | chr10 | 5828013 | - |
| 5CDS-intron | ENST00000397011 | ENST00000479928 | EIF3B | chr7 | 2418877 | + | GDI2 | chr10 | 5828013 | - |
| In-frame | ENST00000360876 | ENST00000380191 | EIF3B | chr7 | 2418877 | + | GDI2 | chr10 | 5828013 | - |
| In-frame | ENST00000397011 | ENST00000380191 | EIF3B | chr7 | 2418877 | + | GDI2 | chr10 | 5828013 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000360876 | EIF3B | chr7 | 2418877 | + | ENST00000380191 | GDI2 | chr10 | 5828013 | - | 4130 | 2397 | 32 | 3346 | 1104 |
| ENST00000397011 | EIF3B | chr7 | 2418877 | + | ENST00000380191 | GDI2 | chr10 | 5828013 | - | 4114 | 2381 | 16 | 3330 | 1104 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000360876 | ENST00000380191 | EIF3B | chr7 | 2418877 | + | GDI2 | chr10 | 5828013 | - | 0.000244122 | 0.9997559 |
| ENST00000397011 | ENST00000380191 | EIF3B | chr7 | 2418877 | + | GDI2 | chr10 | 5828013 | - | 0.000246486 | 0.99975353 |
Top |
Fusion Genomic Features for EIF3B-GDI2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
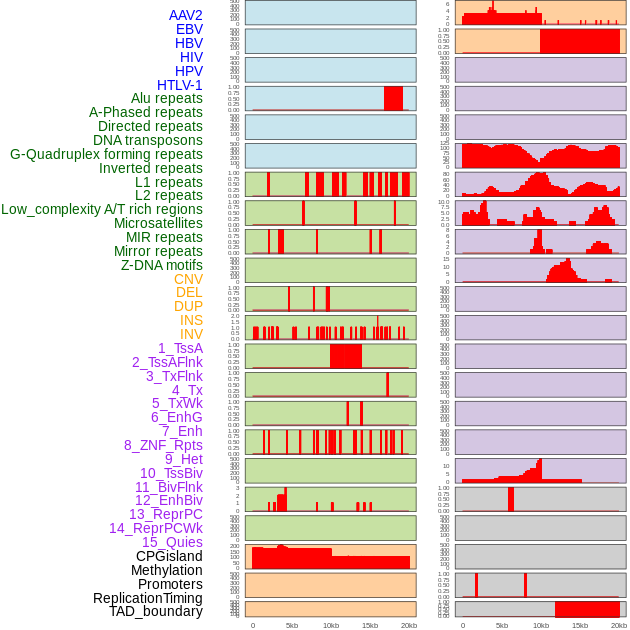 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for EIF3B-GDI2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:2418877/chr10:5828013) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| EIF3B | GDI2 |
| FUNCTION: RNA-binding component of the eukaryotic translation initiation factor 3 (eIF-3) complex, which is required for several steps in the initiation of protein synthesis (PubMed:9388245, PubMed:17581632, PubMed:25849773, PubMed:27462815). The eIF-3 complex associates with the 40S ribosome and facilitates the recruitment of eIF-1, eIF-1A, eIF-2:GTP:methionyl-tRNAi and eIF-5 to form the 43S pre-initiation complex (43S PIC). The eIF-3 complex stimulates mRNA recruitment to the 43S PIC and scanning of the mRNA for AUG recognition. The eIF-3 complex is also required for disassembly and recycling of post-termination ribosomal complexes and subsequently prevents premature joining of the 40S and 60S ribosomal subunits prior to initiation (PubMed:9388245, PubMed:17581632). The eIF-3 complex specifically targets and initiates translation of a subset of mRNAs involved in cell proliferation, including cell cycling, differentiation and apoptosis, and uses different modes of RNA stem-loop binding to exert either translational activation or repression (PubMed:25849773). {ECO:0000255|HAMAP-Rule:MF_03001, ECO:0000269|PubMed:17581632, ECO:0000269|PubMed:25849773, ECO:0000269|PubMed:27462815, ECO:0000269|PubMed:9388245}.; FUNCTION: (Microbial infection) In case of FCV infection, plays a role in the ribosomal termination-reinitiation event leading to the translation of VP2 (PubMed:18056426). {ECO:0000269|PubMed:18056426}. | FUNCTION: Regulates the GDP/GTP exchange reaction of most Rab proteins by inhibiting the dissociation of GDP from them, and the subsequent binding of GTP to them. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000360876 | + | 17 | 19 | 185_268 | 780 | 810.0 | Domain | RRM |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000397011 | + | 17 | 19 | 185_268 | 780 | 791.0 | Domain | RRM |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000360876 | + | 17 | 19 | 285_324 | 780 | 810.0 | Repeat | Note=WD 1 |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000360876 | + | 17 | 19 | 325_365 | 780 | 810.0 | Repeat | Note=WD 2 |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000360876 | + | 17 | 19 | 366_425 | 780 | 810.0 | Repeat | Note=WD 3 |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000360876 | + | 17 | 19 | 426_489 | 780 | 810.0 | Repeat | Note=WD 4 |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000360876 | + | 17 | 19 | 490_553 | 780 | 810.0 | Repeat | Note=WD 5 |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000360876 | + | 17 | 19 | 554_598 | 780 | 810.0 | Repeat | Note=WD 6 |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000360876 | + | 17 | 19 | 599_642 | 780 | 810.0 | Repeat | Note=WD 7 |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000360876 | + | 17 | 19 | 643_685 | 780 | 810.0 | Repeat | Note=WD 8 |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000397011 | + | 17 | 19 | 285_324 | 780 | 791.0 | Repeat | Note=WD 1 |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000397011 | + | 17 | 19 | 325_365 | 780 | 791.0 | Repeat | Note=WD 2 |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000397011 | + | 17 | 19 | 366_425 | 780 | 791.0 | Repeat | Note=WD 3 |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000397011 | + | 17 | 19 | 426_489 | 780 | 791.0 | Repeat | Note=WD 4 |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000397011 | + | 17 | 19 | 490_553 | 780 | 791.0 | Repeat | Note=WD 5 |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000397011 | + | 17 | 19 | 554_598 | 780 | 791.0 | Repeat | Note=WD 6 |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000397011 | + | 17 | 19 | 599_642 | 780 | 791.0 | Repeat | Note=WD 7 |
