
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:EML1-GPI (FusionGDB2 ID:26432) |
Fusion Gene Summary for EML1-GPI |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: EML1-GPI | Fusion gene ID: 26432 | Hgene | Tgene | Gene symbol | EML1 | GPI | Gene ID | 2009 | 10007 |
| Gene name | EMAP like 1 | glucosamine-6-phosphate deaminase 1 | |
| Synonyms | BH|ELP79|EMAP|EMAP-1|EMAPL | GNP1|GNPDA|GNPI|GPI|HLN | |
| Cytomap | 14q32.2 | 5q31.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | echinoderm microtubule-associated protein-like 1echinoderm microtubule associated protein like 1 | glucosamine-6-phosphate isomerase 1GNPDA 1glcN6P deaminase 1oscillin | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | O00423 | Q8IV16 | |
| Ensembl transtripts involved in fusion gene | ENST00000262233, ENST00000334192, ENST00000327921, ENST00000556758, | ENST00000356487, ENST00000415930, ENST00000586425, | |
| Fusion gene scores | * DoF score | 15 X 8 X 11=1320 | 19 X 17 X 10=3230 |
| # samples | 17 | 23 | |
| ** MAII score | log2(17/1320*10)=-2.95693127810811 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(23/3230*10)=-3.81182839863691 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: EML1 [Title/Abstract] AND GPI [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | EML1(100259880)-GPI(34884633), # samples:1 EML1(100259880)-GPI(34884153), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | EML1-GPI seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. EML1-GPI seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. EML1-GPI seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across EML1 (5'-gene) Fusion gene breakpoints across EML1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
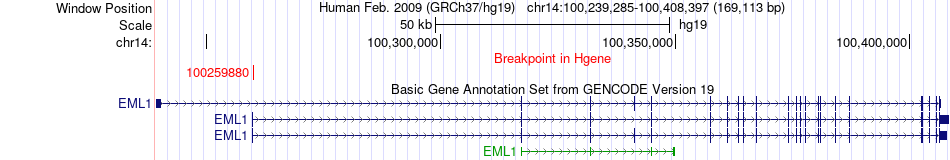 |
 Fusion gene breakpoints across GPI (3'-gene) Fusion gene breakpoints across GPI (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
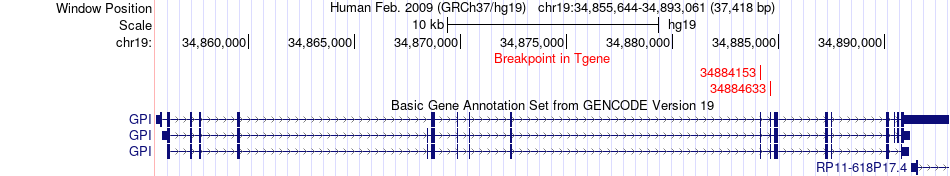 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-RN-AAAQ-01A | EML1 | chr14 | 100259880 | - | GPI | chr19 | 34884153 | + |
| ChimerDB4 | SARC | TCGA-RN-AAAQ-01A | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + |
Top |
Fusion Gene ORF analysis for EML1-GPI |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000262233 | ENST00000356487 | EML1 | chr14 | 100259880 | - | GPI | chr19 | 34884153 | + |
| Frame-shift | ENST00000262233 | ENST00000415930 | EML1 | chr14 | 100259880 | - | GPI | chr19 | 34884153 | + |
| Frame-shift | ENST00000262233 | ENST00000586425 | EML1 | chr14 | 100259880 | - | GPI | chr19 | 34884153 | + |
| Frame-shift | ENST00000334192 | ENST00000356487 | EML1 | chr14 | 100259880 | - | GPI | chr19 | 34884153 | + |
| Frame-shift | ENST00000334192 | ENST00000415930 | EML1 | chr14 | 100259880 | - | GPI | chr19 | 34884153 | + |
| Frame-shift | ENST00000334192 | ENST00000586425 | EML1 | chr14 | 100259880 | - | GPI | chr19 | 34884153 | + |
| In-frame | ENST00000262233 | ENST00000356487 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + |
| In-frame | ENST00000262233 | ENST00000415930 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + |
| In-frame | ENST00000262233 | ENST00000586425 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + |
| In-frame | ENST00000334192 | ENST00000356487 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + |
