
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ENSA-FBP1 (FusionGDB2 ID:26664) |
Fusion Gene Summary for ENSA-FBP1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ENSA-FBP1 | Fusion gene ID: 26664 | Hgene | Tgene | Gene symbol | ENSA | FBP1 | Gene ID | 2029 | 2203 |
| Gene name | endosulfine alpha | fructose-bisphosphatase 1 | |
| Synonyms | ARPP-19e | FBP | |
| Cytomap | 1q21.3 | 9q22.32 | |
| Type of gene | protein-coding | protein-coding | |
| Description | alpha-endosulfine | fructose-1,6-bisphosphatase 1D-fructose-1,6-bisphosphate 1-phosphohydrolase 1FBPase 1growth-inhibiting protein 17liver FBPase | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | P09467 | |
| Ensembl transtripts involved in fusion gene | ENST00000339643, ENST00000361532, ENST00000361631, ENST00000369009, ENST00000369014, ENST00000369016, ENST00000271690, ENST00000354702, ENST00000356527, ENST00000362052, ENST00000503241, ENST00000503345, ENST00000513281, | ENST00000375326, ENST00000415431, | |
| Fusion gene scores | * DoF score | 13 X 11 X 6=858 | 3 X 1 X 2=6 |
| # samples | 17 | 4 | |
| ** MAII score | log2(17/858*10)=-2.33544290136184 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/6*10)=2.73696559416621 | |
| Context | PubMed: ENSA [Title/Abstract] AND FBP1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ENSA(150598117)-FBP1(97382773), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ENSA | GO:0050796 | regulation of insulin secretion | 9653196 |
| Tgene | FBP1 | GO:0006002 | fructose 6-phosphate metabolic process | 8387495|16442285 |
| Tgene | FBP1 | GO:0016311 | dephosphorylation | 8387495|18650089 |
| Tgene | FBP1 | GO:0030308 | negative regulation of cell growth | 19881551 |
| Tgene | FBP1 | GO:0035690 | cellular response to drug | 16442285|18650089 |
| Tgene | FBP1 | GO:0045820 | negative regulation of glycolytic process | 19881551 |
| Tgene | FBP1 | GO:0046580 | negative regulation of Ras protein signal transduction | 19881551 |
| Tgene | FBP1 | GO:0071286 | cellular response to magnesium ion | 10222032 |
 Fusion gene breakpoints across ENSA (5'-gene) Fusion gene breakpoints across ENSA (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
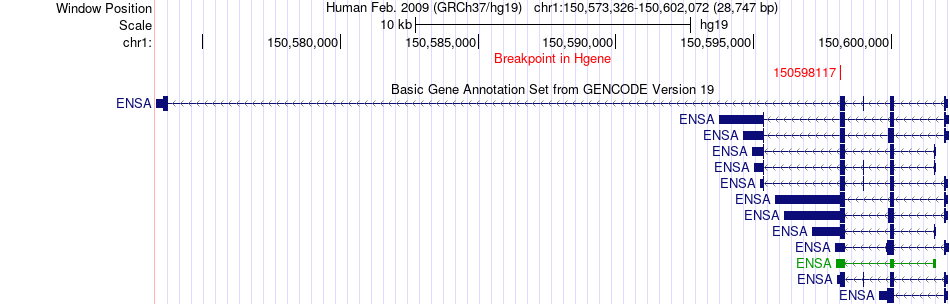 |
 Fusion gene breakpoints across FBP1 (3'-gene) Fusion gene breakpoints across FBP1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
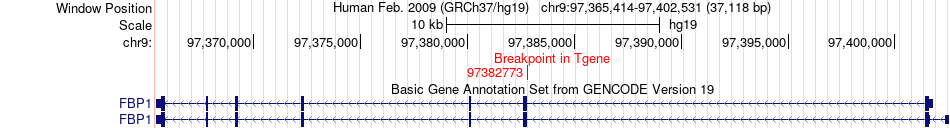 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-A1-A0SF | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
Top |