| Hgene | EIF3B | chr7:2418877 | chr10:5828013 | ENST00000397011 | + | 17 | 19 | 643_685 | 780 | 791.0 | Repeat | Note=WD 8 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Gene Sequence for EIF3B-GDI2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >25782_25782_1_EIF3B-GDI2_EIF3B_chr7_2418877_ENST00000360876_GDI2_chr10_5828013_ENST00000380191_length(transcript)=4130nt_BP=2397nt GGGAGAGTCGGAAGCGCGGCGGCCGCGGAGCCCTGCGAGTAGGCAGCGTTGGGCCCATGCAGGACGCGGAGAACGTGGCGGTGCCCGAGG CGGCCGAGGAGCGCGCCGAGCCCGGCCAGCAGCAGCCGGCCGCCGAGCCGCCGCCAGCCGAGGGGCTGCTGCGGCCCGCGGGGCCCGGCG CTCCGGAGGCCGCGGGGACCGAGGCCTCCAGTGAGGAGGTGGGGATCGCGGAGGCCGGGCCGGAGTCCGAGGTGAGGACCGAGCCGGCGG CCGAGGCAGAGGCGGCCTCCGGCCCGTCCGAGTCGCCCTCGCCGCCGGCCGCCGAGGAGCTGCCCGGGTCGCATGCTGAGCCCCCTGTCC CGGCACAGGGCGAGGCCCCAGGAGAGCAGGCTCGGGACGAGCGCTCCGACAGCCGGGCCCAGGCGGTGTCCGAGGACGCGGGAGGAAACG AGGGCAGAGCGGCCGAGGCCGAACCCCGGGCGCTGGAGAACGGCGACGCGGACGAGCCCTCCTTCAGCGACCCCGAGGACTTCGTGGACG ACGTGAGCGAGGAAGAATTACTGGGAGATGTACTCAAAGATCGGCCCCAGGAAGCAGATGGAATCGATTCGGTGATTGTAGTGGACAATG TCCCTCAGGTGGGACCCGACCGACTTGAGAAACTCAAAAATGTCATCCACAAGATCTTTTCCAAGTTTGGGAAAATCACAAATGATTTTT ATCCTGAAGAGGATGGGAAGACAAAAGGGTATATTTTCCTGGAGTACGCGTCCCCTGCCCACGCTGTGGATGCTGTGAAGAACGCCGACG GCTACAAGCTTGACAAGCAGCACACATTCCGGGTCAACCTCTTTACGGATTTTGACAAGTATATGACGATCAGTGACGAGTGGGATATTC CAGAGAAACAGCCTTTCAAAGACCTGGGGAACTTACGTTACTGGCTTGAAGAGGCAGAATGCAGAGATCAGTACAGTGTGATTTTTGAGA GTGGAGACCGCACTTCCATATTCTGGAATGACGTAAAAGACCCTGTCTCAATTGAAGAAAGAGCGAGATGGACAGAGACGTATGTGCGTT GGTCTCCTAAGGGCACCTACCTGGCTACCTTTCATCAAAGAGGCATTGCTCTATGGGGGGGAGAGAAATTCAAGCAAATTCAGAGATTCA GCCACCAAGGGGTTCAGCTTATTGACTTCTCACCTTGTGAAAGGTACCTGGTGACCTTTAGCCCCCTGATGGACACGCAGGATGACCCTC AGGCCATAATCATCTGGGACATCCTTACGGGGCACAAGAAGAGGGGTTTTCACTGTGAGAGCTCAGCCCATTGGCCTATTTTTAAGTGGA GCCATGATGGCAAATTCTTTGCCAGAATGACCCTGGATACGCTTAGCATCTATGAAACTCCTTCTATGGGTCTTTTGGACAAGAAGAGTT TGAAGATCTCTGGGATAAAAGACTTTTCTTGGTCTCCTGGTGGTAACATAATCGCCTTCTGGGTGCCTGAAGACAAAGATATTCCAGCCA GGGTAACCCTGATGCAGCTCCCTACCAGGCAAGAGATCCGAGTGAGGAACCTGTTCAATGTGGTGGACTGCAAGCTCCATTGGCAGAAGA ACGGAGACTACTTGTGTGTGAAAGTAGATAGGACTCCGAAAGGCACCCAGGGTGTTGTCACAAATTTTGAAATTTTCCGAATGAGGGAGA