
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:EIF3E-PHF20L1 (FusionGDB2 ID:25831) |
Fusion Gene Summary for EIF3E-PHF20L1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: EIF3E-PHF20L1 | Fusion gene ID: 25831 | Hgene | Tgene | Gene symbol | EIF3E | PHF20L1 | Gene ID | 3646 | 51105 |
| Gene name | eukaryotic translation initiation factor 3 subunit E | PHD finger protein 20 like 1 | |
| Synonyms | EIF3-P48|EIF3S6|INT6|eIF3-p46 | CGI-72|TDRD20B|URLC1 | |
| Cytomap | 8q23.1 | 8q24.22 | |
| Type of gene | protein-coding | protein-coding | |
| Description | eukaryotic translation initiation factor 3 subunit EeIF-3 p48eukaryotic translation initiation factor 3 subunit 6eukaryotic translation initiation factor 3 subunit E isoform 2 transcripteukaryotic translation initiation factor 3, subunit 6 (48kD)euka | PHD finger protein 20-like protein 1tudor domain containing 20Btudor domain-containing protein PHF20L1up-regulated in lung cancer 1 | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | P60228 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000220849, ENST00000519030, ENST00000519517, | ENST00000220847, ENST00000395379, ENST00000395390, ENST00000337920, ENST00000395376, ENST00000395386, | |
| Fusion gene scores | * DoF score | 13 X 12 X 7=1092 | 8 X 9 X 7=504 |
| # samples | 16 | 10 | |
| ** MAII score | log2(16/1092*10)=-2.77082904603249 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(10/504*10)=-2.33342373372519 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: EIF3E [Title/Abstract] AND PHF20L1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | EIF3E(109240496)-PHF20L1(133806656), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | EIF3E-PHF20L1 seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. EIF3E-PHF20L1 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. EIF3E-PHF20L1 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. EIF3E-PHF20L1 seems lost the major protein functional domain in Tgene partner, which is a epigenetic factor due to the frame-shifted ORF. EIF3E-PHF20L1 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | EIF3E | GO:0006413 | translational initiation | 17581632 |
 Fusion gene breakpoints across EIF3E (5'-gene) Fusion gene breakpoints across EIF3E (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
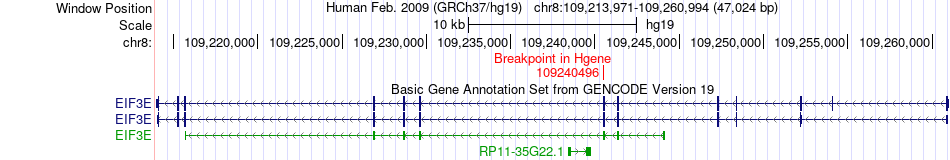 |
 Fusion gene breakpoints across PHF20L1 (3'-gene) Fusion gene breakpoints across PHF20L1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
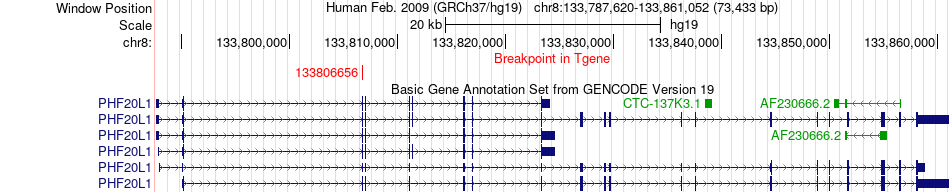 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-YL-A8SH-01B | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| ChimerDB4 | PRAD | TCGA-YL-A8SH | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
Top |