Fusion Gene ORF analysis for ENSA-FBP1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000339643 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| In-frame | ENST00000339643 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| In-frame | ENST00000361532 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| In-frame | ENST00000361532 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| In-frame | ENST00000361631 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| In-frame | ENST00000361631 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| In-frame | ENST00000369009 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| In-frame | ENST00000369009 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| In-frame | ENST00000369014 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| In-frame | ENST00000369014 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| In-frame | ENST00000369016 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| In-frame | ENST00000369016 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| intron-3CDS | ENST00000271690 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| intron-3CDS | ENST00000271690 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| intron-3CDS | ENST00000354702 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| intron-3CDS | ENST00000354702 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| intron-3CDS | ENST00000356527 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| intron-3CDS | ENST00000356527 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| intron-3CDS | ENST00000362052 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| intron-3CDS | ENST00000362052 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| intron-3CDS | ENST00000503241 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| intron-3CDS | ENST00000503241 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| intron-3CDS | ENST00000503345 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| intron-3CDS | ENST00000503345 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| intron-3CDS | ENST00000513281 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
| intron-3CDS | ENST00000513281 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000369016 | ENSA | chr1 | 150598117 | - | ENST00000375326 | FBP1 | chr9 | 97382773 | - | 1589 | 494 | 18 | 1340 | 440 |
| ENST00000369016 | ENSA | chr1 | 150598117 | - | ENST00000415431 | FBP1 | chr9 | 97382773 | - | 1581 | 494 | 18 | 1340 | 440 |