AACAGGTACCTGTGGATGTGGTCGAGATGAAAGAAACCATCATAGCCTTTGCCTGGGAACCAAATGGAAGTAAGTTTGCTGTGCTGCACG GAGAGGCTCCGCGGATATCTGTGTCTTTCTACCACGTCAAAAACAACGGGAAGATTGAACTCATCAAGATGTTCGACAAGCAGCAGGCGA ACACCATCTTCTGGAGCCCCCAAGGACAGTTCGTGGTGTTGGCGGGCCTGAGGAGTATGAACGGTGCCTTAGCGTTTGTGGACACTTCGG ACTGCACGGTCATGAACATCGCAGAGCACTACATGGCTTCCGACGTCGAATGGGATCCTACTGGGCGCTACGTCGTCACCTCTGTGTCCT GGTGGAGCCATAAGGTGGACAACGCGTACTGGCTGTGGACTTTCCAGGGACGCCTCCTGCAGAAGAACAACAAGGACCGCTTCTGCCAGC TGCTGTGGCGGCCCCGGCCTCCCACACTCCTGAGCCAGGAACAGATCAAGCAAATTAAAAAGGATCTGAAGAAATACTCTAAGATCTTTG AACAGAAGGATCGTTTGAGTCAGTCCAAAGCCTCAAAGGAATTGGTGGAGAGAAGGCGCACCATGATGGAAGATTTCCGGAAGTACCGGA AAATGGCCCAGGAGCTCTATATGGAGCAGAAAAACGAGCGCCTGGAGTTGCGAGGAGGCCTAATGGGATTGTTTGAAAAACGTCGCTTCA GGAAATTCCTAGTGTATGTTGCCAACTTCGATGAAAAAGATCCAAGAACTTTTGAAGGCATTGATCCTAAGAAGACCACAATGCGAGATG TGTATAAGAAATTTGATTTGGGTCAAGACGTTATAGATTTTACTGGTCATGCTCTTGCACTTTACAGAACTGATGATTACTTAGATCAAC CGTGTTATGAAACCATTAATAGAATTAAACTTTACAGTGAATCTTTGGCAAGATATGGCAAAAGCCCATACCTTTATCCACTCTATGGCC TTGGAGAACTGCCCCAAGGATTTGCAAGGCTAAGTGCTATTTATGGAGGTACCTATATGCTGAATAAACCCATTGAAGAAATCATTGTAC AGAATGGAAAAGTAATTGGTGTAAAATCTGAAGGAGAAATTGCTCGCTGTAAGCAGCTCATCTGTGACCCCAGCTACGTAAAAGATCGGG TAGAAAAAGTGGGCCAGGTGATCAGAGTTATTTGCATCCTCAGCCACCCCATCAAGAACACCAATGATGCCAACTCCTGCCAGATCATTA TTCCACAGAACCAAGTCAATCGAAAGTCAGATATCTACGTCTGCATGATCTCCTTTGCGCACAATGTAGCAGCACAAGGGAAGTACATTG CTATAGTTAGTACAACTGTGGAAACCAAGGAGCCTGAGAAGGAAATCAGACCAGCTTTGGAGCTCTTGGAACCAATTGAACAGAAATTTG TTAGCATCAGTGACCTCCTGGTACCAAAAGACTTGGGAACAGAAAGCCAGATCTTTATTTCCCGCACATATGATGCCACCACTCATTTTG AGACAACGTGTGATGACATTAAAAACATCTATAAGAGGATGACAGGATCAGAGTTTGACTTTGAGGAAATGAAGCGCAAGAAGAATGACA TCTATGGGGAAGACTAACAGCAGTACATGTTATTATGTAATTAGGACACATTTAAAATTTGGCAAATAATGCATATAATGAAATCAATAT TGTAAGGCCTGCTTTTGTAATGAAAATGGAGAGAATGAAGAGCGCTGTGCCAGTAAATACTCCCCTTCACCTTTCTAATTATTAACTTGT TTTCATGGAGTGGCTATTCAGCATTGGCAGTTACCACATTCTGTTCAATTTAACCAAACTGGCTTTTTTTTTTTCTAGTGAAGTTAAACA AACATTGGGATACCGACACAGACAACTTGAGACAGTTTTTTTAATCTTTTAATCAGTGTAGCTATGTGCTGCTGCTGCTCAAGAGCTGGA TCCATACAGGTTGTGTGTCATCTGTCCTCTTGAGGTTCAGGAGATTCTAAATTGAATATTCCACGGTTTGGGACAGCATCCGGAAGTTTT CCCTATGACTTTATATTTTGTATTATGTCAAATGTTATGGCAGGGCCCAAATAGCATAGCCACAAGTTTGGTTTATGTGGGCATAAAATT CTAACCAAACCCCAGACATAGGGAGTCATTTGGAGAAAGCCTGTATGTGGTGTTTTAACCTAATAAAGTTGATGAGAGAGAAGGGGAGAG GAAGCGAACATAAAGCGGGTCAAGTGTAGTGCATCTTTTGTATCTCAGGCGTGGCTTCTTCAGTGGACCAAGCTGCAATGCAGTATTGAC >25782_25782_1_EIF3B-GDI2_EIF3B_chr7_2418877_ENST00000360876_GDI2_chr10_5828013_ENST00000380191_length(amino acids)=1104AA_BP=788 MRVGSVGPMQDAENVAVPEAAEERAEPGQQQPAAEPPPAEGLLRPAGPGAPEAAGTEASSEEVGIAEAGPESEVRTEPAAEAEAASGPSE SPSPPAAEELPGSHAEPPVPAQGEAPGEQARDERSDSRAQAVSEDAGGNEGRAAEAEPRALENGDADEPSFSDPEDFVDDVSEEELLGDV LKDRPQEADGIDSVIVVDNVPQVGPDRLEKLKNVIHKIFSKFGKITNDFYPEEDGKTKGYIFLEYASPAHAVDAVKNADGYKLDKQHTFR VNLFTDFDKYMTISDEWDIPEKQPFKDLGNLRYWLEEAECRDQYSVIFESGDRTSIFWNDVKDPVSIEERARWTETYVRWSPKGTYLATF HQRGIALWGGEKFKQIQRFSHQGVQLIDFSPCERYLVTFSPLMDTQDDPQAIIIWDILTGHKKRGFHCESSAHWPIFKWSHDGKFFARMT LDTLSIYETPSMGLLDKKSLKISGIKDFSWSPGGNIIAFWVPEDKDIPARVTLMQLPTRQEIRVRNLFNVVDCKLHWQKNGDYLCVKVDR TPKGTQGVVTNFEIFRMREKQVPVDVVEMKETIIAFAWEPNGSKFAVLHGEAPRISVSFYHVKNNGKIELIKMFDKQQANTIFWSPQGQF VVLAGLRSMNGALAFVDTSDCTVMNIAEHYMASDVEWDPTGRYVVTSVSWWSHKVDNAYWLWTFQGRLLQKNNKDRFCQLLWRPRPPTLL SQEQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRKYRKMAQELYMEQKNERLELRGGLMGLFEKRRFRKFLVYVANFD EKDPRTFEGIDPKKTTMRDVYKKFDLGQDVIDFTGHALALYRTDDYLDQPCYETINRIKLYSESLARYGKSPYLYPLYGLGELPQGFARL SAIYGGTYMLNKPIEEIIVQNGKVIGVKSEGEIARCKQLICDPSYVKDRVEKVGQVIRVICILSHPIKNTNDANSCQIIIPQNQVNRKSD IYVCMISFAHNVAAQGKYIAIVSTTVETKEPEKEIRPALELLEPIEQKFVSISDLLVPKDLGTESQIFISRTYDATTHFETTCDDIKNIY -------------------------------------------------------------- >25782_25782_2_EIF3B-GDI2_EIF3B_chr7_2418877_ENST00000397011_GDI2_chr10_5828013_ENST00000380191_length(transcript)=4114nt_BP=2381nt CGGCGGCCGCGGAGCCCTGCGAGTAGGCAGCGTTGGGCCCATGCAGGACGCGGAGAACGTGGCGGTGCCCGAGGCGGCCGAGGAGCGCGC CGAGCCCGGCCAGCAGCAGCCGGCCGCCGAGCCGCCGCCAGCCGAGGGGCTGCTGCGGCCCGCGGGGCCCGGCGCTCCGGAGGCCGCGGG GACCGAGGCCTCCAGTGAGGAGGTGGGGATCGCGGAGGCCGGGCCGGAGTCCGAGGTGAGGACCGAGCCGGCGGCCGAGGCAGAGGCGGC CTCCGGCCCGTCCGAGTCGCCCTCGCCGCCGGCCGCCGAGGAGCTGCCCGGGTCGCATGCTGAGCCCCCTGTCCCGGCACAGGGCGAGGC CCCAGGAGAGCAGGCTCGGGACGAGCGCTCCGACAGCCGGGCCCAGGCGGTGTCCGAGGACGCGGGAGGAAACGAGGGCAGAGCGGCCGA GGCCGAACCCCGGGCGCTGGAGAACGGCGACGCGGACGAGCCCTCCTTCAGCGACCCCGAGGACTTCGTGGACGACGTGAGCGAGGAAGA ATTACTGGGAGATGTACTCAAAGATCGGCCCCAGGAAGCAGATGGAATCGATTCGGTGATTGTAGTGGACAATGTCCCTCAGGTGGGACC CGACCGACTTGAGAAACTCAAAAATGTCATCCACAAGATCTTTTCCAAGTTTGGGAAAATCACAAATGATTTTTATCCTGAAGAGGATGG GAAGACAAAAGGGTATATTTTCCTGGAGTACGCGTCCCCTGCCCACGCTGTGGATGCTGTGAAGAACGCCGACGGCTACAAGCTTGACAA GCAGCACACATTCCGGGTCAACCTCTTTACGGATTTTGACAAGTATATGACGATCAGTGACGAGTGGGATATTCCAGAGAAACAGCCTTT