Fusion Gene ORF analysis for EIF3E-PHF20L1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000220849 | ENST00000220847 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| 5CDS-5UTR | ENST00000220849 | ENST00000395379 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| 5CDS-5UTR | ENST00000220849 | ENST00000395390 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| 5CDS-5UTR | ENST00000519030 | ENST00000220847 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| 5CDS-5UTR | ENST00000519030 | ENST00000395379 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| 5CDS-5UTR | ENST00000519030 | ENST00000395390 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| 5UTR-3CDS | ENST00000519517 | ENST00000337920 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| 5UTR-3CDS | ENST00000519517 | ENST00000395376 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| 5UTR-3CDS | ENST00000519517 | ENST00000395386 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| 5UTR-5UTR | ENST00000519517 | ENST00000220847 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| 5UTR-5UTR | ENST00000519517 | ENST00000395379 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| 5UTR-5UTR | ENST00000519517 | ENST00000395390 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| Frame-shift | ENST00000519030 | ENST00000337920 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| Frame-shift | ENST00000519030 | ENST00000395376 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| Frame-shift | ENST00000519030 | ENST00000395386 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| In-frame | ENST00000220849 | ENST00000337920 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| In-frame | ENST00000220849 | ENST00000395376 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
| In-frame | ENST00000220849 | ENST00000395386 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000220849 | EIF3E | chr8 | 109240496 | - | ENST00000395386 | PHF20L1 | chr8 | 133806656 | + | 6640 | 785 | 63 | 3755 | 1230 |
| ENST00000220849 | EIF3E | chr8 | 109240496 | - | ENST00000337920 | PHF20L1 | chr8 | 133806656 | + | 2742 | 785 | 63 | 1559 | 498 |
| ENST00000220849 | EIF3E | chr8 | 109240496 | - | ENST00000395376 | PHF20L1 | chr8 | 133806656 | + | 2835 | 785 | 63 | 1652 | 529 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000220849 | ENST00000395386 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + | 0.000161241 | 0.9998387 |
| ENST00000220849 | ENST00000337920 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + | 0.000156209 | 0.9998437 |
| ENST00000220849 | ENST00000395376 | EIF3E | chr8 | 109240496 | - | PHF20L1 | chr8 | 133806656 | + | 0.000136058 | 0.999864 |
Top |
Fusion Genomic Features for EIF3E-PHF20L1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| EIF3E | chr8 | 109240495 | - | PHF20L1 | chr8 | 133806655 | + | 0.003738333 | 0.99626166 |
| EIF3E | chr8 | 109240495 | - | PHF20L1 | chr8 | 133806655 | + | 0.003738333 | 0.99626166 |
| EIF3E | chr8 | 109240495 | - | PHF20L1 | chr8 | 133806655 | + | 0.003738333 | 0.99626166 |
| EIF3E | chr8 | 109240495 | - | PHF20L1 | chr8 | 133806655 | + | 0.003738333 | 0.99626166 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
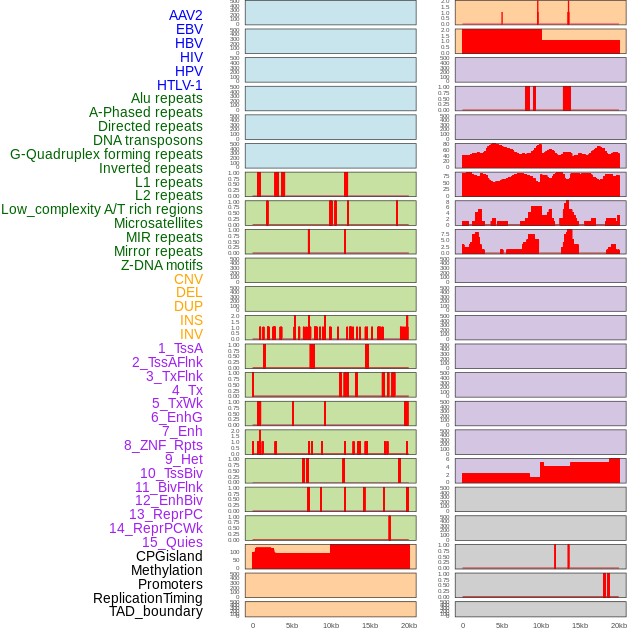 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
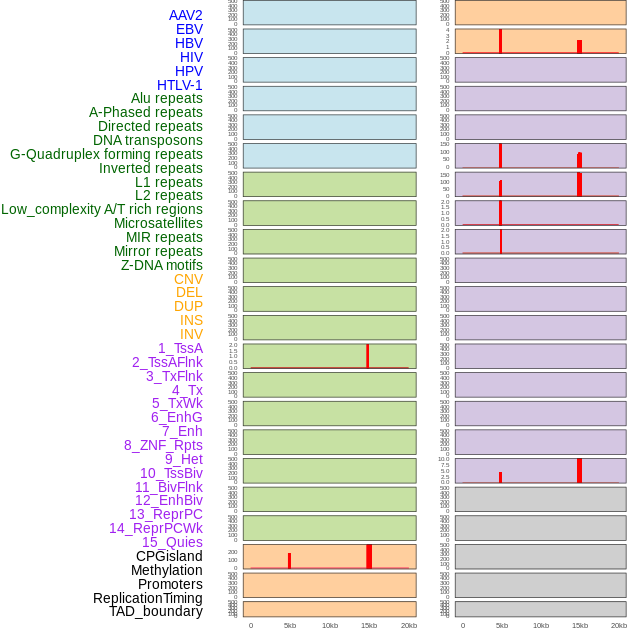 |
Top |
Fusion Protein Features for EIF3E-PHF20L1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:109240496/chr8:133806656) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| EIF3E | . |