| In-frame | ENST00000334192 | ENST00000415930 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + |
| In-frame | ENST00000334192 | ENST00000586425 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + |
| intron-3CDS | ENST00000327921 | ENST00000356487 | EML1 | chr14 | 100259880 | - | GPI | chr19 | 34884153 | + |
| intron-3CDS | ENST00000327921 | ENST00000356487 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + |
| intron-3CDS | ENST00000327921 | ENST00000415930 | EML1 | chr14 | 100259880 | - | GPI | chr19 | 34884153 | + |
| intron-3CDS | ENST00000327921 | ENST00000415930 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + |
| intron-3CDS | ENST00000327921 | ENST00000586425 | EML1 | chr14 | 100259880 | - | GPI | chr19 | 34884153 | + |
| intron-3CDS | ENST00000327921 | ENST00000586425 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + |
| intron-3CDS | ENST00000556758 | ENST00000356487 | EML1 | chr14 | 100259880 | - | GPI | chr19 | 34884153 | + |
| intron-3CDS | ENST00000556758 | ENST00000356487 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + |
| intron-3CDS | ENST00000556758 | ENST00000415930 | EML1 | chr14 | 100259880 | - | GPI | chr19 | 34884153 | + |
| intron-3CDS | ENST00000556758 | ENST00000415930 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + |
| intron-3CDS | ENST00000556758 | ENST00000586425 | EML1 | chr14 | 100259880 | - | GPI | chr19 | 34884153 | + |
| intron-3CDS | ENST00000556758 | ENST00000586425 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000262233 | EML1 | chr14 | 100259880 | + | ENST00000415930 | GPI | chr19 | 34884633 | + | 3138 | 206 | 139 | 1017 | 292 |
| ENST00000262233 | EML1 | chr14 | 100259880 | + | ENST00000356487 | GPI | chr19 | 34884633 | + | 1310 | 206 | 1035 | 73 | 320 |
| ENST00000262233 | EML1 | chr14 | 100259880 | + | ENST00000586425 | GPI | chr19 | 34884633 | + | 1115 | 206 | 792 | 73 | 239 |
| ENST00000334192 | EML1 | chr14 | 100259880 | + | ENST00000415930 | GPI | chr19 | 34884633 | + | 3133 | 201 | 134 | 1012 | 292 |
| ENST00000334192 | EML1 | chr14 | 100259880 | + | ENST00000356487 | GPI | chr19 | 34884633 | + | 1305 | 201 | 1030 | 68 | 320 |
| ENST00000334192 | EML1 | chr14 | 100259880 | + | ENST00000586425 | GPI | chr19 | 34884633 | + | 1110 | 201 | 787 | 68 | 239 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000262233 | ENST00000415930 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + | 0.011992403 | 0.98800755 |
| ENST00000262233 | ENST00000356487 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + | 0.018494299 | 0.9815057 |
| ENST00000262233 | ENST00000586425 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + | 0.029555589 | 0.97044444 |
| ENST00000334192 | ENST00000415930 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + | 0.012441185 | 0.98755884 |
| ENST00000334192 | ENST00000356487 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + | 0.019712595 | 0.98028743 |
| ENST00000334192 | ENST00000586425 | EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884633 | + | 0.02994336 | 0.9700566 |
Top |
Fusion Genomic Features for EML1-GPI |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884632 | + | 2.79E-09 | 1 |
| EML1 | chr14 | 100259880 | + | GPI | chr19 | 34884632 | + | 2.79E-09 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
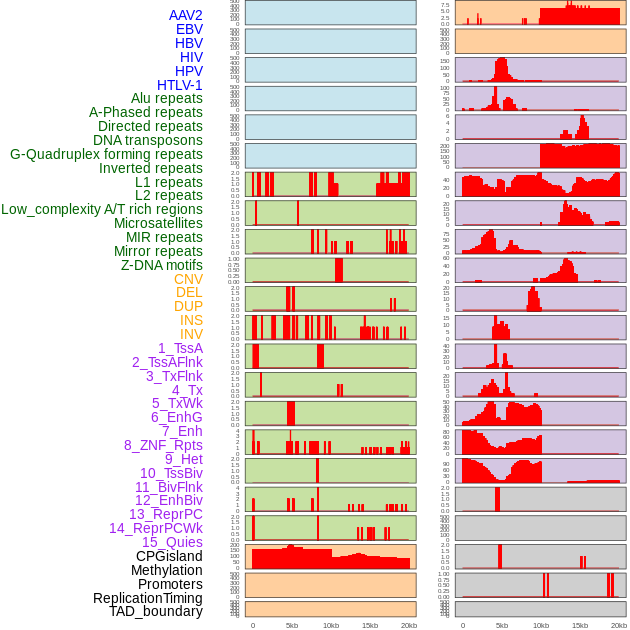 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
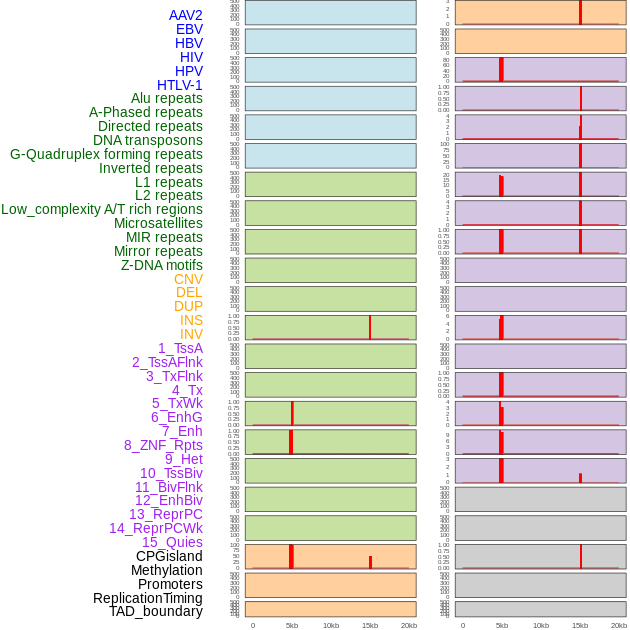 |