CAAAGACCTGGGGAACTTACGTTACTGGCTTGAAGAGGCAGAATGCAGAGATCAGTACAGTGTGATTTTTGAGAGTGGAGACCGCACTTC CATATTCTGGAATGACGTAAAAGACCCTGTCTCAATTGAAGAAAGAGCGAGATGGACAGAGACGTATGTGCGTTGGTCTCCTAAGGGCAC CTACCTGGCTACCTTTCATCAAAGAGGCATTGCTCTATGGGGGGGAGAGAAATTCAAGCAAATTCAGAGATTCAGCCACCAAGGGGTTCA GCTTATTGACTTCTCACCTTGTGAAAGGTACCTGGTGACCTTTAGCCCCCTGATGGACACGCAGGATGACCCTCAGGCCATAATCATCTG GGACATCCTTACGGGGCACAAGAAGAGGGGTTTTCACTGTGAGAGCTCAGCCCATTGGCCTATTTTTAAGTGGAGCCATGATGGCAAATT CTTTGCCAGAATGACCCTGGATACGCTTAGCATCTATGAAACTCCTTCTATGGGTCTTTTGGACAAGAAGAGTTTGAAGATCTCTGGGAT AAAAGACTTTTCTTGGTCTCCTGGTGGTAACATAATCGCCTTCTGGGTGCCTGAAGACAAAGATATTCCAGCCAGGGTAACCCTGATGCA GCTCCCTACCAGGCAAGAGATCCGAGTGAGGAACCTGTTCAATGTGGTGGACTGCAAGCTCCATTGGCAGAAGAACGGAGACTACTTGTG TGTGAAAGTAGATAGGACTCCGAAAGGCACCCAGGGTGTTGTCACAAATTTTGAAATTTTCCGAATGAGGGAGAAACAGGTACCTGTGGA TGTGGTCGAGATGAAAGAAACCATCATAGCCTTTGCCTGGGAACCAAATGGAAGTAAGTTTGCTGTGCTGCACGGAGAGGCTCCGCGGAT ATCTGTGTCTTTCTACCACGTCAAAAACAACGGGAAGATTGAACTCATCAAGATGTTCGACAAGCAGCAGGCGAACACCATCTTCTGGAG CCCCCAAGGACAGTTCGTGGTGTTGGCGGGCCTGAGGAGTATGAACGGTGCCTTAGCGTTTGTGGACACTTCGGACTGCACGGTCATGAA CATCGCAGAGCACTACATGGCTTCCGACGTCGAATGGGATCCTACTGGGCGCTACGTCGTCACCTCTGTGTCCTGGTGGAGCCATAAGGT GGACAACGCGTACTGGCTGTGGACTTTCCAGGGACGCCTCCTGCAGAAGAACAACAAGGACCGCTTCTGCCAGCTGCTGTGGCGGCCCCG GCCTCCCACACTCCTGAGCCAGGAACAGATCAAGCAAATTAAAAAGGATCTGAAGAAATACTCTAAGATCTTTGAACAGAAGGATCGTTT GAGTCAGTCCAAAGCCTCAAAGGAATTGGTGGAGAGAAGGCGCACCATGATGGAAGATTTCCGGAAGTACCGGAAAATGGCCCAGGAGCT CTATATGGAGCAGAAAAACGAGCGCCTGGAGTTGCGAGGAGGCCTAATGGGATTGTTTGAAAAACGTCGCTTCAGGAAATTCCTAGTGTA TGTTGCCAACTTCGATGAAAAAGATCCAAGAACTTTTGAAGGCATTGATCCTAAGAAGACCACAATGCGAGATGTGTATAAGAAATTTGA TTTGGGTCAAGACGTTATAGATTTTACTGGTCATGCTCTTGCACTTTACAGAACTGATGATTACTTAGATCAACCGTGTTATGAAACCAT TAATAGAATTAAACTTTACAGTGAATCTTTGGCAAGATATGGCAAAAGCCCATACCTTTATCCACTCTATGGCCTTGGAGAACTGCCCCA AGGATTTGCAAGGCTAAGTGCTATTTATGGAGGTACCTATATGCTGAATAAACCCATTGAAGAAATCATTGTACAGAATGGAAAAGTAAT TGGTGTAAAATCTGAAGGAGAAATTGCTCGCTGTAAGCAGCTCATCTGTGACCCCAGCTACGTAAAAGATCGGGTAGAAAAAGTGGGCCA GGTGATCAGAGTTATTTGCATCCTCAGCCACCCCATCAAGAACACCAATGATGCCAACTCCTGCCAGATCATTATTCCACAGAACCAAGT CAATCGAAAGTCAGATATCTACGTCTGCATGATCTCCTTTGCGCACAATGTAGCAGCACAAGGGAAGTACATTGCTATAGTTAGTACAAC TGTGGAAACCAAGGAGCCTGAGAAGGAAATCAGACCAGCTTTGGAGCTCTTGGAACCAATTGAACAGAAATTTGTTAGCATCAGTGACCT CCTGGTACCAAAAGACTTGGGAACAGAAAGCCAGATCTTTATTTCCCGCACATATGATGCCACCACTCATTTTGAGACAACGTGTGATGA