| FUNCTION: Component of the eukaryotic translation initiation factor 3 (eIF-3) complex, which is required for several steps in the initiation of protein synthesis (PubMed:17581632, PubMed:25849773, PubMed:27462815). The eIF-3 complex associates with the 40S ribosome and facilitates the recruitment of eIF-1, eIF-1A, eIF-2:GTP:methionyl-tRNAi and eIF-5 to form the 43S pre-initiation complex (43S PIC). The eIF-3 complex stimulates mRNA recruitment to the 43S PIC and scanning of the mRNA for AUG recognition. The eIF-3 complex is also required for disassembly and recycling of post-termination ribosomal complexes and subsequently prevents premature joining of the 40S and 60S ribosomal subunits prior to initiation (PubMed:17581632). The eIF-3 complex specifically targets and initiates translation of a subset of mRNAs involved in cell proliferation, including cell cycling, differentiation and apoptosis, and uses different modes of RNA stem-loop binding to exert either translational activation or repression (PubMed:25849773). Required for nonsense-mediated mRNA decay (NMD); may act in conjunction with UPF2 to divert mRNAs from translation to the NMD pathway (PubMed:17468741). May interact with MCM7 and EPAS1 and regulate the proteasome-mediated degradation of these proteins (PubMed:17310990, PubMed:17324924). {ECO:0000255|HAMAP-Rule:MF_03004, ECO:0000269|PubMed:17310990, ECO:0000269|PubMed:17324924, ECO:0000269|PubMed:17468741, ECO:0000269|PubMed:17581632, ECO:0000269|PubMed:25849773, ECO:0000269|PubMed:27462815}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | PHF20L1 | chr8:109240496 | chr8:133806656 | ENST00000220847 | 1 | 20 | 530_579 | 0 | 405.0 | Compositional bias | Note=Lys-rich | |
| Tgene | PHF20L1 | chr8:109240496 | chr8:133806656 | ENST00000337920 | 1 | 8 | 530_579 | 27 | 286.0 | Compositional bias | Note=Lys-rich | |
| Tgene | PHF20L1 | chr8:109240496 | chr8:133806656 | ENST00000395386 | 1 | 21 | 530_579 | 27 | 1018.0 | Compositional bias | Note=Lys-rich | |
| Tgene | PHF20L1 | chr8:109240496 | chr8:133806656 | ENST00000220847 | 1 | 20 | 11_71 | 0 | 405.0 | Domain | Note=Tudor 1 | |
| Tgene | PHF20L1 | chr8:109240496 | chr8:133806656 | ENST00000220847 | 1 | 20 | 85_141 | 0 | 405.0 | Domain | Note=Tudor 2 | |
| Tgene | PHF20L1 | chr8:109240496 | chr8:133806656 | ENST00000337920 | 1 | 8 | 85_141 | 27 | 286.0 | Domain | Note=Tudor 2 | |
| Tgene | PHF20L1 | chr8:109240496 | chr8:133806656 | ENST00000395386 | 1 | 21 | 85_141 | 27 | 1018.0 | Domain | Note=Tudor 2 | |
| Tgene | PHF20L1 | chr8:109240496 | chr8:133806656 | ENST00000220847 | 1 | 20 | 681_729 | 0 | 405.0 | Zinc finger | Note=PHD-type | |
| Tgene | PHF20L1 | chr8:109240496 | chr8:133806656 | ENST00000337920 | 1 | 8 | 681_729 | 27 | 286.0 | Zinc finger | Note=PHD-type | |
| Tgene | PHF20L1 | chr8:109240496 | chr8:133806656 | ENST00000395386 | 1 | 21 | 681_729 | 27 | 1018.0 | Zinc finger | Note=PHD-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EIF3E | chr8:109240496 | chr8:133806656 | ENST00000220849 | - | 7 | 13 | 221_398 | 240 | 446.0 | Domain | PCI |
| Tgene | PHF20L1 | chr8:109240496 | chr8:133806656 | ENST00000337920 | 1 | 8 | 11_71 | 27 | 286.0 | Domain | Note=Tudor 1 | |
