
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:AFF1-DHRS13 (FusionGDB2 ID:2652) |
Fusion Gene Summary for AFF1-DHRS13 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: AFF1-DHRS13 | Fusion gene ID: 2652 | Hgene | Tgene | Gene symbol | AFF1 | DHRS13 | Gene ID | 4299 | 147015 |
| Gene name | AF4/FMR2 family member 1 | dehydrogenase/reductase 13 | |
| Synonyms | AF4|MLLT2|PBM1 | SDR7C5 | |
| Cytomap | 4q21.3-q22.1 | 17q11.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | AF4/FMR2 family member 1ALL1-fused gene from chromosome 4 proteinmyeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 2pre-B-cell monocytic leukemia partner 1proto-oncogene AF4 | dehydrogenase/reductase SDR family member 13dehydrogenase/reductase (SDR family) member 13short chain dehydrogenase/reductase family 7C member 5 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P51825 | Q6UX07 | |
| Ensembl transtripts involved in fusion gene | ENST00000307808, ENST00000395146, ENST00000544085, ENST00000511996, | ENST00000581974, ENST00000378895, ENST00000394901, ENST00000426464, | |
| Fusion gene scores | * DoF score | 27 X 51 X 13=17901 | 1 X 1 X 1=1 |
| # samples | 68 | 1 | |
| ** MAII score | log2(68/17901*10)=-4.71836162613835 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(1/1*10)=3.32192809488736 | |
| Context | PubMed: AFF1 [Title/Abstract] AND DHRS13 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | AFF1(88048269)-DHRS13(27225910), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | AFF1-DHRS13 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. AFF1-DHRS13 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across AFF1 (5'-gene) Fusion gene breakpoints across AFF1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
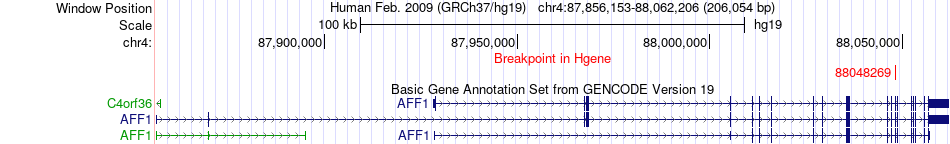 |
 Fusion gene breakpoints across DHRS13 (3'-gene) Fusion gene breakpoints across DHRS13 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
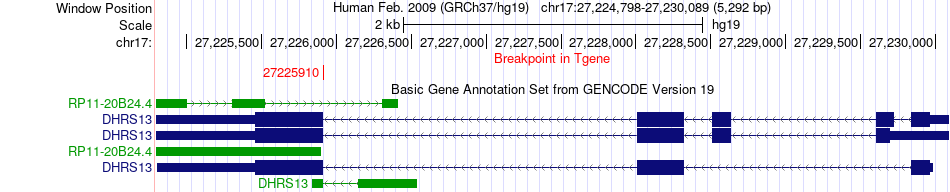 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-LN-A4A2 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - |
Top |
Fusion Gene ORF analysis for AFF1-DHRS13 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000307808 | ENST00000581974 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - |
| 5CDS-5UTR | ENST00000395146 | ENST00000581974 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - |
| 5CDS-5UTR | ENST00000544085 | ENST00000581974 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - |
| Frame-shift | ENST00000307808 | ENST00000378895 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - |
| Frame-shift | ENST00000307808 | ENST00000394901 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - |
| Frame-shift | ENST00000395146 | ENST00000378895 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - |
| Frame-shift | ENST00000395146 | ENST00000394901 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - |
| Frame-shift | ENST00000544085 | ENST00000378895 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - |
| Frame-shift | ENST00000544085 | ENST00000394901 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - |
| In-frame | ENST00000307808 | ENST00000426464 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - |
| In-frame | ENST00000395146 | ENST00000426464 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - |
| In-frame | ENST00000544085 | ENST00000426464 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - |
| intron-3CDS | ENST00000511996 | ENST00000378895 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - |
| intron-3CDS | ENST00000511996 | ENST00000394901 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - |
| intron-3CDS | ENST00000511996 | ENST00000426464 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - |
| intron-5UTR | ENST00000511996 | ENST00000581974 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000395146 | AFF1 | chr4 | 88048269 | + | ENST00000426464 | DHRS13 | chr17 | 27225910 | - | 4287 | 3178 | 266 | 3454 | 1062 |
| ENST00000307808 | AFF1 | chr4 | 88048269 | + | ENST00000426464 | DHRS13 | chr17 | 27225910 | - | 4411 | 3302 | 420 | 3578 | 1052 |
| ENST00000544085 | AFF1 | chr4 | 88048269 | + | ENST00000426464 | DHRS13 | chr17 | 27225910 | - | 3109 | 2000 | 132 | 2276 | 714 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000395146 | ENST00000426464 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - | 0.006082023 | 0.99391794 |
| ENST00000307808 | ENST00000426464 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - | 0.006499409 | 0.99350053 |
| ENST00000544085 | ENST00000426464 | AFF1 | chr4 | 88048269 | + | DHRS13 | chr17 | 27225910 | - | 0.024018932 | 0.97598106 |
Top |
Fusion Genomic Features for AFF1-DHRS13 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
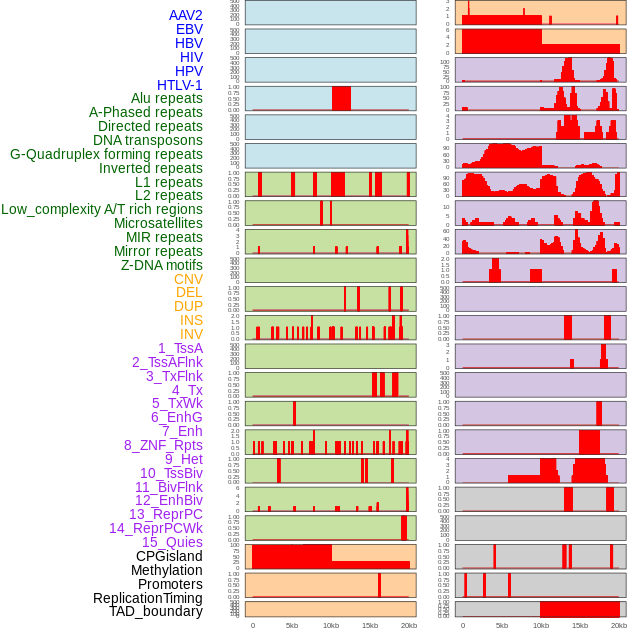 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for AFF1-DHRS13 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr4:88048269/chr17:27225910) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| AFF1 | DHRS13 |
| FUNCTION: Putative oxidoreductase. {ECO:0000305}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AFF1 | chr4:88048269 | chr17:27225910 | ENST00000307808 | + | 14 | 20 | 483_492 | 960 | 1211.0 | Compositional bias | Note=Poly-Ser |
| Hgene | AFF1 | chr4:88048269 | chr17:27225910 | ENST00000307808 | + | 14 | 20 | 835_843 | 960 | 1211.0 | Compositional bias | Note=Poly-Ser |
| Hgene | AFF1 | chr4:88048269 | chr17:27225910 | ENST00000307808 | + | 14 | 20 | 866_869 | 960 | 1211.0 | Compositional bias | Note=Poly-Pro |