Top |
Fusion Protein Features for EML1-GPI |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr14:100259880/chr19:34884633) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| EML1 | GPI |
| FUNCTION: Modulates the assembly and organization of the microtubule cytoskeleton, and probably plays a role in regulating the orientation of the mitotic spindle and the orientation of the plane of cell division. Required for normal proliferation of neuronal progenitor cells in the developing brain and for normal brain development. Does not affect neuron migration per se. {ECO:0000250|UniProtKB:Q05BC3}. | FUNCTION: Mediates the transport of lipoprotein lipase LPL from the basolateral to the apical surface of endothelial cells in capillaries (By similarity). Anchors LPL on the surface of endothelial cells in the lumen of blood capillaries (By similarity). Protects LPL against loss of activity, and against ANGPTL4-mediated unfolding (PubMed:27929370, PubMed:29899144). Thereby, plays an important role in lipolytic processing of chylomicrons by LPL, triglyceride metabolism and lipid homeostasis (PubMed:19304573, PubMed:21314738). Binds chylomicrons and phospholipid particles that contain APOA5 (PubMed:17997385, PubMed:19304573). Binds high-density lipoprotein (HDL) and plays a role in the uptake of lipids from HDL (By similarity). {ECO:0000250|UniProtKB:Q9D1N2, ECO:0000269|PubMed:17997385, ECO:0000269|PubMed:19304573, ECO:0000269|PubMed:21314738, ECO:0000269|PubMed:27929370, ECO:0000269|PubMed:29899144}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000262233 | + | 1 | 22 | 160_166 | 22 | 816.0 | Compositional bias | Note=Poly-Ser |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000334192 | + | 1 | 23 | 160_166 | 22 | 835.0 | Compositional bias | Note=Poly-Ser |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000262233 | + | 1 | 22 | 176_815 | 22 | 816.0 | Region | Note=Tandem atypical propeller in EMLs |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000334192 | + | 1 | 23 | 176_815 | 22 | 835.0 | Region | Note=Tandem atypical propeller in EMLs |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000262233 | + | 1 | 22 | 261_310 | 22 | 816.0 | Repeat | Note=WD 1 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000262233 | + | 1 | 22 | 315_358 | 22 | 816.0 | Repeat | Note=WD 2 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000262233 | + | 1 | 22 | 363_400 | 22 | 816.0 | Repeat | Note=WD 3 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000262233 | + | 1 | 22 | 409_446 | 22 | 816.0 | Repeat | Note=WD 4 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000262233 | + | 1 | 22 | 450_489 | 22 | 816.0 | Repeat | Note=WD 5 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000262233 | + | 1 | 22 | 493_530 | 22 | 816.0 | Repeat | Note=WD 6 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000262233 | + | 1 | 22 | 535_572 | 22 | 816.0 | Repeat | Note=WD 7 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000262233 | + | 1 | 22 | 578_613 | 22 | 816.0 | Repeat | Note=WD 8 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000262233 | + | 1 | 22 | 617_655 | 22 | 816.0 | Repeat | Note=WD 9 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000262233 | + | 1 | 22 | 664_701 | 22 | 816.0 | Repeat | Note=WD 10 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000262233 | + | 1 | 22 | 709_768 | 22 | 816.0 | Repeat | Note=WD 11 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000262233 | + | 1 | 22 | 775_814 | 22 | 816.0 | Repeat | Note=WD 12 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000334192 | + | 1 | 23 | 261_310 | 22 | 835.0 | Repeat | Note=WD 1 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000334192 | + | 1 | 23 | 315_358 | 22 | 835.0 | Repeat | Note=WD 2 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000334192 | + | 1 | 23 | 363_400 | 22 | 835.0 | Repeat | Note=WD 3 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000334192 | + | 1 | 23 | 409_446 | 22 | 835.0 | Repeat | Note=WD 4 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000334192 | + | 1 | 23 | 450_489 | 22 | 835.0 | Repeat | Note=WD 5 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000334192 | + | 1 | 23 | 493_530 | 22 | 835.0 | Repeat | Note=WD 6 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000334192 | + | 1 | 23 | 535_572 | 22 | 835.0 | Repeat | Note=WD 7 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000334192 | + | 1 | 23 | 578_613 | 22 | 835.0 | Repeat | Note=WD 8 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000334192 | + | 1 | 23 | 617_655 | 22 | 835.0 | Repeat | Note=WD 9 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000334192 | + | 1 | 23 | 664_701 | 22 | 835.0 | Repeat | Note=WD 10 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000334192 | + | 1 | 23 | 709_768 | 22 | 835.0 | Repeat | Note=WD 11 |
| Hgene | EML1 | chr14:100259880 | chr19:34884633 | ENST00000334192 | + | 1 | 23 | 775_814 | 22 | 835.0 | Repeat | Note=WD 12 |
| Tgene | GPI | chr14:100259880 | chr19:34884633 | ENST00000356487 | 9 | 18 | 159_160 | 288 | 559.0 | Region | Glucose-6-phosphate binding | |
| Tgene | GPI | chr14:100259880 | chr19:34884633 | ENST00000356487 | 9 | 18 | 210_215 | 288 | 559.0 | Region | Glucose-6-phosphate binding | |
| Tgene | GPI | chr14:100259880 | chr19:34884633 | ENST00000415930 | 9 | 18 | 159_160 | 299 | 570.0 | Region | Glucose-6-phosphate binding | |
| Tgene | GPI | chr14:100259880 | chr19:34884633 | ENST00000415930 | 9 | 18 | 210_215 | 299 | 570.0 | Region | Glucose-6-phosphate binding |
Top |
Fusion Gene Sequence for EML1-GPI |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >26432_26432_1_EML1-GPI_EML1_chr14_100259880_ENST00000262233_GPI_chr19_34884633_ENST00000356487_length(transcript)=1310nt_BP=206nt ATGGTAACCGGCAGCAGCAGCCGCCACAGCAGGGCCGGCCCCAGCGCCAGCGCCGGTCGCGGTCGGGCTCAGCTCAGTGTGTGGTGAGCG GCGGCGGCGCGGCCGGGCCGGGGAGCGGGCGCGGCCCGGCGGCCTCAGCATGGAGGACGGCTTCTCCAGCTACAGCAGCCTGTACGACAC GTCCTCGCTGCTCCAGTTCTGCAACGGTTTTGACAACTTCGAGCAGCTGCTCTCGGGGGCTCACTGGATGGACCAGCACTTCCGCACGAC GCCCCTGGAGAAGAACGCCCCCGTCTTGCTGGCCCTGCTGGGTATCTGGTACATCAACTGCTTTGGGTGTGAGACACACGCCATGCTGCC CTATGACCAGTACCTGCACCGCTTTGCTGCGTACTTCCAGCAGGGCGACATGGAGTCCAATGGGAAATACATCACCAAATCTGGAACCCG TGTGGACCACCAGACAGGCCCCATTGTGTGGGGGGAGCCAGGGACCAATGGCCAGCATGCTTTTTACCAGCTCATCCACCAAGGCACCAA GATGATACCCTGTGACTTCCTCATCCCGGTCCAGACCCAGCACCCCATACGGAAGGGTCTGCATCACAAGATCCTCCTGGCCAACTTCTT GGCCCAGACAGAGGCCCTGATGAGGGGAAAATCGACGGAGGAGGCCCGAAAGGAGCTCCAGGCTGCGGGCAAGAGTCCAGAGGACCTTGA GAGGCTGCTGCCACATAAGGTCTTTGAAGGAAATCGCCCAACCAACTCTATTGTGTTCACCAAGCTCACACCATTCATGCTTGGAGCCTT GGTCGCCATGTATGAGCACAAGATCTTCGTTCAGGGCATCATCTGGGACATCAACAGCTTTGACCAGTGGGGAGTGGAGCTGGGAAAGCA GCTGGCTAAGAAAATAGAGCCTGAGCTTGATGGCAGTGCTCAAGTGACCTCTCACGACGCTTCTACCAATGGGCTCATCAACTTCATCAA