| ENST00000369014 | ENSA | chr1 | 150598117 | - | ENST00000375326 | FBP1 | chr9 | 97382773 | - | 1571 | 476 | 48 | 1322 | 424 |
| ENST00000369014 | ENSA | chr1 | 150598117 | - | ENST00000415431 | FBP1 | chr9 | 97382773 | - | 1563 | 476 | 48 | 1322 | 424 |
| ENST00000369009 | ENSA | chr1 | 150598117 | - | ENST00000375326 | FBP1 | chr9 | 97382773 | - | 1640 | 545 | 48 | 1391 | 447 |
| ENST00000369009 | ENSA | chr1 | 150598117 | - | ENST00000415431 | FBP1 | chr9 | 97382773 | - | 1632 | 545 | 48 | 1391 | 447 |
| ENST00000339643 | ENSA | chr1 | 150598117 | - | ENST00000375326 | FBP1 | chr9 | 97382773 | - | 1599 | 504 | 28 | 1350 | 440 |
| ENST00000339643 | ENSA | chr1 | 150598117 | - | ENST00000415431 | FBP1 | chr9 | 97382773 | - | 1591 | 504 | 28 | 1350 | 440 |
| ENST00000361532 | ENSA | chr1 | 150598117 | - | ENST00000375326 | FBP1 | chr9 | 97382773 | - | 1468 | 373 | 35 | 1219 | 394 |
| ENST00000361532 | ENSA | chr1 | 150598117 | - | ENST00000415431 | FBP1 | chr9 | 97382773 | - | 1460 | 373 | 35 | 1219 | 394 |
| ENST00000361631 | ENSA | chr1 | 150598117 | - | ENST00000375326 | FBP1 | chr9 | 97382773 | - | 1504 | 409 | 23 | 1255 | 410 |
| ENST00000361631 | ENSA | chr1 | 150598117 | - | ENST00000415431 | FBP1 | chr9 | 97382773 | - | 1496 | 409 | 23 | 1255 | 410 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000369016 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - | 0.001230558 | 0.99876946 |
| ENST00000369016 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - | 0.001317902 | 0.9986821 |
| ENST00000369014 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - | 0.00133341 | 0.99866664 |
| ENST00000369014 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - | 0.001431753 | 0.9985682 |
| ENST00000369009 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - | 0.000786576 | 0.99921346 |
| ENST00000369009 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - | 0.000859727 | 0.99914026 |
| ENST00000339643 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - | 0.00119686 | 0.99880314 |
| ENST00000339643 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - | 0.001291379 | 0.9987086 |
| ENST00000361532 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - | 0.000746256 | 0.9992537 |
| ENST00000361532 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - | 0.000814577 | 0.9991854 |
| ENST00000361631 | ENST00000375326 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - | 0.000819487 | 0.9991805 |
| ENST00000361631 | ENST00000415431 | ENSA | chr1 | 150598117 | - | FBP1 | chr9 | 97382773 | - | 0.000900955 | 0.999099 |
Top |