| Hgene | AFF1 | chr4:88048269 | chr17:27225910 | ENST00000307808 | + | 14 | 20 | 871_874 | 960 | 1211.0 | Compositional bias | Note=Poly-Ser |
| Hgene | AFF1 | chr4:88048269 | chr17:27225910 | ENST00000395146 | + | 15 | 21 | 483_492 | 967 | 1219.0 | Compositional bias | Note=Poly-Ser |
| Hgene | AFF1 | chr4:88048269 | chr17:27225910 | ENST00000395146 | + | 15 | 21 | 835_843 | 967 | 1219.0 | Compositional bias | Note=Poly-Ser |
| Hgene | AFF1 | chr4:88048269 | chr17:27225910 | ENST00000395146 | + | 15 | 21 | 866_869 | 967 | 1219.0 | Compositional bias | Note=Poly-Pro |
| Hgene | AFF1 | chr4:88048269 | chr17:27225910 | ENST00000395146 | + | 15 | 21 | 871_874 | 967 | 1219.0 | Compositional bias | Note=Poly-Ser |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | DHRS13 | chr4:88048269 | chr17:27225910 | ENST00000378895 | 3 | 5 | 43_49 | 227 | 378.0 | Nucleotide binding | NAD or NADP | |
| Tgene | DHRS13 | chr4:88048269 | chr17:27225910 | ENST00000394901 | 2 | 4 | 43_49 | 177 | 328.0 | Nucleotide binding | NAD or NADP |
Top |
Fusion Gene Sequence for AFF1-DHRS13 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >2652_2652_1_AFF1-DHRS13_AFF1_chr4_88048269_ENST00000307808_DHRS13_chr17_27225910_ENST00000426464_length(transcript)=4411nt_BP=3302nt GGCAATTTCTTTTCCTTTCTAACTGTGGCCCGCGTTGTGCTGTTGCTGGGCAGGCGTTGGGCGCCGGCGGTCTTCGAGCGTGGGGGCCCG CTGGCTTTCCCTTCTCAGAAACTGCGCCGGGGGCGCTCGCTTGCCCCGGATTCGGACGCGGCGCTCCCCGGGCTCGTCTGAAGTGCAGAT CGCCGCAGAGGCCCCAGTGCCCGGATGTCCATCAGGATTAGCGCGAGCCAATACGGGCCGAGCCCGGGGCTGCGCCGAGGACGCCCGGGG CTCGAGAGCAGGTAGTCCCGTAACATCGGGGCGCCGCGCCGGGACGCGTCCCCGCCCGGCTCCGCCAAATGGTGAGCGCGGCGCTGGCAG CAGGGCCCGCGGGGTGAAGGCGCTCATGGACGGAAGACCCCTGGCTCTATAAGCTGAATTATGGCAGCCCAGTCAAGTTTGTACAATGAC GACAGAAACCTGCTTCGAATTAGAGAGAAGGAAAGACGCAACCAGGAAGCCCACCAAGAGAAAGAGGCATTTCCTGAAAAGATTCCCCTT TTTGGAGAGCCCTACAAGACAGCAAAAGGTGATGAGCTGTCTAGTCGAATACAGAACATGTTGGGAAACTACGAAGAAGTGAAGGAGTTC CTTAGTACTAAGTCTCACACTCATCGCCTGGATGCTTCTGAAAATAGGTTGGGAAAGCCGAAATATCCTTTAATTCCTGACAAAGGGAGC AGCATTCCATCCAGCTCCTTCCACACTAGTGTCCACCACCAGTCCATTCACACTCCTGCGTCTGGACCACTTTCTGTTGGCAACATTAGC CACAATCCAAAGATGGCGCAGCCAAGAACTGAACCAATGCCAAGTCTCCATGCCAAAAGCTGCGGCCCACCGGACAGCCAGCACCTGACC CAGGATCGCCTTGGTCAGGAGGGGTTCGGCTCTAGTCATCACAAGAAAGGTGACCGAAGAGCTGACGGAGACCACTGTGCTTCGGTGACA GATTCGGCTCCAGAGAGGGAGCTTTCTCCCTTAATCTCTTTGCCTTCCCCAGTTCCCCCTTTGTCACCTATACATTCCAACCAGCAAACT CTTCCCCGGACGCAAGGAAGCAGCAAGGTTCATGGCAGCAGCAATAACAGTAAAGGCTATTGCCCAGCCAAATCTCCCAAGGACCTAGCA GTGAAAGTCCATGATAAAGAGACCCCTCAAGACAGTTTGGTGGCCCCTGCCCAGCCGCCTTCTCAGACATTTCCACCTCCCTCCCTCCCC TCAAAAAGTGTTGCAATGCAGCAGAAGCCCACGGCTTATGTCCGGCCCATGGATGGTCAAGATCAGGCCCCTAGTGAATCCCCTGAACTG AAACCACTGCCGGAGGACTATCGACAGCAGACCTTTGAAAAAACAGACTTGAAAGTGCCTGCCAAAGCCAAGCTCACCAAACTGAAGATG CCTTCTCAGTCAGTTGAGCAGACCTACTCCAATGAAGTCCATTGTGTTGAAGAGATTCTGAAGGAAATGACCCATTCATGGCCGCCTCCT