GCAGCAGCGCGAGGCCAGAGTCCAATAAACTCGTGCTCATCTGCAGCCTCCTCTGTGACTCCCCTTTCTCTTCTCGTCCCTCCTCCCCGG AGCCGGCACTGCATGTTCCTGGACACCACCCAGAGCACCCTCTGGTTGTGGGCTTGGACCACGAGCCCTTAGCAGGGAAGGCTGGTCTCC CCCAGCCTAACCCCCAGCCCCTCCATGTCTATGCTCCCTCTGTGTTAGAATTGGCTGAAGTGTTTTTGTGCAGCTGACTTTTCTGACCCA >26432_26432_1_EML1-GPI_EML1_chr14_100259880_ENST00000262233_GPI_chr19_34884633_ENST00000356487_length(amino acids)=320AA_BP=1 MQMSTSLLDSGLALLLDEVDEPIGRSVVRGHLSTAIKLRLYFLSQLLSQLHSPLVKAVDVPDDALNEDLVLIHGDQGSKHEWCELGEHNR VGWAISFKDLMWQQPLKVLWTLARSLELLSGLLRRFSPHQGLCLGQEVGQEDLVMQTLPYGVLGLDRDEEVTGYHLGALVDELVKSMLAI GPWLPPHNGACLVVHTGSRFGDVFPIGLHVALLEVRSKAVQVLVIGQHGVCLTPKAVDVPDTQQGQQDGGVLLQGRRAEVLVHPVSPREQ -------------------------------------------------------------- >26432_26432_2_EML1-GPI_EML1_chr14_100259880_ENST00000262233_GPI_chr19_34884633_ENST00000415930_length(transcript)=3138nt_BP=206nt ATGGTAACCGGCAGCAGCAGCCGCCACAGCAGGGCCGGCCCCAGCGCCAGCGCCGGTCGCGGTCGGGCTCAGCTCAGTGTGTGGTGAGCG GCGGCGGCGCGGCCGGGCCGGGGAGCGGGCGCGGCCCGGCGGCCTCAGCATGGAGGACGGCTTCTCCAGCTACAGCAGCCTGTACGACAC GTCCTCGCTGCTCCAGTTCTGCAACGGTTTTGACAACTTCGAGCAGCTGCTCTCGGGGGCTCACTGGATGGACCAGCACTTCCGCACGAC GCCCCTGGAGAAGAACGCCCCCGTCTTGCTGGCCCTGCTGGGTATCTGGTACATCAACTGCTTTGGGTGTGAGACACACGCCATGCTGCC CTATGACCAGTACCTGCACCGCTTTGCTGCGTACTTCCAGCAGGGCGACATGGAGTCCAATGGGAAATACATCACCAAATCTGGAACCCG TGTGGACCACCAGACAGGCCCCATTGTGTGGGGGGAGCCAGGGACCAATGGCCAGCATGCTTTTTACCAGCTCATCCACCAAGGCACCAA GATGATACCCTGTGACTTCCTCATCCCGGTCCAGACCCAGCACCCCATACGGAAGGGTCTGCATCACAAGATCCTCCTGGCCAACTTCTT GGCCCAGACAGAGGCCCTGATGAGGGGAAAATCGACGGAGGAGGCCCGAAAGGAGCTCCAGGCTGCGGGCAAGAGTCCAGAGGACCTTGA GAGGCTGCTGCCACATAAGGTCTTTGAAGGAAATCGCCCAACCAACTCTATTGTGTTCACCAAGCTCACACCATTCATGCTTGGAGCCTT GGTCGCCATGTATGAGCACAAGATCTTCGTTCAGGGCATCATCTGGGACATCAACAGCTTTGACCAGTGGGGAGTGGAGCTGGGAAAGCA GCTGGCTAAGAAAATAGAGCCTGAGCTTGATGGCAGTGCTCAAGTGACCTCTCACGACGCTTCTACCAATGGGCTCATCAACTTCATCAA GCAGCAGCGCGAGGCCAGAGTCCAATAAACTCGTGCTCATCTGCAGCCTCCTCTGTGACTCCCCTTTCTCTTCTCGTCCCTCCTCCCCGG AGCCGGCACTGCATGTTCCTGGACACCACCCAGAGCACCCTCTGGTTGTGGGCTTGGACCACGAGCCCTTAGCAGGGAAGGCTGGTCTCC CCCAGCCTAACCCCCAGCCCCTCCATGTCTATGCTCCCTCTGTGTTAGAATTGGCTGAAGTGTTTTTGTGCAGCTGACTTTTCTGACCCA TGTTCACGTTGTTCACATCCCATGTAGAAAAATAAAGATGCCACGGAGGAGGTTGTAGGCTCAGCCTCTGATTTTTTTTTTCCTGTGATG GTGCTTTATGTAGCAGAGGGCAGGAGCGCTCAGCAGGACGCAGGCTGTGCCTCTGCGGACACTTAACACTAAGTGGTGAGCGGGTCTAGA GTGGAGCAAGGTGCCCTGAGAAGACAATAGTGGGGTGGGGGCACAATCAGTCAGGACGGCAACTTGGCCTGTGTCACCAAATCCCAAGAC TGTTTTCCACTCCTCACCTCTGTGACTGCAGAAATTGGATACTCTGTTCACTCGATGGTTCTAAAAACTGCATTGAGATTATGTTTGTTT CGGGTGAATTCCTGGACAAGACCGAGGATGACTGCCATCTCCTGGCAAGACGCTCAGGTAGTTCTTTTGCTTTAAAAGGCAGATATTGAA AACTGGAATTTTTTTTTTTTGAGTCTCGCTCTGTCACCCAGACTGGAGTGCAGTGGTGCAATCTCGGCTCACTGCAACCTCCGCCTCCCG GGTTCAAGCTATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGATTACACGGCGCACACCACCATACCCAGCTAATTTTTGTATTTTTAGT AGTGAAGGGGTTTTACCATGTTGGGCAGGCTGGTCTTGAACTCCTGACCTCAGGTGATCTGCCCGCCTCAGCCTCCCACAGTGCTGGGAT TACAGGTATGAGCCACCACGCCCGGCCCATTTTTTTTTTTTTTTTGACAACTTTTTTTTTTTTTTGAGACAGGGTCTTGTTCCATTGCCC AGACTGGAGTGCAGTGGCATGATCACAGCTCACTGCAGCCAGTAATCCTCTTGCCTCAGCCTCCCAAGTAGTTGAGACTACAGGTTGTAC CACTATGCCCTGCTAGTTTTTTCATTTTTTGTAGAGAGACGGGTCTTTTTTTTTTTTGAGACGGAGTCTCGCTCTGTCGCCCAAGCTGGA GTGCAGTAGCACGGTCTCAGCTCATTGCAAGCTCCGCCTCCCAGGTTCACGCCATTCTCCTGCCTCAGACTCCTGTGTAGCTGGGAGTAC AGGCACCTGCCACCATGCCCGGCTAATTTTTTATATATTTTTTTAGCAGAGACAGTGTCTCACTGTGTTAGTCAGGATGGTCTCGATCTC