CATTAAAAACATCTATAAGAGGATGACAGGATCAGAGTTTGACTTTGAGGAAATGAAGCGCAAGAAGAATGACATCTATGGGGAAGACTA ACAGCAGTACATGTTATTATGTAATTAGGACACATTTAAAATTTGGCAAATAATGCATATAATGAAATCAATATTGTAAGGCCTGCTTTT GTAATGAAAATGGAGAGAATGAAGAGCGCTGTGCCAGTAAATACTCCCCTTCACCTTTCTAATTATTAACTTGTTTTCATGGAGTGGCTA TTCAGCATTGGCAGTTACCACATTCTGTTCAATTTAACCAAACTGGCTTTTTTTTTTTCTAGTGAAGTTAAACAAACATTGGGATACCGA CACAGACAACTTGAGACAGTTTTTTTAATCTTTTAATCAGTGTAGCTATGTGCTGCTGCTGCTCAAGAGCTGGATCCATACAGGTTGTGT GTCATCTGTCCTCTTGAGGTTCAGGAGATTCTAAATTGAATATTCCACGGTTTGGGACAGCATCCGGAAGTTTTCCCTATGACTTTATAT TTTGTATTATGTCAAATGTTATGGCAGGGCCCAAATAGCATAGCCACAAGTTTGGTTTATGTGGGCATAAAATTCTAACCAAACCCCAGA CATAGGGAGTCATTTGGAGAAAGCCTGTATGTGGTGTTTTAACCTAATAAAGTTGATGAGAGAGAAGGGGAGAGGAAGCGAACATAAAGC GGGTCAAGTGTAGTGCATCTTTTGTATCTCAGGCGTGGCTTCTTCAGTGGACCAAGCTGCAATGCAGTATTGACTTGACAGAGCCTCTAC >25782_25782_2_EIF3B-GDI2_EIF3B_chr7_2418877_ENST00000397011_GDI2_chr10_5828013_ENST00000380191_length(amino acids)=1104AA_BP=788 MRVGSVGPMQDAENVAVPEAAEERAEPGQQQPAAEPPPAEGLLRPAGPGAPEAAGTEASSEEVGIAEAGPESEVRTEPAAEAEAASGPSE SPSPPAAEELPGSHAEPPVPAQGEAPGEQARDERSDSRAQAVSEDAGGNEGRAAEAEPRALENGDADEPSFSDPEDFVDDVSEEELLGDV LKDRPQEADGIDSVIVVDNVPQVGPDRLEKLKNVIHKIFSKFGKITNDFYPEEDGKTKGYIFLEYASPAHAVDAVKNADGYKLDKQHTFR VNLFTDFDKYMTISDEWDIPEKQPFKDLGNLRYWLEEAECRDQYSVIFESGDRTSIFWNDVKDPVSIEERARWTETYVRWSPKGTYLATF HQRGIALWGGEKFKQIQRFSHQGVQLIDFSPCERYLVTFSPLMDTQDDPQAIIIWDILTGHKKRGFHCESSAHWPIFKWSHDGKFFARMT LDTLSIYETPSMGLLDKKSLKISGIKDFSWSPGGNIIAFWVPEDKDIPARVTLMQLPTRQEIRVRNLFNVVDCKLHWQKNGDYLCVKVDR TPKGTQGVVTNFEIFRMREKQVPVDVVEMKETIIAFAWEPNGSKFAVLHGEAPRISVSFYHVKNNGKIELIKMFDKQQANTIFWSPQGQF VVLAGLRSMNGALAFVDTSDCTVMNIAEHYMASDVEWDPTGRYVVTSVSWWSHKVDNAYWLWTFQGRLLQKNNKDRFCQLLWRPRPPTLL SQEQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRKYRKMAQELYMEQKNERLELRGGLMGLFEKRRFRKFLVYVANFD EKDPRTFEGIDPKKTTMRDVYKKFDLGQDVIDFTGHALALYRTDDYLDQPCYETINRIKLYSESLARYGKSPYLYPLYGLGELPQGFARL SAIYGGTYMLNKPIEEIIVQNGKVIGVKSEGEIARCKQLICDPSYVKDRVEKVGQVIRVICILSHPIKNTNDANSCQIIIPQNQVNRKSD IYVCMISFAHNVAAQGKYIAIVSTTVETKEPEKEIRPALELLEPIEQKFVSISDLLVPKDLGTESQIFISRTYDATTHFETTCDDIKNIY -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for EIF3B-GDI2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for EIF3B-GDI2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for EIF3B-GDI2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