TTGACAGCAATACATACGCCTAGTACAGCTGAGCCATCCAAGTTTCCTTTCCCTACAAAGGACTCTCAGCATGTCAGTTCTGTAACCCAA AACCAAAAACAATATGATACATCTTCAAAAACTCACTCAAATTCTCAGCAAGGAACGTCATCCATGCTCGAAGACGACCTTCAGCTCAGT GACAGTGAGGACAGTGACAGTGAACAAACCCCAGAGAAGCCTCCCTCCTCATCTGCACCTCCAAGTGCTCCACAGTCCCTTCCAGAACCA GTGGCATCAGCACATTCCAGCAGTGCAGAGTCAGAAAGCACCAGTGACTCAGACAGTTCCTCAGACTCAGAGAGCGAGAGCAGTTCAAGT GACAGCGAAGAAAATGAGCCCCTAGAAACCCCAGCTCCGGAGCCTGAGCCTCCAACAACAAACAAATGGCAGCTGGACAACTGGCTGACC AAAGTCAGCCAGCCAGCTGCGCCACCAGAGGGCCCCAGGAGCACAGAGCCCCCACGGCGGCACCCAGAGAGTAAGGGCAGCAGCGACAGT GCCACGAGTCAGGAGCATTCTGAATCCAAAGATCCTCCCCCTAAAAGCTCCAGCAAAGCCCCCCGGGCCCCACCCGAAGCCCCCCACCCC GGAAAGAGGAGCTGTCAGAAGTCTCCGGCACAGCAGGAGCCCCCACAAAGGCAAACCGTTGGAACCAAACAACCCAAAAAACCTGTCAAG GCCTCTGCCCGGGCAGGTTCACGGACCAGCCTGCAGGGGGAAAGGGAGCCAGGGCTTCTTCCCTATGGCTCCCGAGACCAGACTTCCAAA GACAAGCCCAAGGTGAAGACGAAAGGACGGCCCCGGGCCGCAGCAAGCAACGAACCCAAGCCAGCAGTGCCCCCCTCCAGTGAGAAGAAG AAGCACAAGAGCTCCCTCCCTGCCCCCTCTAAGGCTCTCTCAGGCCCAGAACCCGCGAAGGACAATGTGGAGGACAGGACCCCTGAGCAC TTTGCTCTTGTTCCCCTGACTGAGAGCCAGGGCCCACCCCACAGTGGCAGCGGCAGCAGGACTAGTGGCTGCCGCCAAGCCGTGGTGGTC CAGGAGGACAGCCGCAAAGACAGACTCCCATTGCCTTTGAGAGACACCAAGCTGCTCTCACCGCTCAGGGACACTCCTCCCCCACAAAGC TTGATGGTGAAGATCACCCTAGACCTGCTCTCTCGGATACCCCAGCCTCCCGGGAAGGGGAGCCGCCAGAGGAAAGCAGAAGATAAACAG CCGCCCGCAGGGAAGAAGCACAGCTCTGAGAAGAGGAGCTCAGACAGCTCAAGCAAGTTGGCCAAAAAGAGAAAGGGTGAAGCAGAAAGA GACTGTGATAACAAGAAAATCAGACTGGAGAAGGAAATCAAATCACAGTCATCTTCATCTTCATCCTCCCACAAAGAATCTTCTAAAACA AAGCCCTCCAGGCCCTCCTCACAGTCCTCAAAGAAGGAAATGCTCCCCCCGCCACCCGTGTCCTCGTCCTCCCAGAAGCCAGCCAAGCCT GCACTTAAGAGGTCAAGGCGGGAAGCAGACACCTGTGGCCAGGACCCTCCCAAAAGTGCCAGCAGTACCAAGAGCAACCACAAAGACTCT TCCATTCCCAAGCAGAGAAGAGTAGAGGGGAAGGGCTCCAGAAGCTCCTCGGAGCACAAGGGTTCTTCCGGAGATACTGCAAATCCTTTT CCAGTGCCTTCTTTGCCAAATGGTAACTCTAAACCAGGGAAGCCTCAAGTGAAGTTTGACAAGGCCTGTGAACTCGGAGCTGTTCCTGCG CCATGTTCCTGGATGGCTGCGCCCACTTTTGCGCCCATTGGCTTGGCTGGTGCTCCGGGCACCAAGAGGGGGTGCCCAGACACCCCTGTA TTGTGCTCTACAAGAGGGCATCGAGCCCCTCAGTGGGAGATATTTTGCCAACTGCCATGTGGAAGAGGTGCCTCCAGCTGCCCGAGACGA CCGGGCAGCCCATCGGCTATGGGAGGCCAGCAAGAGGCTGGCAGGGCTTGGGCCTGGGGAGGATGCTGAACCCGATGAAGACCCCCAGTC TGAGGACTCAGAGGCCCCATCTTCTCTAAGCACCCCCCACCCTGAGGAGCCCACAGTTTCTCAACCTTACCCCAGCCCTCAGAGCTCACC AGATTTGTCTAAGATGACGCACCGAATTCAGGCTAAAGTTGAGCCTGAGATCCAGCTCTCCTAACCCTCAGGCCAGGATGCTTGCCATGG CACTTCATGGTCCTTGAAAACCTCGGATGTGTGCGAGGCCATGCCCTGGACACTGACGGGTTTGTGATCTTGACCTCCGTGGTTACTTTC TGGGGCCCCAAGCTGTGCCCTGGACATCTCTTTTCCTGGTTGAAGGAATAATGGGTGATTATTTCTTCCTGAGAGTGACAGTAACCCCAG ATGGAGAGATAGGGGTATGCTAGACACTGTGCTTCTCGGAAATTTGGATGTAGTATTTTCAGGCCCCACCCTTATTGATTCTGATCAGCT CTGGAGCAGAGGCAGGGAGTTTGCAATGTGATGCACTGCCAACATTGAGAATTAGTGAACTGATCCCTTTGCAACCGTCTAGCTAGGTAG TTAAATTACCCCCATGTTAATGAAGCGGAATTAGGCTCCCGAGCTAAGGGACTCGCCTAGGGTCTCACAGTGAGTAGGAGGAGGGCCTGG GATCTGAACCCAAGGGTCTGAGGCCAGGGCCGACTGCCGTAAGATGGGTGCTGAGAAGTGAGTCAGGGCAGGGCAGCTGGTATCGAGGTG