| Tgene | PHF20L1 | chr8:109240496 | chr8:133806656 | ENST00000395386 | 1 | 21 | 11_71 | 27 | 1018.0 | Domain | Note=Tudor 1 |
Top |
Fusion Gene Sequence for EIF3E-PHF20L1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >25831_25831_1_EIF3E-PHF20L1_EIF3E_chr8_109240496_ENST00000220849_PHF20L1_chr8_133806656_ENST00000337920_length(transcript)=2742nt_BP=785nt TGTTCGTTTGTCGGCGCTACCAATAAAGTTTTAGTGAGCACAGACTCCCTTTTCTTTGGCAAGATGGCGGAGTACGACTTGACTACTCGC ATCGCGCACTTTTTGGATCGGCATCTAGTCTTTCCGCTTCTTGAATTTCTCTCTGTAAAGGAGATATATAATGAAAAGGAATTATTACAA GGTAAATTGGACCTTCTTAGTGATACCAACATGGTAGACTTTGCTATGGATGTATACAAAAACCTTTATTCTGATGATATTCCTCATGCT TTGAGAGAGAAAAGAACCACAGTGGTTGCACAACTGAAACAGCTTCAGGCAGAAACAGAACCAATTGTGAAGATGTTTGAAGATCCAGAA ACTACAAGGCAAATGCAGTCAACCAGGGATGGTAGGATGCTCTTTGACTACCTGGCGGACAAGCATGGTTTTAGGCAGGAATATTTAGAT ACACTCTACAGATATGCAAAATTCCAGTACGAATGTGGGAATTACTCAGGAGCAGCAGAATATCTTTATTTTTTTAGAGTGCTGGTTCCA GCAACAGATAGAAATGCTTTAAGTTCACTCTGGGGAAAGCTGGCCTCTGAAATCTTAATGCAGAATTGGGATGCAGCCATGGAAGACCTT ACACGGTTAAAAGAGACCATAGATAATAATTCTGTGAGTTCTCCACTTCAGTCTCTTCAGCAGAGAACATGGCTCATTCACTGGTCTCTG TTTGTTTTCTTCAATCACCCCAAAGGTCGCGATAATATTATTGACCTCTTCCTTTATCAGCCACAGTATCCATCACGAATTGAAAAAATT GACTATGAGGAGGGCAAGATGTTGGTCCATTTTGAGCGCTGGAGTCATCGTTATGATGAGTGGATTTACTGGGATAGCAATAGATTGCGA CCCCTTGAGAGACCAGCACTAAGAAAAGAAGGGCTAAAAGATGAGGAAGATTTCTTTGATTTTAAAGCTGGAGAAGAAGTTCTGGCTCGT TGGACAGACTGTCGCTATTACCCTGCCAAGATTGAAGCAATTAACAAAGAAGGAACATTTACAGTTCAGTTTTATGATGGAGTAATTCGT TGTTTAAAAAGAATGCACATTAAAGCCATGCCCGAGGATGCTAAGGGGCAGGATTGGATAGCTTTAGTCAAAGCAGCTGCTGCAGCTGCA GCCAAGAACAAAACAGGGAGTAAACCTCGAACCAGCGCTAACAGCAATAAAGATAAGGATAAAGATGAGAGAAAGTGGTTTAAAGTACCT TCAAAGAAGGAGGAAACTTCAACTTGTATAGCCACACCAGACGTAGAGAAGAAGGAAGATCTGCCTACATCTAGTGAAACATTTGGACTT CATGTAGAGAACGTTCCAAAGATGGTCTTTCCACAGCCAGAGAGCACATTATCAAACAAGAGGAAAAATAATCAAGGCAACTCGTTTCAG GCAAAGAGAGCTCGACTTAACAAGATTACTGGTTTGTTGGCATCCAAAGCTGTTGGGGTTGATGGTGCTGAAAAAAAGGAAGACTACAAT GAAACAGCTCCAATGCTGGAGCAGGTATGAAATGGTAGCATTTGATTTTTTTCAAGGTTCCCACTGGAATGATCACGGAGCTCTAGTATC TACGTAATTGAAATTCCCCTTTTGATCAGATATGGGAATAGGAAGCTAATTAATTGATGATATATTTTTAACTCTGCCTTCTCCAAGGTA TACAGATAACTACAGACTTCATTAGTGTTTGATTTCATAGACTGGCATTAAAGCCTTTGTCTCCAAAATTTGAAGGCCTGGAGACAGGCA TCCCAGGGTATAAGCTTCCAGCTGGATCACCTTTAACTAGCTTACAAAAAGACCCTTACACAAATACTTTCCATTTGTGAGTTAAAACAG GGTTTTTTGACATCGAATAAAGCTTCTTTTGTAAGAAGTTTCTTTTATATTGTAACAATTGGTGTTGTTAGACTGACTATGTGTTTTATG AATTGATCAATGAGCCATACATTTTTGCTAGGATGTATATTTTAATAACTACAAAAAAAGAGAGTTTAAAAAACTATTAAGGGTCTGTCT TTAAACCACCTAATAGAGGAATTTAAAAGTGAAGGTTGCAGTTGCTAGATTTTGCCTTGGCTACCTTCAGATTCCCACAATACTTGTACA TAATGAACTGTGCATGCTTTTACATTCTCTAAGGGCTTAACTTCTTAGGCACTGTAAGACTATAAGAACGCAGTAATAGCCATAACTGGA TTTCCTTGGTTCTCCTGTTTTAAAATCTGAGCCTTAATGTAGTTTTAATATATCCTTTTTGTATTACTTTTCTTTATTAGTAGCCTAATA AAGAAGACTTTGAGTATCTTTTGCTCTCCTCTAGATGCGTTTTAAAAACTATTCCAAATTTAATTGATATTGTATTGAGTTTTTCAGGGT TATTTTAGGCCTGATTTGATAACAATGCGTTGTTTTGGTGCCTCCATTAATGTTGTATTAAATCAATGGGTCTTTTAAATTGATTAATAT GTCATTGAATCATACCTTCTGATTTTCTGTGATCATCCAACGGTTAAGTCTTTGCCGTTATTGTCAATATATCAACCAAATTTTAAATAA CTTTTTTTACTGTGTATTACTATATTCAACGTCCCAGATATAACAATATTTTGTATTTTCACCACTGTTTATGGTATTCTCAGAATGCGA >25831_25831_1_EIF3E-PHF20L1_EIF3E_chr8_109240496_ENST00000220849_PHF20L1_chr8_133806656_ENST00000337920_length(amino acids)=498AA_BP=241 MAEYDLTTRIAHFLDRHLVFPLLEFLSVKEIYNEKELLQGKLDLLSDTNMVDFAMDVYKNLYSDDIPHALREKRTTVVAQLKQLQAETEP IVKMFEDPETTRQMQSTRDGRMLFDYLADKHGFRQEYLDTLYRYAKFQYECGNYSGAAEYLYFFRVLVPATDRNALSSLWGKLASEILMQ NWDAAMEDLTRLKETIDNNSVSSPLQSLQQRTWLIHWSLFVFFNHPKGRDNIIDLFLYQPQYPSRIEKIDYEEGKMLVHFERWSHRYDEW IYWDSNRLRPLERPALRKEGLKDEEDFFDFKAGEEVLARWTDCRYYPAKIEAINKEGTFTVQFYDGVIRCLKRMHIKAMPEDAKGQDWIA LVKAAAAAAAKNKTGSKPRTSANSNKDKDKDERKWFKVPSKKEETSTCIATPDVEKKEDLPTSSETFGLHVENVPKMVFPQPESTLSNKR -------------------------------------------------------------- >25831_25831_2_EIF3E-PHF20L1_EIF3E_chr8_109240496_ENST00000220849_PHF20L1_chr8_133806656_ENST00000395376_length(transcript)=2835nt_BP=785nt TGTTCGTTTGTCGGCGCTACCAATAAAGTTTTAGTGAGCACAGACTCCCTTTTCTTTGGCAAGATGGCGGAGTACGACTTGACTACTCGC ATCGCGCACTTTTTGGATCGGCATCTAGTCTTTCCGCTTCTTGAATTTCTCTCTGTAAAGGAGATATATAATGAAAAGGAATTATTACAA GGTAAATTGGACCTTCTTAGTGATACCAACATGGTAGACTTTGCTATGGATGTATACAAAAACCTTTATTCTGATGATATTCCTCATGCT TTGAGAGAGAAAAGAACCACAGTGGTTGCACAACTGAAACAGCTTCAGGCAGAAACAGAACCAATTGTGAAGATGTTTGAAGATCCAGAA ACTACAAGGCAAATGCAGTCAACCAGGGATGGTAGGATGCTCTTTGACTACCTGGCGGACAAGCATGGTTTTAGGCAGGAATATTTAGAT ACACTCTACAGATATGCAAAATTCCAGTACGAATGTGGGAATTACTCAGGAGCAGCAGAATATCTTTATTTTTTTAGAGTGCTGGTTCCA GCAACAGATAGAAATGCTTTAAGTTCACTCTGGGGAAAGCTGGCCTCTGAAATCTTAATGCAGAATTGGGATGCAGCCATGGAAGACCTT ACACGGTTAAAAGAGACCATAGATAATAATTCTGTGAGTTCTCCACTTCAGTCTCTTCAGCAGAGAACATGGCTCATTCACTGGTCTCTG TTTGTTTTCTTCAATCACCCCAAAGGTCGCGATAATATTATTGACCTCTTCCTTTATCAGCCACAGTATCCATCACGAATTGAAAAAATT GACTATGAGGAGGGCAAGATGTTGGTCCATTTTGAGCGCTGGAGTCATCGTTATGATGAGTGGATTTACTGGGATAGCAATAGATTGCGA CCCCTTGAGAGACCAGCACTAAGAAAAGAAGGGCTAAAAGATGAGGAAGATTTCTTTGATTTTAAAGCTGGAGAAGAAGTTCTGGCTCGT TGGACAGACTGTCGCTATTACCCTGCCAAGATTGAAGCAATTAACAAAGAAGGAACATTTACAGTTCAGTTTTATGATGGAGTAATTCGT TGTTTAAAAAGAATGCACATTAAAGCCATGCCCGAGGATGCTAAGGGGCAGTTTCTGTTCCAGGTGAAATCCCAGCATCCACTAAGCTGG TGTTGTCCTATCGACCCAGCTGGATCGTGTAACCAGTCTATGGGAAGTGAGGATTGGATAGCTTTAGTCAAAGCAGCTGCTGCAGCTGCA GCCAAGAACAAAACAGGGAGTAAACCTCGAACCAGCGCTAACAGCAATAAAGATAAGGATAAAGATGAGAGAAAGTGGTTTAAAGTACCT TCAAAGAAGGAGGAAACTTCAACTTGTATAGCCACACCAGACGTAGAGAAGAAGGAAGATCTGCCTACATCTAGTGAAACATTTGTAGGA CTTCATGTAGAGAACGTTCCAAAGATGGTCTTTCCACAGCCAGAGAGCACATTATCAAACAAGAGGAAAAATAATCAAGGCAACTCGTTT CAGGCAAAGAGAGCTCGACTTAACAAGATTACTGGTTTGTTGGCATCCAAAGCTGTTGGGGTTGATGGTGCTGAAAAAAAGGAAGACTAC AATGAAACAGCTCCAATGCTGGAGCAGGTATGAAATGGTAGCATTTGATTTTTTTCAAGGTTCCCACTGGAATGATCACGGAGCTCTAGT ATCTACGTAATTGAAATTCCCCTTTTGATCAGATATGGGAATAGGAAGCTAATTAATTGATGATATATTTTTAACTCTGCCTTCTCCAAG GTATACAGATAACTACAGACTTCATTAGTGTTTGATTTCATAGACTGGCATTAAAGCCTTTGTCTCCAAAATTTGAAGGCCTGGAGACAG GCATCCCAGGGTATAAGCTTCCAGCTGGATCACCTTTAACTAGCTTACAAAAAGACCCTTACACAAATACTTTCCATTTGTGAGTTAAAA CAGGGTTTTTTGACATCGAATAAAGCTTCTTTTGTAAGAAGTTTCTTTTATATTGTAACAATTGGTGTTGTTAGACTGACTATGTGTTTT ATGAATTGATCAATGAGCCATACATTTTTGCTAGGATGTATATTTTAATAACTACAAAAAAAGAGAGTTTAAAAAACTATTAAGGGTCTG TCTTTAAACCACCTAATAGAGGAATTTAAAAGTGAAGGTTGCAGTTGCTAGATTTTGCCTTGGCTACCTTCAGATTCCCACAATACTTGT ACATAATGAACTGTGCATGCTTTTACATTCTCTAAGGGCTTAACTTCTTAGGCACTGTAAGACTATAAGAACGCAGTAATAGCCATAACT