Fusion Genomic Features for ENSA-FBP1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
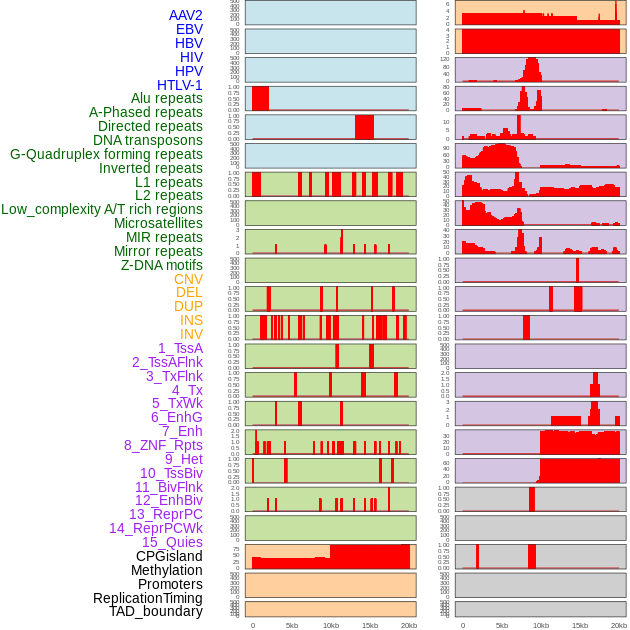 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ENSA-FBP1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:150598117/chr9:97382773) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | FBP1 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Catalyzes the hydrolysis of fructose 1,6-bisphosphate to fructose 6-phosphate in the presence of divalent cations, acting as a rate-limiting enzyme in gluconeogenesis. Plays a role in regulating glucose sensing and insulin secretion of pancreatic beta-cells. Appears to modulate glycerol gluconeogenesis in liver. Important regulator of appetite and adiposity; increased expression of the protein in liver after nutrient excess increases circulating satiety hormones and reduces appetite-stimulating neuropeptides and thus seems to provide a feedback mechanism to limit weight gain. {ECO:0000269|PubMed:16497803, ECO:0000269|PubMed:18375435, ECO:0000269|PubMed:22517657}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | FBP1 | chr1:150598117 | chr9:97382773 | ENST00000375326 | 0 | 7 | 113_114 | 56 | 339.0 | Nucleotide binding | AMP | |
| Tgene | FBP1 | chr1:150598117 | chr9:97382773 | ENST00000415431 | 1 | 8 | 113_114 | 56 | 339.0 | Nucleotide binding | AMP | |
| Tgene | FBP1 | chr1:150598117 | chr9:97382773 | ENST00000375326 | 0 | 7 | 122_125 | 56 | 339.0 | Region | Substrate binding | |
| Tgene | FBP1 | chr1:150598117 | chr9:97382773 | ENST00000375326 | 0 | 7 | 213_216 | 56 | 339.0 | Region | Substrate binding | |
| Tgene | FBP1 | chr1:150598117 | chr9:97382773 | ENST00000375326 | 0 | 7 | 244_249 | 56 | 339.0 | Region | Substrate binding | |
| Tgene | FBP1 | chr1:150598117 | chr9:97382773 | ENST00000415431 | 1 | 8 | 122_125 | 56 | 339.0 | Region | Substrate binding | |
| Tgene | FBP1 | chr1:150598117 | chr9:97382773 | ENST00000415431 | 1 | 8 | 213_216 | 56 | 339.0 | Region | Substrate binding | |
| Tgene | FBP1 | chr1:150598117 | chr9:97382773 | ENST00000415431 | 1 | 8 | 244_249 | 56 | 339.0 | Region | Substrate binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | FBP1 | chr1:150598117 | chr9:97382773 | ENST00000375326 | 0 | 7 | 18_22 | 56 | 339.0 | Nucleotide binding | AMP | |
| Tgene | FBP1 | chr1:150598117 | chr9:97382773 | ENST00000375326 | 0 | 7 | 28_32 | 56 | 339.0 | Nucleotide binding | AMP | |
| Tgene | FBP1 | chr1:150598117 | chr9:97382773 | ENST00000415431 | 1 | 8 | 18_22 | 56 | 339.0 | Nucleotide binding | AMP | |