CCCCATGGGAGTAAGGGGACGCCTTCCGGGCGGATGCAGGGCTGGGGTCATCTGTATCTGAAGCCCCTCGGAATAAAGCGCGTTGACCGC >2652_2652_1_AFF1-DHRS13_AFF1_chr4_88048269_ENST00000307808_DHRS13_chr17_27225910_ENST00000426464_length(amino acids)=1052AA_BP=444 MAAQSSLYNDDRNLLRIREKERRNQEAHQEKEAFPEKIPLFGEPYKTAKGDELSSRIQNMLGNYEEVKEFLSTKSHTHRLDASENRLGKP KYPLIPDKGSSIPSSSFHTSVHHQSIHTPASGPLSVGNISHNPKMAQPRTEPMPSLHAKSCGPPDSQHLTQDRLGQEGFGSSHHKKGDRR ADGDHCASVTDSAPERELSPLISLPSPVPPLSPIHSNQQTLPRTQGSSKVHGSSNNSKGYCPAKSPKDLAVKVHDKETPQDSLVAPAQPP SQTFPPPSLPSKSVAMQQKPTAYVRPMDGQDQAPSESPELKPLPEDYRQQTFEKTDLKVPAKAKLTKLKMPSQSVEQTYSNEVHCVEEIL KEMTHSWPPPLTAIHTPSTAEPSKFPFPTKDSQHVSSVTQNQKQYDTSSKTHSNSQQGTSSMLEDDLQLSDSEDSDSEQTPEKPPSSSAP PSAPQSLPEPVASAHSSSAESESTSDSDSSSDSESESSSSDSEENEPLETPAPEPEPPTTNKWQLDNWLTKVSQPAAPPEGPRSTEPPRR HPESKGSSDSATSQEHSESKDPPPKSSSKAPRAPPEAPHPGKRSCQKSPAQQEPPQRQTVGTKQPKKPVKASARAGSRTSLQGEREPGLL PYGSRDQTSKDKPKVKTKGRPRAAASNEPKPAVPPSSEKKKHKSSLPAPSKALSGPEPAKDNVEDRTPEHFALVPLTESQGPPHSGSGSR TSGCRQAVVVQEDSRKDRLPLPLRDTKLLSPLRDTPPPQSLMVKITLDLLSRIPQPPGKGSRQRKAEDKQPPAGKKHSSEKRSSDSSSKL AKKRKGEAERDCDNKKIRLEKEIKSQSSSSSSSHKESSKTKPSRPSSQSSKKEMLPPPPVSSSSQKPAKPALKRSRREADTCGQDPPKSA SSTKSNHKDSSIPKQRRVEGKGSRSSSEHKGSSGDTANPFPVPSLPNGNSKPGKPQVKFDKACELGAVPAPCSWMAAPTFAPIGLAGAPG -------------------------------------------------------------- >2652_2652_2_AFF1-DHRS13_AFF1_chr4_88048269_ENST00000395146_DHRS13_chr17_27225910_ENST00000426464_length(transcript)=4287nt_BP=3178nt GAGCTTGCCAGCCAGCTAGCGAGCGACGGGCGCGCGCGGCCCCTGCGCACTCGGCCGCCGCCTGCGCGCGTTGCGGCCGCAGCTGCACCT TTGCCTCCGCGGTTCCGCAGCCGAGCCCCGGGAGGAGGCGCTCCCCTGCGTCGTCCGCACTTGCGCCCTTTCCCACCGCAACAGGTCAGG GGAGGGCGGAGGTGACGAGGGCGCCCGACCCGGGACTAGAGGACGGAAGGGTCTGGAAGATGAACAGACTAGCCACTTTGCATTGACTGG AAACAATGGCATTTACAGAAAGAGTCAACAGCAGTGGCAACAGTTTGTACAATGACGACAGAAACCTGCTTCGAATTAGAGAGAAGGAAA GACGCAACCAGGAAGCCCACCAAGAGAAAGAGGCATTTCCTGAAAAGATTCCCCTTTTTGGAGAGCCCTACAAGACAGCAAAAGGTGATG AGCTGTCTAGTCGAATACAGAACATGTTGGGAAACTACGAAGAAGTGAAGGAGTTCCTTAGTACTAAGTCTCACACTCATCGCCTGGATG CTTCTGAAAATAGGTTGGGAAAGCCGAAATATCCTTTAATTCCTGACAAAGGGAGCAGCATTCCATCCAGCTCCTTCCACACTAGTGTCC ACCACCAGTCCATTCACACTCCTGCGTCTGGACCACTTTCTGTTGGCAACATTAGCCACAATCCAAAGATGGCGCAGCCAAGAACTGAAC CAATGCCAAGTCTCCATGCCAAAAGCTGCGGCCCACCGGACAGCCAGCACCTGACCCAGGATCGCCTTGGTCAGGAGGGGTTCGGCTCTA GTCATCACAAGAAAGGTGACCGAAGAGCTGACGGAGACCACTGTGCTTCGGTGACAGATTCGGCTCCAGAGAGGGAGCTTTCTCCCTTAA TCTCTTTGCCTTCCCCAGTTCCCCCTTTGTCACCTATACATTCCAACCAGCAAACTCTTCCCCGGACGCAAGGAAGCAGCAAGGTTCATG GCAGCAGCAATAACAGTAAAGGCTATTGCCCAGCCAAATCTCCCAAGGACCTAGCAGTGAAAGTCCATGATAAAGAGACCCCTCAAGACA GTTTGGTGGCCCCTGCCCAGCCGCCTTCTCAGACATTTCCACCTCCCTCCCTCCCCTCAAAAAGTGTTGCAATGCAGCAGAAGCCCACGG CTTATGTCCGGCCCATGGATGGTCAAGATCAGGCCCCTAGTGAATCCCCTGAACTGAAACCACTGCCGGAGGACTATCGACAGCAGACCT TTGAAAAAACAGACTTGAAAGTGCCTGCCAAAGCCAAGCTCACCAAACTGAAGATGCCTTCTCAGTCAGTTGAGCAGACCTACTCCAATG