GGATTTCCTTGGTTCTCCTGTTTTAAAATCTGAGCCTTAATGTAGTTTTAATATATCCTTTTTGTATTACTTTTCTTTATTAGTAGCCTA ATAAAGAAGACTTTGAGTATCTTTTGCTCTCCTCTAGATGCGTTTTAAAAACTATTCCAAATTTAATTGATATTGTATTGAGTTTTTCAG GGTTATTTTAGGCCTGATTTGATAACAATGCGTTGTTTTGGTGCCTCCATTAATGTTGTATTAAATCAATGGGTCTTTTAAATTGATTAA TATGTCATTGAATCATACCTTCTGATTTTCTGTGATCATCCAACGGTTAAGTCTTTGCCGTTATTGTCAATATATCAACCAAATTTTAAA TAACTTTTTTTACTGTGTATTACTATATTCAACGTCCCAGATATAACAATATTTTGTATTTTCACCACTGTTTATGGTATTCTCAGAATG >25831_25831_2_EIF3E-PHF20L1_EIF3E_chr8_109240496_ENST00000220849_PHF20L1_chr8_133806656_ENST00000395376_length(amino acids)=529AA_BP=241 MAEYDLTTRIAHFLDRHLVFPLLEFLSVKEIYNEKELLQGKLDLLSDTNMVDFAMDVYKNLYSDDIPHALREKRTTVVAQLKQLQAETEP IVKMFEDPETTRQMQSTRDGRMLFDYLADKHGFRQEYLDTLYRYAKFQYECGNYSGAAEYLYFFRVLVPATDRNALSSLWGKLASEILMQ NWDAAMEDLTRLKETIDNNSVSSPLQSLQQRTWLIHWSLFVFFNHPKGRDNIIDLFLYQPQYPSRIEKIDYEEGKMLVHFERWSHRYDEW IYWDSNRLRPLERPALRKEGLKDEEDFFDFKAGEEVLARWTDCRYYPAKIEAINKEGTFTVQFYDGVIRCLKRMHIKAMPEDAKGQFLFQ VKSQHPLSWCCPIDPAGSCNQSMGSEDWIALVKAAAAAAAKNKTGSKPRTSANSNKDKDKDERKWFKVPSKKEETSTCIATPDVEKKEDL -------------------------------------------------------------- >25831_25831_3_EIF3E-PHF20L1_EIF3E_chr8_109240496_ENST00000220849_PHF20L1_chr8_133806656_ENST00000395386_length(transcript)=6640nt_BP=785nt TGTTCGTTTGTCGGCGCTACCAATAAAGTTTTAGTGAGCACAGACTCCCTTTTCTTTGGCAAGATGGCGGAGTACGACTTGACTACTCGC ATCGCGCACTTTTTGGATCGGCATCTAGTCTTTCCGCTTCTTGAATTTCTCTCTGTAAAGGAGATATATAATGAAAAGGAATTATTACAA GGTAAATTGGACCTTCTTAGTGATACCAACATGGTAGACTTTGCTATGGATGTATACAAAAACCTTTATTCTGATGATATTCCTCATGCT TTGAGAGAGAAAAGAACCACAGTGGTTGCACAACTGAAACAGCTTCAGGCAGAAACAGAACCAATTGTGAAGATGTTTGAAGATCCAGAA ACTACAAGGCAAATGCAGTCAACCAGGGATGGTAGGATGCTCTTTGACTACCTGGCGGACAAGCATGGTTTTAGGCAGGAATATTTAGAT ACACTCTACAGATATGCAAAATTCCAGTACGAATGTGGGAATTACTCAGGAGCAGCAGAATATCTTTATTTTTTTAGAGTGCTGGTTCCA GCAACAGATAGAAATGCTTTAAGTTCACTCTGGGGAAAGCTGGCCTCTGAAATCTTAATGCAGAATTGGGATGCAGCCATGGAAGACCTT ACACGGTTAAAAGAGACCATAGATAATAATTCTGTGAGTTCTCCACTTCAGTCTCTTCAGCAGAGAACATGGCTCATTCACTGGTCTCTG TTTGTTTTCTTCAATCACCCCAAAGGTCGCGATAATATTATTGACCTCTTCCTTTATCAGCCACAGTATCCATCACGAATTGAAAAAATT GACTATGAGGAGGGCAAGATGTTGGTCCATTTTGAGCGCTGGAGTCATCGTTATGATGAGTGGATTTACTGGGATAGCAATAGATTGCGA CCCCTTGAGAGACCAGCACTAAGAAAAGAAGGGCTAAAAGATGAGGAAGATTTCTTTGATTTTAAAGCTGGAGAAGAAGTTCTGGCTCGT TGGACAGACTGTCGCTATTACCCTGCCAAGATTGAAGCAATTAACAAAGAAGGAACATTTACAGTTCAGTTTTATGATGGAGTAATTCGT TGTTTAAAAAGAATGCACATTAAAGCCATGCCCGAGGATGCTAAGGGGCAGGTGAAATCCCAGCATCCACTAAGCTGGTGTTGTCCTATC GACCCAGCTGGATCGTGTAACCAGTCTATGGGAAGTGAGGATTGGATAGCTTTAGTCAAAGCAGCTGCTGCAGCTGCAGCCAAGAACAAA ACAGGGAGTAAACCTCGAACCAGCGCTAACAGCAATAAAGATAAGGATAAAGATGAGAGAAAGTGGTTTAAAGTACCTTCAAAGAAGGAG GAAACTTCAACTTGTATAGCCACACCAGACGTAGAGAAGAAGGAAGATCTGCCTACATCTAGTGAAACATTTGGACTTCATGTAGAGAAC GTTCCAAAGATGGTCTTTCCACAGCCAGAGAGCACATTATCAAACAAGAGGAAAAATAATCAAGGCAACTCGTTTCAGGCAAAGAGAGCT CGACTTAACAAGATTACTGGTTTGTTGGCATCCAAAGCTGTTGGGGTTGATGGTGCTGAAAAAAAGGAAGACTACAATGAAACAGCTCCA ATGCTGGAGCAGGCGATTTCACCTAAACCTCAAAGTCAGAAAAAAAATGAAGCTGACATTAGCAGTTCTGCCAACACTCAGAAACCTGCA CTGTTATCCTCAACTTTGTCTTCAGGGAAGGCTCGCAGCAAGAAATGCAAACATGAATCTGGAGATTCTTCTGGGTGTATAAAACCCCCT AAATCACCACTTTCCCCAGAATTAATACAAGTCGAGGATTTGACGCTTGTATCTCAGCTTTCTTCTTCAGTGATAAATAAAACTAGTCCT