| Tgene | FBP1 | chr1:150598117 | chr9:97382773 | ENST00000415431 | 1 | 8 | 28_32 | 56 | 339.0 | Nucleotide binding | AMP |
Top |
Fusion Gene Sequence for ENSA-FBP1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >26664_26664_1_ENSA-FBP1_ENSA_chr1_150598117_ENST00000339643_FBP1_chr9_97382773_ENST00000375326_length(transcript)=1599nt_BP=504nt GTGACAGGAGCCGAAGCAGCAGCGCAGGTTGTCCCCGTTTCCCCTCCCCCTTCCCTTCTCCGGTTGCCTTCCCGGGCCCCTTACACTCCA CAGTCCCGGTCCCGCCATGTCCCAGAAACAAGAAGAAGAGAACCCTGCGGAGGAGACCGGCGAGGAGAAGCAGGACACGCAGGAGAAAGA AGGTATTCTGCCTGAGAGAGCTGAAGAGGCAAAGCTAAAGGCCAAATACCCAAGCCTAGGACAAAAGCCTGGAGGCTCCGACTTCCTCAT GAAGAGACTCCAGAAAGGGGATTATAAATCATTACATTGGAGTGTGCTTCTCTGTGCGGATGAAATGCAAAAGTACTTTGACTCAGGAGA CTACAACATGGCCAAAGCCAAGATGAAGAATAAGCAGCTGCCAAGTGCAGGACCAGACAAGAACCTGGTGACTGGTGATCACATCCCCAC CCCACAGGATCTGCCCCAGAGAAAGTCCTCGCTCGTCACCAGCAAGCTTGCGGGCTATGGCATTGCTGGTTCTACCAACGTGACAGGTGA TCAAGTTAAGAAGCTGGACGTCCTCTCCAACGACCTGGTTATGAACATGTTAAAGTCATCCTTTGCCACGTGTGTTCTCGTGTCAGAAGA AGATAAACACGCCATCATAGTGGAACCGGAGAAAAGGGGTAAATATGTGGTCTGTTTTGATCCCCTTGATGGATCTTCCAACATCGATTG CCTTGTGTCCGTTGGAACCATTTTTGGCATCTATAGAAAGAAATCAACTGATGAGCCTTCTGAGAAGGATGCTCTGCAACCAGGCCGGAA CCTGGTGGCAGCCGGCTACGCACTGTATGGCAGTGCCACCATGCTGGTCCTTGCCATGGACTGTGGGGTCAACTGCTTCATGCTGGACCC GGCCATCGGGGAGTTCATTTTGGTGGACAAGGATGTGAAGATAAAAAAGAAAGGTAAAATCTACAGCCTTAACGAGGGCTACGCCAGGGA CTTTGACCCTGCCGTCACTGAGTACATCCAGAGGAAGAAGTTCCCCCCAGATAATTCAGCTCCTTATGGGGCCCGGTATGTGGGCTCCAT GGTGGCTGATGTTCATCGCACTCTGGTCTACGGAGGGATATTTCTGTACCCCGCTAACAAGAAGAGCCCCAATGGAAAGCTGAGACTGCT GTACGAATGCAACCCCATGGCCTACGTCATGGAGAAGGCTGGGGGAATGGCCACCACTGGGAAGGAGGCCGTGTTAGACGTCATTCCCAC AGACATTCACCAGAGGGCGCCGGTGATCTTGGGATCCCCCGACGACGTGCTCGAGTTCCTGAAGGTGTATGAGAAGCACTCTGCCCAGTG AGCACCTGCCCTGCCTGCATCCGGAGAATTGCCTCTACCTGGACCTTTTGTCTCACACAGCAGTACCCTGACCTGCTGTGCACCTTACAT TCCTAGAGAGCAGAAATAAAAAGCATGACTATTTCCACCATCAAATGCTGTAGAATGCTTGGCACTCCCTAACCAAATGCTGTCTCCATA >26664_26664_1_ENSA-FBP1_ENSA_chr1_150598117_ENST00000339643_FBP1_chr9_97382773_ENST00000375326_length(amino acids)=440AA_BP=149 MSPFPLPLPFSGCLPGPLTLHSPGPAMSQKQEEENPAEETGEEKQDTQEKEGILPERAEEAKLKAKYPSLGQKPGGSDFLMKRLQKGDYK SLHWSVLLCADEMQKYFDSGDYNMAKAKMKNKQLPSAGPDKNLVTGDHIPTPQDLPQRKSSLVTSKLAGYGIAGSTNVTGDQVKKLDVLS NDLVMNMLKSSFATCVLVSEEDKHAIIVEPEKRGKYVVCFDPLDGSSNIDCLVSVGTIFGIYRKKSTDEPSEKDALQPGRNLVAAGYALY GSATMLVLAMDCGVNCFMLDPAIGEFILVDKDVKIKKKGKIYSLNEGYARDFDPAVTEYIQRKKFPPDNSAPYGARYVGSMVADVHRTLV -------------------------------------------------------------- >26664_26664_2_ENSA-FBP1_ENSA_chr1_150598117_ENST00000339643_FBP1_chr9_97382773_ENST00000415431_length(transcript)=1591nt_BP=504nt