CTGACCTCGTGATCCGCCCGCCTCAGCCTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACCGCGCCCAGCAAGGCGGGTCTTGCTGTGT TTCCCAGACTAGACTGGTCTTGAATTCCAGGGCTCAAGAGATCTCCCACCTCAGCCTCCCACAGTGCTGGGATTACAGGCGTGAGCCGCC ACACCCAGCCTATTCAAAATTTTTTTTTCTTAGAGACAGGGTCTTTGTTGCCCAGGCTGGACTGCAGTGATACAATCATAGCTGACTGAA GCCTCAAATTCCCAGGCTAAGGTGATCTTCTCACCTCAGCCTTCCAAGTAGCTGGGTCCGCAGATGCATGCCAGTACACCCAGCTCATTT AAAAAAAAATTTTTTTCTTTTTTGAGAGTCTTGCTTTGTTGCCCAGGCTGGAGTGCAGTGGTGTGATCTCGGCTCACTGCAAGCTCCACC TCCCGGCTTCACGCCATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGACTACAGGTGCCCGCCACCACACCCGGCTAATTTTTTGTATTT TTAGTAGAGACGGGGTTTCACCGTGTTAGCCAGGATGGTCTTGATCTCCTGACCTCATGATCCGCCTGCTTTGGCCTCCCAAAGTGCTGG >26432_26432_2_EML1-GPI_EML1_chr14_100259880_ENST00000262233_GPI_chr19_34884633_ENST00000415930_length(amino acids)=292AA_BP=22 MEDGFSSYSSLYDTSSLLQFCNGFDNFEQLLSGAHWMDQHFRTTPLEKNAPVLLALLGIWYINCFGCETHAMLPYDQYLHRFAAYFQQGD MESNGKYITKSGTRVDHQTGPIVWGEPGTNGQHAFYQLIHQGTKMIPCDFLIPVQTQHPIRKGLHHKILLANFLAQTEALMRGKSTEEAR KELQAAGKSPEDLERLLPHKVFEGNRPTNSIVFTKLTPFMLGALVAMYEHKIFVQGIIWDINSFDQWGVELGKQLAKKIEPELDGSAQVT -------------------------------------------------------------- >26432_26432_3_EML1-GPI_EML1_chr14_100259880_ENST00000262233_GPI_chr19_34884633_ENST00000586425_length(transcript)=1115nt_BP=206nt ATGGTAACCGGCAGCAGCAGCCGCCACAGCAGGGCCGGCCCCAGCGCCAGCGCCGGTCGCGGTCGGGCTCAGCTCAGTGTGTGGTGAGCG GCGGCGGCGCGGCCGGGCCGGGGAGCGGGCGCGGCCCGGCGGCCTCAGCATGGAGGACGGCTTCTCCAGCTACAGCAGCCTGTACGACAC GTCCTCGCTGCTCCAGTTCTGCAACGGTTTTGACAACTTCGAGCAGCTGCTCTCGGGGGCTCACTGGATGGACCAGCACTTCCGCACGAC GCCCCTGGAGAAGAACGCCCCCGTCTTGCTGGCCCTGCTGGGTATCTGGTACATCAACTGCTTTGGGTGTGAGACACACGCCATGCTGCC CTATGACCAGTACCTGCACCGCTTTGCTGCGTACTTCCAGCAGGGCGACATGGAGTCCAATGGGAAATACATCACCAAATCTGGAACCCG TGTGGACCACCAGACAGGCCCCATTGTGTGGGGGGAGCCAGGGACCAATGGCCAGCATGCTTTTTACCAGCTCATCCACCAAGGCACCAA GATGATACCCTGTGACTTCCTCATCCCGGTCCAGACCCAGCACCCCATACGGAAGGGTCTGCATCACAAGATCCTCCTGGCCAACTTCTT GGCCCAGACAGAGGCCCTGATGAGGGGAAAATCGACGGAGGAGGCCCGAAAGGAGCTCCAGGCTGCGGGCAAGAGTCCAGAGGACCTTGA GAGGCTGCTGCCACATAAGAGTGGAGCTGGGAAAGCAGCTGGCTAAGAAAATAGAGCCTGAGCTTGATGGCAGTGCTCAAGTGACCTCTC ACGACGCTTCTACCAATGGGCTCATCAACTTCATCAAGCAGCAGCGCGAGGCCAGAGTCCAATAAACTCGTGCTCATCTGCAGCCTCCTC TGTGACTCCCCTTTCTCTTCTCGTCCCTCCTCCCCGGAGCCGGCACTGCATGTTCCTGGACACCACCCAGAGCACCCTCTGGTTGTGGGC TTGGACCACGAGCCCTTAGCAGGGAAGGCTGGTCTCCCCCAGCCTAACCCCCAGCCCCTCCATGTCTATGCTCCCTCTGTGTTAGAATTG >26432_26432_3_EML1-GPI_EML1_chr14_100259880_ENST00000262233_GPI_chr19_34884633_ENST00000586425_length(amino acids)=239AA_BP=1 MPSSSGSIFLASCFPSSTLMWQQPLKVLWTLARSLELLSGLLRRFSPHQGLCLGQEVGQEDLVMQTLPYGVLGLDRDEEVTGYHLGALVD ELVKSMLAIGPWLPPHNGACLVVHTGSRFGDVFPIGLHVALLEVRSKAVQVLVIGQHGVCLTPKAVDVPDTQQGQQDGGVLLQGRRAEVL -------------------------------------------------------------- >26432_26432_4_EML1-GPI_EML1_chr14_100259880_ENST00000334192_GPI_chr19_34884633_ENST00000356487_length(transcript)=1305nt_BP=201nt AACCGGCAGCAGCAGCCGCCACAGCAGGGCCGGCCCCAGCGCCAGCGCCGGTCGCGGTCGGGCTCAGCTCAGTGTGTGGTGAGCGGCGGC GGCGCGGCCGGGCCGGGGAGCGGGCGCGGCCCGGCGGCCTCAGCATGGAGGACGGCTTCTCCAGCTACAGCAGCCTGTACGACACGTCCT CGCTGCTCCAGTTCTGCAACGGTTTTGACAACTTCGAGCAGCTGCTCTCGGGGGCTCACTGGATGGACCAGCACTTCCGCACGACGCCCC TGGAGAAGAACGCCCCCGTCTTGCTGGCCCTGCTGGGTATCTGGTACATCAACTGCTTTGGGTGTGAGACACACGCCATGCTGCCCTATG ACCAGTACCTGCACCGCTTTGCTGCGTACTTCCAGCAGGGCGACATGGAGTCCAATGGGAAATACATCACCAAATCTGGAACCCGTGTGG ACCACCAGACAGGCCCCATTGTGTGGGGGGAGCCAGGGACCAATGGCCAGCATGCTTTTTACCAGCTCATCCACCAAGGCACCAAGATGA