AAGTCCATTGTGTTGAAGAGATTCTGAAGGAAATGACCCATTCATGGCCGCCTCCTTTGACAGCAATACATACGCCTAGTACAGCTGAGC CATCCAAGTTTCCTTTCCCTACAAAGGACTCTCAGCATGTCAGTTCTGTAACCCAAAACCAAAAACAATATGATACATCTTCAAAAACTC ACTCAAATTCTCAGCAAGGAACGTCATCCATGCTCGAAGACGACCTTCAGCTCAGTGACAGTGAGGACAGTGACAGTGAACAAACCCCAG AGAAGCCTCCCTCCTCATCTGCACCTCCAAGTGCTCCACAGTCCCTTCCAGAACCAGTGGCATCAGCACATTCCAGCAGTGCAGAGTCAG AAAGCACCAGTGACTCAGACAGTTCCTCAGACTCAGAGAGCGAGAGCAGTTCAAGTGACAGCGAAGAAAATGAGCCCCTAGAAACCCCAG CTCCGGAGCCTGAGCCTCCAACAACAAACAAATGGCAGCTGGACAACTGGCTGACCAAAGTCAGCCAGCCAGCTGCGCCACCAGAGGGCC CCAGGAGCACAGAGCCCCCACGGCGGCACCCAGAGAGTAAGGGCAGCAGCGACAGTGCCACGAGTCAGGAGCATTCTGAATCCAAAGATC CTCCCCCTAAAAGCTCCAGCAAAGCCCCCCGGGCCCCACCCGAAGCCCCCCACCCCGGAAAGAGGAGCTGTCAGAAGTCTCCGGCACAGC AGGAGCCCCCACAAAGGCAAACCGTTGGAACCAAACAACCCAAAAAACCTGTCAAGGCCTCTGCCCGGGCAGGTTCACGGACCAGCCTGC AGGGGGAAAGGGAGCCAGGGCTTCTTCCCTATGGCTCCCGAGACCAGACTTCCAAAGACAAGCCCAAGGTGAAGACGAAAGGACGGCCCC GGGCCGCAGCAAGCAACGAACCCAAGCCAGCAGTGCCCCCCTCCAGTGAGAAGAAGAAGCACAAGAGCTCCCTCCCTGCCCCCTCTAAGG CTCTCTCAGGCCCAGAACCCGCGAAGGACAATGTGGAGGACAGGACCCCTGAGCACTTTGCTCTTGTTCCCCTGACTGAGAGCCAGGGCC CACCCCACAGTGGCAGCGGCAGCAGGACTAGTGGCTGCCGCCAAGCCGTGGTGGTCCAGGAGGACAGCCGCAAAGACAGACTCCCATTGC CTTTGAGAGACACCAAGCTGCTCTCACCGCTCAGGGACACTCCTCCCCCACAAAGCTTGATGGTGAAGATCACCCTAGACCTGCTCTCTC GGATACCCCAGCCTCCCGGGAAGGGGAGCCGCCAGAGGAAAGCAGAAGATAAACAGCCGCCCGCAGGGAAGAAGCACAGCTCTGAGAAGA GGAGCTCAGACAGCTCAAGCAAGTTGGCCAAAAAGAGAAAGGGTGAAGCAGAAAGAGACTGTGATAACAAGAAAATCAGACTGGAGAAGG AAATCAAATCACAGTCATCTTCATCTTCATCCTCCCACAAAGAATCTTCTAAAACAAAGCCCTCCAGGCCCTCCTCACAGTCCTCAAAGA AGGAAATGCTCCCCCCGCCACCCGTGTCCTCGTCCTCCCAGAAGCCAGCCAAGCCTGCACTTAAGAGGTCAAGGCGGGAAGCAGACACCT GTGGCCAGGACCCTCCCAAAAGTGCCAGCAGTACCAAGAGCAACCACAAAGACTCTTCCATTCCCAAGCAGAGAAGAGTAGAGGGGAAGG GCTCCAGAAGCTCCTCGGAGCACAAGGGTTCTTCCGGAGATACTGCAAATCCTTTTCCAGTGCCTTCTTTGCCAAATGGTAACTCTAAAC CAGGGAAGCCTCAAGTGAAGTTTGACAAGGCCTGTGAACTCGGAGCTGTTCCTGCGCCATGTTCCTGGATGGCTGCGCCCACTTTTGCGC CCATTGGCTTGGCTGGTGCTCCGGGCACCAAGAGGGGGTGCCCAGACACCCCTGTATTGTGCTCTACAAGAGGGCATCGAGCCCCTCAGT GGGAGATATTTTGCCAACTGCCATGTGGAAGAGGTGCCTCCAGCTGCCCGAGACGACCGGGCAGCCCATCGGCTATGGGAGGCCAGCAAG AGGCTGGCAGGGCTTGGGCCTGGGGAGGATGCTGAACCCGATGAAGACCCCCAGTCTGAGGACTCAGAGGCCCCATCTTCTCTAAGCACC CCCCACCCTGAGGAGCCCACAGTTTCTCAACCTTACCCCAGCCCTCAGAGCTCACCAGATTTGTCTAAGATGACGCACCGAATTCAGGCT AAAGTTGAGCCTGAGATCCAGCTCTCCTAACCCTCAGGCCAGGATGCTTGCCATGGCACTTCATGGTCCTTGAAAACCTCGGATGTGTGC GAGGCCATGCCCTGGACACTGACGGGTTTGTGATCTTGACCTCCGTGGTTACTTTCTGGGGCCCCAAGCTGTGCCCTGGACATCTCTTTT CCTGGTTGAAGGAATAATGGGTGATTATTTCTTCCTGAGAGTGACAGTAACCCCAGATGGAGAGATAGGGGTATGCTAGACACTGTGCTT CTCGGAAATTTGGATGTAGTATTTTCAGGCCCCACCCTTATTGATTCTGATCAGCTCTGGAGCAGAGGCAGGGAGTTTGCAATGTGATGC ACTGCCAACATTGAGAATTAGTGAACTGATCCCTTTGCAACCGTCTAGCTAGGTAGTTAAATTACCCCCATGTTAATGAAGCGGAATTAG GCTCCCGAGCTAAGGGACTCGCCTAGGGTCTCACAGTGAGTAGGAGGAGGGCCTGGGATCTGAACCCAAGGGTCTGAGGCCAGGGCCGAC