CCACAGCCTGTGAATCCCCCTAGACCTTTCAAGCATAGTGAGCGGAGAAGAAGATCTCAGCGTTTAGCCACCTTACCCATGCCTGATGAT TCTGTAGAAAAGGTTTCTTCTCCCTCTCCAGCCACTGATGGGAAAGTATTCTCCATCAGTTCTCAAAATCAGCAAGAATCTTCAGTACCA GAGGTGCCTGATGTTGCACATTTGCCACTTGAGAAGCTGGGACCCTGTCTCCCTCTTGACTTAAGTCGTGGTTCAGAAGTTACAGCACCG GTAGCCTCAGATTCCTCTTACCGTAATGAATGTCCCAGGGCAGAAAAAGAGGATACACAGATGCTTCCAAATCCTTCTTCCAAAGCAATA GCTGATGGAAGAGGAGCTCCAGCAGCAGCAGGAATATCGAAAACAGAAAAAAAAGTGAAATTGGAAGACAAAAGCTCAACAGCATTTGGT AAGAGAAAAGAAAAAGATAAGGAAAGAAGAGAGAAGAGAGACAAAGATCACTACAGACCAAAACAGAAGAAGAAGAAAAAAAAGAAAAAG AAATCTAAGCAACATGACTATTCAGACTATGAAGACAGTTCCCTCGAATTTTTGGAAAGGTGCTCTTCTCCACTAACTCGATCTTCTGGG AGTTCTCTGGCTTCACGAAGCATGTTTACGGAGAAAACTACAACCTATCAGTACCCAAGGGCAATTCTATCCGTTGATCTTAGTGGTGAA AACTTATCAGATGTAGACTTCCTAGATGATTCTTCAACGGAGAGTTTGCTTCTGAGTGGGGATGAATACAATCAGGACTTTGATTCAACC AATTTTGAGGAATCTCAGGATGAGGATGATGCTCTTAATGAAATTGTGCGATGTATTTGTGAGATGGATGAGGAGAATGGCTTCATGATC CAGTGTGAAGAGTGCTTGTGTTGGCAACACAGCGTGTGCATGGGGCTGCTGGAGGAGAGCATTCCAGAGCAGTACATCTGCTATATCTGC CGGGACCCACCAGGTCAGAGGTGGAGTGCAAAATATCGTTATGATAAGGAGTGGTTGAATAATGGGAGAATGTGCGGGTTATCATTTTTC AAAGAAAATTATTCTCATCTCAATGCCAAAAAGATAGTTTCTACACATCACCTGCTTGCTGATGTCTATGGTGTTACAGAAGTGCTACAC GGGCTACAGCTGAAGATTGGAATACTAAAGAATAAACATCATCCTGACCTTCATCTCTGGGCTTGTTCCGGGAAGCGAAAAGACCAAGAT CAAATAATAGCTGGGGTGGAGAAAAAAATAGCTCAAGACACAGTTAATCGAGAAGAAAAGAAATATGTACAGAACCATAAAGAACCACCT CGTTTGCCCCTAAAAATGGAAGGAACTTATATAACAAGTGAGCATAGCTATCAAAAGCCACAAAGTTTTGGTCAGGACTGTAAATCTCTC GCAGACCCTGGGAGCTCAGATGATGATGATGTTAGTAGTTTGGAAGAAGAACAAGAATTCCACATGAGAAGTAAAAACAGTTTACAGTAC TCAGCAAAAGAACATGGAATGCCTGAAAAGAATCCAGCTGAAGGGAATACAGTATTTGTTTATAATGATAAAAAGGGCACCGAAGACCCA GGAGACTCACATCTTCAGTGGCAGCTCAATCTCCTTACACACATAGAAAATGTGCAGAACGAAGTTACCAGCAGGATGGACCTAATAGAA AAAGAAGTCGATGTTCTGGAAAGCTGGCTTGATTTCACAGGGGAGTTGGAGCCACCAGATCCTCTTGCAAGATTGCCCCAACTTAAACGC CACATAAAACAGCTCCTAATTGACATGGGCAAAGTACAGCAGATAGCAACTCTTTGCTCTGTATGACAACAGTGAACACTTAATGAAAGA ATGTGGCTTTCTTCAGTCAAAGCATTTTTATTATCCACGTGATGGCTAAGTGGATAATTTAAAAGCTTAGTAATGTCTGGTCATTCACTG ATTTGTGATGTCAATAGGATGGCACCTTGGAAAGAAAAATGAAGAACAACTTTATCAAGGAAGCTAGTATTTAAAAACAAATTCATGAGC AAGCTGCAAATGAGAATGTGTTATATGCCAAGGAACAATGAAGTAGAATATAATGTATACTAAGGGATTTCAAGTTCTCAGAATTTTTGA GTAGTTGCTTACGTGAAGCTCAAGATACCTGTAGAAAGAAATATGGTATATTTGTATAGTTTTTAATAGAAAGATCTATGTTTATAAACC AGCACTTGGCCAAAAACAAAATTGTAAAGGAAATTTAAATTCTGGAGAATTCTACAGGGTTGCTCTAAGAACTGTCTTCTCAGCAGTTGA TCCAGCTGTACGGAAATTTAGGGTATTTAAACTTTTAAAGGATCATGAGCTGTTTCTTGGGCGATGAATGTTCTCAATCAGAAAACTGAC AGTAGAAATCTCACTTCTGGGGAAAACAGTTGTGGAATTCTTACTTCATTATGAATGTATTTAAAAAACAAACACCAAATAATTGGAATA TATTGCAGGCATTAAGCTCATTAAAAACAAACTGGCTTGCAGAAGGGTCCGATGTGCCAAGTGATCATGATTCTGCTGGAAAGAGGATTT TAAATATTGTGGGAGTTCTCCCACCCTAAGTCTTACATAATGCCACCAGTCCATCCAAAACCTATATATCACCTATACTATATATATCAT ATATATAGTTGAATGGCAGTATTCAGGCTCAACGTACAGTTTGATCCTGAGTATGCTTGGTGTTTGCCTTCAGAAAAAAAAAAATACATT GTAAATAACCTCAGCTGGGATGAGGAGTGACAGAATATCAAAATAATTTGTGGCTGTGGATTTTTTTAACTGCTAGTAGTGGAATACTGG AAAAGCTTCATTTCTGAAGATGAATTTTATTTTTAAAAAATACATGCACACTCAAAACTTTTAGCTTTGATCACAAGTGGACAAATTTCT GAAACCAAAGGCAACTAAGTTGCTGTGTTAGCTCTTGCTGGATTTTGAGCCTAGGTCCTACTGTCTGCCAGTACTCATGTGAGTTGTATG TGCCCCCAGTGCTACATACGCAGGTATGCGTAAGTGTGTATGCTTGTTTTAAACAAACACTCAACGTACATATGTACATAATCTACACAT