TACCCTGTGACTTCCTCATCCCGGTCCAGACCCAGCACCCCATACGGAAGGGTCTGCATCACAAGATCCTCCTGGCCAACTTCTTGGCCC AGACAGAGGCCCTGATGAGGGGAAAATCGACGGAGGAGGCCCGAAAGGAGCTCCAGGCTGCGGGCAAGAGTCCAGAGGACCTTGAGAGGC TGCTGCCACATAAGGTCTTTGAAGGAAATCGCCCAACCAACTCTATTGTGTTCACCAAGCTCACACCATTCATGCTTGGAGCCTTGGTCG CCATGTATGAGCACAAGATCTTCGTTCAGGGCATCATCTGGGACATCAACAGCTTTGACCAGTGGGGAGTGGAGCTGGGAAAGCAGCTGG CTAAGAAAATAGAGCCTGAGCTTGATGGCAGTGCTCAAGTGACCTCTCACGACGCTTCTACCAATGGGCTCATCAACTTCATCAAGCAGC AGCGCGAGGCCAGAGTCCAATAAACTCGTGCTCATCTGCAGCCTCCTCTGTGACTCCCCTTTCTCTTCTCGTCCCTCCTCCCCGGAGCCG GCACTGCATGTTCCTGGACACCACCCAGAGCACCCTCTGGTTGTGGGCTTGGACCACGAGCCCTTAGCAGGGAAGGCTGGTCTCCCCCAG CCTAACCCCCAGCCCCTCCATGTCTATGCTCCCTCTGTGTTAGAATTGGCTGAAGTGTTTTTGTGCAGCTGACTTTTCTGACCCATGTTC >26432_26432_4_EML1-GPI_EML1_chr14_100259880_ENST00000334192_GPI_chr19_34884633_ENST00000356487_length(amino acids)=320AA_BP=1 MQMSTSLLDSGLALLLDEVDEPIGRSVVRGHLSTAIKLRLYFLSQLLSQLHSPLVKAVDVPDDALNEDLVLIHGDQGSKHEWCELGEHNR VGWAISFKDLMWQQPLKVLWTLARSLELLSGLLRRFSPHQGLCLGQEVGQEDLVMQTLPYGVLGLDRDEEVTGYHLGALVDELVKSMLAI GPWLPPHNGACLVVHTGSRFGDVFPIGLHVALLEVRSKAVQVLVIGQHGVCLTPKAVDVPDTQQGQQDGGVLLQGRRAEVLVHPVSPREQ -------------------------------------------------------------- >26432_26432_5_EML1-GPI_EML1_chr14_100259880_ENST00000334192_GPI_chr19_34884633_ENST00000415930_length(transcript)=3133nt_BP=201nt AACCGGCAGCAGCAGCCGCCACAGCAGGGCCGGCCCCAGCGCCAGCGCCGGTCGCGGTCGGGCTCAGCTCAGTGTGTGGTGAGCGGCGGC GGCGCGGCCGGGCCGGGGAGCGGGCGCGGCCCGGCGGCCTCAGCATGGAGGACGGCTTCTCCAGCTACAGCAGCCTGTACGACACGTCCT CGCTGCTCCAGTTCTGCAACGGTTTTGACAACTTCGAGCAGCTGCTCTCGGGGGCTCACTGGATGGACCAGCACTTCCGCACGACGCCCC TGGAGAAGAACGCCCCCGTCTTGCTGGCCCTGCTGGGTATCTGGTACATCAACTGCTTTGGGTGTGAGACACACGCCATGCTGCCCTATG ACCAGTACCTGCACCGCTTTGCTGCGTACTTCCAGCAGGGCGACATGGAGTCCAATGGGAAATACATCACCAAATCTGGAACCCGTGTGG ACCACCAGACAGGCCCCATTGTGTGGGGGGAGCCAGGGACCAATGGCCAGCATGCTTTTTACCAGCTCATCCACCAAGGCACCAAGATGA TACCCTGTGACTTCCTCATCCCGGTCCAGACCCAGCACCCCATACGGAAGGGTCTGCATCACAAGATCCTCCTGGCCAACTTCTTGGCCC AGACAGAGGCCCTGATGAGGGGAAAATCGACGGAGGAGGCCCGAAAGGAGCTCCAGGCTGCGGGCAAGAGTCCAGAGGACCTTGAGAGGC TGCTGCCACATAAGGTCTTTGAAGGAAATCGCCCAACCAACTCTATTGTGTTCACCAAGCTCACACCATTCATGCTTGGAGCCTTGGTCG CCATGTATGAGCACAAGATCTTCGTTCAGGGCATCATCTGGGACATCAACAGCTTTGACCAGTGGGGAGTGGAGCTGGGAAAGCAGCTGG CTAAGAAAATAGAGCCTGAGCTTGATGGCAGTGCTCAAGTGACCTCTCACGACGCTTCTACCAATGGGCTCATCAACTTCATCAAGCAGC AGCGCGAGGCCAGAGTCCAATAAACTCGTGCTCATCTGCAGCCTCCTCTGTGACTCCCCTTTCTCTTCTCGTCCCTCCTCCCCGGAGCCG GCACTGCATGTTCCTGGACACCACCCAGAGCACCCTCTGGTTGTGGGCTTGGACCACGAGCCCTTAGCAGGGAAGGCTGGTCTCCCCCAG CCTAACCCCCAGCCCCTCCATGTCTATGCTCCCTCTGTGTTAGAATTGGCTGAAGTGTTTTTGTGCAGCTGACTTTTCTGACCCATGTTC ACGTTGTTCACATCCCATGTAGAAAAATAAAGATGCCACGGAGGAGGTTGTAGGCTCAGCCTCTGATTTTTTTTTTCCTGTGATGGTGCT TTATGTAGCAGAGGGCAGGAGCGCTCAGCAGGACGCAGGCTGTGCCTCTGCGGACACTTAACACTAAGTGGTGAGCGGGTCTAGAGTGGA GCAAGGTGCCCTGAGAAGACAATAGTGGGGTGGGGGCACAATCAGTCAGGACGGCAACTTGGCCTGTGTCACCAAATCCCAAGACTGTTT TCCACTCCTCACCTCTGTGACTGCAGAAATTGGATACTCTGTTCACTCGATGGTTCTAAAAACTGCATTGAGATTATGTTTGTTTCGGGT GAATTCCTGGACAAGACCGAGGATGACTGCCATCTCCTGGCAAGACGCTCAGGTAGTTCTTTTGCTTTAAAAGGCAGATATTGAAAACTG GAATTTTTTTTTTTTGAGTCTCGCTCTGTCACCCAGACTGGAGTGCAGTGGTGCAATCTCGGCTCACTGCAACCTCCGCCTCCCGGGTTC AAGCTATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGATTACACGGCGCACACCACCATACCCAGCTAATTTTTGTATTTTTAGTAGTGA AGGGGTTTTACCATGTTGGGCAGGCTGGTCTTGAACTCCTGACCTCAGGTGATCTGCCCGCCTCAGCCTCCCACAGTGCTGGGATTACAG