TGCCGTAAGATGGGTGCTGAGAAGTGAGTCAGGGCAGGGCAGCTGGTATCGAGGTGCCCCATGGGAGTAAGGGGACGCCTTCCGGGCGGA >2652_2652_2_AFF1-DHRS13_AFF1_chr4_88048269_ENST00000395146_DHRS13_chr17_27225910_ENST00000426464_length(amino acids)=1062AA_BP=454 METMAFTERVNSSGNSLYNDDRNLLRIREKERRNQEAHQEKEAFPEKIPLFGEPYKTAKGDELSSRIQNMLGNYEEVKEFLSTKSHTHRL DASENRLGKPKYPLIPDKGSSIPSSSFHTSVHHQSIHTPASGPLSVGNISHNPKMAQPRTEPMPSLHAKSCGPPDSQHLTQDRLGQEGFG SSHHKKGDRRADGDHCASVTDSAPERELSPLISLPSPVPPLSPIHSNQQTLPRTQGSSKVHGSSNNSKGYCPAKSPKDLAVKVHDKETPQ DSLVAPAQPPSQTFPPPSLPSKSVAMQQKPTAYVRPMDGQDQAPSESPELKPLPEDYRQQTFEKTDLKVPAKAKLTKLKMPSQSVEQTYS NEVHCVEEILKEMTHSWPPPLTAIHTPSTAEPSKFPFPTKDSQHVSSVTQNQKQYDTSSKTHSNSQQGTSSMLEDDLQLSDSEDSDSEQT PEKPPSSSAPPSAPQSLPEPVASAHSSSAESESTSDSDSSSDSESESSSSDSEENEPLETPAPEPEPPTTNKWQLDNWLTKVSQPAAPPE GPRSTEPPRRHPESKGSSDSATSQEHSESKDPPPKSSSKAPRAPPEAPHPGKRSCQKSPAQQEPPQRQTVGTKQPKKPVKASARAGSRTS LQGEREPGLLPYGSRDQTSKDKPKVKTKGRPRAAASNEPKPAVPPSSEKKKHKSSLPAPSKALSGPEPAKDNVEDRTPEHFALVPLTESQ GPPHSGSGSRTSGCRQAVVVQEDSRKDRLPLPLRDTKLLSPLRDTPPPQSLMVKITLDLLSRIPQPPGKGSRQRKAEDKQPPAGKKHSSE KRSSDSSSKLAKKRKGEAERDCDNKKIRLEKEIKSQSSSSSSSHKESSKTKPSRPSSQSSKKEMLPPPPVSSSSQKPAKPALKRSRREAD TCGQDPPKSASSTKSNHKDSSIPKQRRVEGKGSRSSSEHKGSSGDTANPFPVPSLPNGNSKPGKPQVKFDKACELGAVPAPCSWMAAPTF -------------------------------------------------------------- >2652_2652_3_AFF1-DHRS13_AFF1_chr4_88048269_ENST00000544085_DHRS13_chr17_27225910_ENST00000426464_length(transcript)=3109nt_BP=2000nt GTAGTCCCGTAACATCGGGGCGCCGCGCCGGGACGCGTCCCCGCCCGGCTCCGCCAAATGGTGAGCGCGGCGCTGGCAGCAGGGCCCGCG GGGTGAAGGCGCTCATGGACGGAAGACCCCTGGCTCTATAAGCTGAATTATGGCAGCCCAGTCAAGCAGACCTACTCCAATGAAGTCCAT TGTGTTGAAGAGATTCTGAAGGAAATGACCCATTCATGGCCGCCTCCTTTGACAGCAATACATACGCCTAGTACAGCTGAGCCATCCAAG TTTCCTTTCCCTACAAAGGACTCTCAGCATGTCAGTTCTGTAACCCAAAACCAAAAACAATATGATACATCTTCAAAAACTCACTCAAAT TCTCAGCAAGGAACGTCATCCATGCTCGAAGACGACCTTCAGCTCAGTGACAGTGAGGACAGTGACAGTGAACAAACCCCAGAGAAGCCT CCCTCCTCATCTGCACCTCCAAGTGCTCCACAGTCCCTTCCAGAACCAGTGGCATCAGCACATTCCAGCAGTGCAGAGTCAGAAAGCACC AGTGACTCAGACAGTTCCTCAGACTCAGAGAGCGAGAGCAGTTCAAGTGACAGCGAAGAAAATGAGCCCCTAGAAACCCCAGCTCCGGAG CCTGAGCCTCCAACAACAAACAAATGGCAGCTGGACAACTGGCTGACCAAAGTCAGCCAGCCAGCTGCGCCACCAGAGGGCCCCAGGAGC ACAGAGCCCCCACGGCGGCACCCAGAGAGTAAGGGCAGCAGCGACAGTGCCACGAGTCAGGAGCATTCTGAATCCAAAGATCCTCCCCCT AAAAGCTCCAGCAAAGCCCCCCGGGCCCCACCCGAAGCCCCCCACCCCGGAAAGAGGAGCTGTCAGAAGTCTCCGGCACAGCAGGAGCCC CCACAAAGGCAAACCGTTGGAACCAAACAACCCAAAAAACCTGTCAAGGCCTCTGCCCGGGCAGGTTCACGGACCAGCCTGCAGGGGGAA AGGGAGCCAGGGCTTCTTCCCTATGGCTCCCGAGACCAGACTTCCAAAGACAAGCCCAAGGTGAAGACGAAAGGACGGCCCCGGGCCGCA GCAAGCAACGAACCCAAGCCAGCAGTGCCCCCCTCCAGTGAGAAGAAGAAGCACAAGAGCTCCCTCCCTGCCCCCTCTAAGGCTCTCTCA GGCCCAGAACCCGCGAAGGACAATGTGGAGGACAGGACCCCTGAGCACTTTGCTCTTGTTCCCCTGACTGAGAGCCAGGGCCCACCCCAC AGTGGCAGCGGCAGCAGGACTAGTGGCTGCCGCCAAGCCGTGGTGGTCCAGGAGGACAGCCGCAAAGACAGACTCCCATTGCCTTTGAGA