ATTTATATCACATATCTAGTTTTATTACTATAGACTATACGAATTGGTGGTTAACATGAAATGTTACCTTTTAACAGACTGTTTTTAAAA ATTAAAAATGTATGTATAGGTTTTGAAATTTTTTTAAAAGGGGAGAAAGACTGTTAAGAGGAGGCTATTTGATGACATAACACTTGAATA TTTTATGCCTCATTCTGTTTATCAGTTCTCGCAATCTGTATAAATGCATTTTAGAACTGATAGACAGTAAACTTGAATTTATCTTTGATA AGAATACATGCCACTGTACATTCAGATATTATTTAAATTTGCAAACACATTGTTCTATATGTAAGGGTACTGTATGTAAAACTCTGTATT AAAACTATTCCACATATCCTAAAATTTCAGCTTGCCTTTTTGCGGCCTTATATTTTGATGTAAAGATTAAAAGAATGTGCAAAACAGTCA GAAATTTTTATTGCCTTTTGAGATTCTCCAACTTGACAAATGTGCCAAAGATCAACAGACAGAAAATATCATCCTGTGTATTTACTTGTC ATACTTTAACTTTGTGAAAGATCTTACTGATAAATGAAAAGCTTTAGCAGAGGTGGTATTGTGGGGTAATTGCTTAAATTTACATATCAA AGTAAAAAAGTAGTGCCTGTATTCCATGTGTGGAGTAAATTTGCTAATGTCTATATTTTATGATTGATACTATTATTTTTCCTTTGCATT TTAAAATAGTGGCTCTAATATTTTCCCTGTCATGGCTACATTCTAGACATTGAATTTATATCTTCTTTTTTTCAGATACGGAGCAAATAG CATCCTTAACCTATGAATAATGGTGTTGCCTAGATTCTTGTCACACACACAGAAGACCCTTTGTACCTAGTAACATTCATCCTCTTGATT CCTGGTGAACACGGTTAAATTCATGCACATTTGTTCTTGTAGTTTCTAAAAATTAGATCAATTTATTTGTTAGCCAGCAAATTGAAAATT CCATTATTAGATTAATGAAATTTTTGCTCTGCTTATATGTATACGAACTGGAAATCTGAATTTTTAAATTTAGATCTTTAAATCAAATTA TTTTTATGCATATTTTCATTTAATATAGAGTATACCAATCGATTGAAGCCTTTCACAAGTAGTGCGCTGAGCTTTTCTTATTGAAGAGAG TGAATTAGTTTCTGAGAAGTCAGTCTATTGTGAAAAGTTTCAGATGAGATTATTTTCTTTTAGTCTTTTTAAATATCACTATATGTATTT AAATAGAAGCAATAAACTAGAGAGAGTGTTTCTTGAAAAGACAATGTTTTATTTGCCACCAGGCAGAACAGAATGAGCATTGGTAGAGAA GGCTCCCTCTTGGTATGTTACTGATAAACTTGTACTTCCTCATACTTGTTTGTGGTTCTACTGACTTATTTCCAAACTTAAGAGTAATGT ACTTCGAAAACAACTTGGTTATTCTTTAAATTTCAGGTAAAACCCCTTTTCCTAACACTCATCTTATTTCCACTGTCTTATTTCCTCTCA >25831_25831_3_EIF3E-PHF20L1_EIF3E_chr8_109240496_ENST00000220849_PHF20L1_chr8_133806656_ENST00000395386_length(amino acids)=1230AA_BP=241 MAEYDLTTRIAHFLDRHLVFPLLEFLSVKEIYNEKELLQGKLDLLSDTNMVDFAMDVYKNLYSDDIPHALREKRTTVVAQLKQLQAETEP IVKMFEDPETTRQMQSTRDGRMLFDYLADKHGFRQEYLDTLYRYAKFQYECGNYSGAAEYLYFFRVLVPATDRNALSSLWGKLASEILMQ NWDAAMEDLTRLKETIDNNSVSSPLQSLQQRTWLIHWSLFVFFNHPKGRDNIIDLFLYQPQYPSRIEKIDYEEGKMLVHFERWSHRYDEW IYWDSNRLRPLERPALRKEGLKDEEDFFDFKAGEEVLARWTDCRYYPAKIEAINKEGTFTVQFYDGVIRCLKRMHIKAMPEDAKGQVKSQ HPLSWCCPIDPAGSCNQSMGSEDWIALVKAAAAAAAKNKTGSKPRTSANSNKDKDKDERKWFKVPSKKEETSTCIATPDVEKKEDLPTSS ETFGLHVENVPKMVFPQPESTLSNKRKNNQGNSFQAKRARLNKITGLLASKAVGVDGAEKKEDYNETAPMLEQAISPKPQSQKKNEADIS SSANTQKPALLSSTLSSGKARSKKCKHESGDSSGCIKPPKSPLSPELIQVEDLTLVSQLSSSVINKTSPPQPVNPPRPFKHSERRRRSQR LATLPMPDDSVEKVSSPSPATDGKVFSISSQNQQESSVPEVPDVAHLPLEKLGPCLPLDLSRGSEVTAPVASDSSYRNECPRAEKEDTQM LPNPSSKAIADGRGAPAAAGISKTEKKVKLEDKSSTAFGKRKEKDKERREKRDKDHYRPKQKKKKKKKKKSKQHDYSDYEDSSLEFLERC SSPLTRSSGSSLASRSMFTEKTTTYQYPRAILSVDLSGENLSDVDFLDDSSTESLLLSGDEYNQDFDSTNFEESQDEDDALNEIVRCICE MDEENGFMIQCEECLCWQHSVCMGLLEESIPEQYICYICRDPPGQRWSAKYRYDKEWLNNGRMCGLSFFKENYSHLNAKKIVSTHHLLAD VYGVTEVLHGLQLKIGILKNKHHPDLHLWACSGKRKDQDQIIAGVEKKIAQDTVNREEKKYVQNHKEPPRLPLKMEGTYITSEHSYQKPQ SFGQDCKSLADPGSSDDDDVSSLEEEQEFHMRSKNSLQYSAKEHGMPEKNPAEGNTVFVYNDKKGTEDPGDSHLQWQLNLLTHIENVQNE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for EIF3E-PHF20L1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for EIF3E-PHF20L1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for EIF3E-PHF20L1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