GTATGAGCCACCACGCCCGGCCCATTTTTTTTTTTTTTTTGACAACTTTTTTTTTTTTTTGAGACAGGGTCTTGTTCCATTGCCCAGACT GGAGTGCAGTGGCATGATCACAGCTCACTGCAGCCAGTAATCCTCTTGCCTCAGCCTCCCAAGTAGTTGAGACTACAGGTTGTACCACTA TGCCCTGCTAGTTTTTTCATTTTTTGTAGAGAGACGGGTCTTTTTTTTTTTTGAGACGGAGTCTCGCTCTGTCGCCCAAGCTGGAGTGCA GTAGCACGGTCTCAGCTCATTGCAAGCTCCGCCTCCCAGGTTCACGCCATTCTCCTGCCTCAGACTCCTGTGTAGCTGGGAGTACAGGCA CCTGCCACCATGCCCGGCTAATTTTTTATATATTTTTTTAGCAGAGACAGTGTCTCACTGTGTTAGTCAGGATGGTCTCGATCTCCTGAC CTCGTGATCCGCCCGCCTCAGCCTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACCGCGCCCAGCAAGGCGGGTCTTGCTGTGTTTCCC AGACTAGACTGGTCTTGAATTCCAGGGCTCAAGAGATCTCCCACCTCAGCCTCCCACAGTGCTGGGATTACAGGCGTGAGCCGCCACACC CAGCCTATTCAAAATTTTTTTTTCTTAGAGACAGGGTCTTTGTTGCCCAGGCTGGACTGCAGTGATACAATCATAGCTGACTGAAGCCTC AAATTCCCAGGCTAAGGTGATCTTCTCACCTCAGCCTTCCAAGTAGCTGGGTCCGCAGATGCATGCCAGTACACCCAGCTCATTTAAAAA AAAATTTTTTTCTTTTTTGAGAGTCTTGCTTTGTTGCCCAGGCTGGAGTGCAGTGGTGTGATCTCGGCTCACTGCAAGCTCCACCTCCCG GCTTCACGCCATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGACTACAGGTGCCCGCCACCACACCCGGCTAATTTTTTGTATTTTTAGT AGAGACGGGGTTTCACCGTGTTAGCCAGGATGGTCTTGATCTCCTGACCTCATGATCCGCCTGCTTTGGCCTCCCAAAGTGCTGGGATTA >26432_26432_5_EML1-GPI_EML1_chr14_100259880_ENST00000334192_GPI_chr19_34884633_ENST00000415930_length(amino acids)=292AA_BP=22 MEDGFSSYSSLYDTSSLLQFCNGFDNFEQLLSGAHWMDQHFRTTPLEKNAPVLLALLGIWYINCFGCETHAMLPYDQYLHRFAAYFQQGD MESNGKYITKSGTRVDHQTGPIVWGEPGTNGQHAFYQLIHQGTKMIPCDFLIPVQTQHPIRKGLHHKILLANFLAQTEALMRGKSTEEAR KELQAAGKSPEDLERLLPHKVFEGNRPTNSIVFTKLTPFMLGALVAMYEHKIFVQGIIWDINSFDQWGVELGKQLAKKIEPELDGSAQVT -------------------------------------------------------------- >26432_26432_6_EML1-GPI_EML1_chr14_100259880_ENST00000334192_GPI_chr19_34884633_ENST00000586425_length(transcript)=1110nt_BP=201nt AACCGGCAGCAGCAGCCGCCACAGCAGGGCCGGCCCCAGCGCCAGCGCCGGTCGCGGTCGGGCTCAGCTCAGTGTGTGGTGAGCGGCGGC GGCGCGGCCGGGCCGGGGAGCGGGCGCGGCCCGGCGGCCTCAGCATGGAGGACGGCTTCTCCAGCTACAGCAGCCTGTACGACACGTCCT CGCTGCTCCAGTTCTGCAACGGTTTTGACAACTTCGAGCAGCTGCTCTCGGGGGCTCACTGGATGGACCAGCACTTCCGCACGACGCCCC TGGAGAAGAACGCCCCCGTCTTGCTGGCCCTGCTGGGTATCTGGTACATCAACTGCTTTGGGTGTGAGACACACGCCATGCTGCCCTATG ACCAGTACCTGCACCGCTTTGCTGCGTACTTCCAGCAGGGCGACATGGAGTCCAATGGGAAATACATCACCAAATCTGGAACCCGTGTGG ACCACCAGACAGGCCCCATTGTGTGGGGGGAGCCAGGGACCAATGGCCAGCATGCTTTTTACCAGCTCATCCACCAAGGCACCAAGATGA TACCCTGTGACTTCCTCATCCCGGTCCAGACCCAGCACCCCATACGGAAGGGTCTGCATCACAAGATCCTCCTGGCCAACTTCTTGGCCC AGACAGAGGCCCTGATGAGGGGAAAATCGACGGAGGAGGCCCGAAAGGAGCTCCAGGCTGCGGGCAAGAGTCCAGAGGACCTTGAGAGGC TGCTGCCACATAAGAGTGGAGCTGGGAAAGCAGCTGGCTAAGAAAATAGAGCCTGAGCTTGATGGCAGTGCTCAAGTGACCTCTCACGAC GCTTCTACCAATGGGCTCATCAACTTCATCAAGCAGCAGCGCGAGGCCAGAGTCCAATAAACTCGTGCTCATCTGCAGCCTCCTCTGTGA CTCCCCTTTCTCTTCTCGTCCCTCCTCCCCGGAGCCGGCACTGCATGTTCCTGGACACCACCCAGAGCACCCTCTGGTTGTGGGCTTGGA CCACGAGCCCTTAGCAGGGAAGGCTGGTCTCCCCCAGCCTAACCCCCAGCCCCTCCATGTCTATGCTCCCTCTGTGTTAGAATTGGCTGA >26432_26432_6_EML1-GPI_EML1_chr14_100259880_ENST00000334192_GPI_chr19_34884633_ENST00000586425_length(amino acids)=239AA_BP=1 MPSSSGSIFLASCFPSSTLMWQQPLKVLWTLARSLELLSGLLRRFSPHQGLCLGQEVGQEDLVMQTLPYGVLGLDRDEEVTGYHLGALVD ELVKSMLAIGPWLPPHNGACLVVHTGSRFGDVFPIGLHVALLEVRSKAVQVLVIGQHGVCLTPKAVDVPDTQQGQQDGGVLLQGRRAEVL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for EML1-GPI |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for EML1-GPI |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for EML1-GPI |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