GACACCAAGCTGCTCTCACCGCTCAGGGACACTCCTCCCCCACAAAGCTTGATGGTGAAGATCACCCTAGACCTGCTCTCTCGGATACCC CAGCCTCCCGGGAAGGGGAGCCGCCAGAGGAAAGCAGAAGATAAACAGCCGCCCGCAGGGAAGAAGCACAGCTCTGAGAAGAGGAGCTCA GACAGCTCAAGCAAGTTGGCCAAAAAGAGAAAGGGTGAAGCAGAAAGAGACTGTGATAACAAGAAAATCAGACTGGAGAAGGAAATCAAA TCACAGTCATCTTCATCTTCATCCTCCCACAAAGAATCTTCTAAAACAAAGCCCTCCAGGCCCTCCTCACAGTCCTCAAAGAAGGAAATG CTCCCCCCGCCACCCGTGTCCTCGTCCTCCCAGAAGCCAGCCAAGCCTGCACTTAAGAGGTCAAGGCGGGAAGCAGACACCTGTGGCCAG GACCCTCCCAAAAGTGCCAGCAGTACCAAGAGCAACCACAAAGACTCTTCCATTCCCAAGCAGAGAAGAGTAGAGGGGAAGGGCTCCAGA AGCTCCTCGGAGCACAAGGGTTCTTCCGGAGATACTGCAAATCCTTTTCCAGTGCCTTCTTTGCCAAATGGTAACTCTAAACCAGGGAAG CCTCAAGTGAAGTTTGACAAGGCCTGTGAACTCGGAGCTGTTCCTGCGCCATGTTCCTGGATGGCTGCGCCCACTTTTGCGCCCATTGGC TTGGCTGGTGCTCCGGGCACCAAGAGGGGGTGCCCAGACACCCCTGTATTGTGCTCTACAAGAGGGCATCGAGCCCCTCAGTGGGAGATA TTTTGCCAACTGCCATGTGGAAGAGGTGCCTCCAGCTGCCCGAGACGACCGGGCAGCCCATCGGCTATGGGAGGCCAGCAAGAGGCTGGC AGGGCTTGGGCCTGGGGAGGATGCTGAACCCGATGAAGACCCCCAGTCTGAGGACTCAGAGGCCCCATCTTCTCTAAGCACCCCCCACCC TGAGGAGCCCACAGTTTCTCAACCTTACCCCAGCCCTCAGAGCTCACCAGATTTGTCTAAGATGACGCACCGAATTCAGGCTAAAGTTGA GCCTGAGATCCAGCTCTCCTAACCCTCAGGCCAGGATGCTTGCCATGGCACTTCATGGTCCTTGAAAACCTCGGATGTGTGCGAGGCCAT GCCCTGGACACTGACGGGTTTGTGATCTTGACCTCCGTGGTTACTTTCTGGGGCCCCAAGCTGTGCCCTGGACATCTCTTTTCCTGGTTG AAGGAATAATGGGTGATTATTTCTTCCTGAGAGTGACAGTAACCCCAGATGGAGAGATAGGGGTATGCTAGACACTGTGCTTCTCGGAAA TTTGGATGTAGTATTTTCAGGCCCCACCCTTATTGATTCTGATCAGCTCTGGAGCAGAGGCAGGGAGTTTGCAATGTGATGCACTGCCAA CATTGAGAATTAGTGAACTGATCCCTTTGCAACCGTCTAGCTAGGTAGTTAAATTACCCCCATGTTAATGAAGCGGAATTAGGCTCCCGA GCTAAGGGACTCGCCTAGGGTCTCACAGTGAGTAGGAGGAGGGCCTGGGATCTGAACCCAAGGGTCTGAGGCCAGGGCCGACTGCCGTAA GATGGGTGCTGAGAAGTGAGTCAGGGCAGGGCAGCTGGTATCGAGGTGCCCCATGGGAGTAAGGGGACGCCTTCCGGGCGGATGCAGGGC >2652_2652_3_AFF1-DHRS13_AFF1_chr4_88048269_ENST00000544085_DHRS13_chr17_27225910_ENST00000426464_length(amino acids)=714AA_BP=106 MNYGSPVKQTYSNEVHCVEEILKEMTHSWPPPLTAIHTPSTAEPSKFPFPTKDSQHVSSVTQNQKQYDTSSKTHSNSQQGTSSMLEDDLQ LSDSEDSDSEQTPEKPPSSSAPPSAPQSLPEPVASAHSSSAESESTSDSDSSSDSESESSSSDSEENEPLETPAPEPEPPTTNKWQLDNW LTKVSQPAAPPEGPRSTEPPRRHPESKGSSDSATSQEHSESKDPPPKSSSKAPRAPPEAPHPGKRSCQKSPAQQEPPQRQTVGTKQPKKP VKASARAGSRTSLQGEREPGLLPYGSRDQTSKDKPKVKTKGRPRAAASNEPKPAVPPSSEKKKHKSSLPAPSKALSGPEPAKDNVEDRTP EHFALVPLTESQGPPHSGSGSRTSGCRQAVVVQEDSRKDRLPLPLRDTKLLSPLRDTPPPQSLMVKITLDLLSRIPQPPGKGSRQRKAED KQPPAGKKHSSEKRSSDSSSKLAKKRKGEAERDCDNKKIRLEKEIKSQSSSSSSSHKESSKTKPSRPSSQSSKKEMLPPPPVSSSSQKPA KPALKRSRREADTCGQDPPKSASSTKSNHKDSSIPKQRRVEGKGSRSSSEHKGSSGDTANPFPVPSLPNGNSKPGKPQVKFDKACELGAV -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for AFF1-DHRS13 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for AFF1-DHRS13 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for AFF1-DHRS13 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