GTGACAGGAGCCGAAGCAGCAGCGCAGGTTGTCCCCGTTTCCCCTCCCCCTTCCCTTCTCCGGTTGCCTTCCCGGGCCCCTTACACTCCA CAGTCCCGGTCCCGCCATGTCCCAGAAACAAGAAGAAGAGAACCCTGCGGAGGAGACCGGCGAGGAGAAGCAGGACACGCAGGAGAAAGA AGGTATTCTGCCTGAGAGAGCTGAAGAGGCAAAGCTAAAGGCCAAATACCCAAGCCTAGGACAAAAGCCTGGAGGCTCCGACTTCCTCAT GAAGAGACTCCAGAAAGGGGATTATAAATCATTACATTGGAGTGTGCTTCTCTGTGCGGATGAAATGCAAAAGTACTTTGACTCAGGAGA CTACAACATGGCCAAAGCCAAGATGAAGAATAAGCAGCTGCCAAGTGCAGGACCAGACAAGAACCTGGTGACTGGTGATCACATCCCCAC CCCACAGGATCTGCCCCAGAGAAAGTCCTCGCTCGTCACCAGCAAGCTTGCGGGCTATGGCATTGCTGGTTCTACCAACGTGACAGGTGA TCAAGTTAAGAAGCTGGACGTCCTCTCCAACGACCTGGTTATGAACATGTTAAAGTCATCCTTTGCCACGTGTGTTCTCGTGTCAGAAGA AGATAAACACGCCATCATAGTGGAACCGGAGAAAAGGGGTAAATATGTGGTCTGTTTTGATCCCCTTGATGGATCTTCCAACATCGATTG CCTTGTGTCCGTTGGAACCATTTTTGGCATCTATAGAAAGAAATCAACTGATGAGCCTTCTGAGAAGGATGCTCTGCAACCAGGCCGGAA CCTGGTGGCAGCCGGCTACGCACTGTATGGCAGTGCCACCATGCTGGTCCTTGCCATGGACTGTGGGGTCAACTGCTTCATGCTGGACCC GGCCATCGGGGAGTTCATTTTGGTGGACAAGGATGTGAAGATAAAAAAGAAAGGTAAAATCTACAGCCTTAACGAGGGCTACGCCAGGGA CTTTGACCCTGCCGTCACTGAGTACATCCAGAGGAAGAAGTTCCCCCCAGATAATTCAGCTCCTTATGGGGCCCGGTATGTGGGCTCCAT GGTGGCTGATGTTCATCGCACTCTGGTCTACGGAGGGATATTTCTGTACCCCGCTAACAAGAAGAGCCCCAATGGAAAGCTGAGACTGCT GTACGAATGCAACCCCATGGCCTACGTCATGGAGAAGGCTGGGGGAATGGCCACCACTGGGAAGGAGGCCGTGTTAGACGTCATTCCCAC AGACATTCACCAGAGGGCGCCGGTGATCTTGGGATCCCCCGACGACGTGCTCGAGTTCCTGAAGGTGTATGAGAAGCACTCTGCCCAGTG AGCACCTGCCCTGCCTGCATCCGGAGAATTGCCTCTACCTGGACCTTTTGTCTCACACAGCAGTACCCTGACCTGCTGTGCACCTTACAT TCCTAGAGAGCAGAAATAAAAAGCATGACTATTTCCACCATCAAATGCTGTAGAATGCTTGGCACTCCCTAACCAAATGCTGTCTCCATA >26664_26664_2_ENSA-FBP1_ENSA_chr1_150598117_ENST00000339643_FBP1_chr9_97382773_ENST00000415431_length(amino acids)=440AA_BP=149 MSPFPLPLPFSGCLPGPLTLHSPGPAMSQKQEEENPAEETGEEKQDTQEKEGILPERAEEAKLKAKYPSLGQKPGGSDFLMKRLQKGDYK SLHWSVLLCADEMQKYFDSGDYNMAKAKMKNKQLPSAGPDKNLVTGDHIPTPQDLPQRKSSLVTSKLAGYGIAGSTNVTGDQVKKLDVLS NDLVMNMLKSSFATCVLVSEEDKHAIIVEPEKRGKYVVCFDPLDGSSNIDCLVSVGTIFGIYRKKSTDEPSEKDALQPGRNLVAAGYALY GSATMLVLAMDCGVNCFMLDPAIGEFILVDKDVKIKKKGKIYSLNEGYARDFDPAVTEYIQRKKFPPDNSAPYGARYVGSMVADVHRTLV -------------------------------------------------------------- >26664_26664_3_ENSA-FBP1_ENSA_chr1_150598117_ENST00000361532_FBP1_chr9_97382773_ENST00000375326_length(transcript)=1468nt_BP=373nt GGCGGGTGGTTGGAAAGACGGTGAATGGAAAGGGGATGGCTGGTGGTCTTGGGTGTGATGTGTGTTATTGGTTTGTAGAGGACACGCAGG AGAAAGAAGGTATTCTGCCTGAGAGAGCTGAAGAGGCAAAGCTAAAGGCCAAATACCCAAGCCTAGGACAAAAGCCTGGAGGCTCCGACT TCCTCATGAAGAGACTCCAGAAAGGGCAAAAGTACTTTGACTCAGGAGACTACAACATGGCCAAAGCCAAGATGAAGAATAAGCAGCTGC CAAGTGCAGGACCAGACAAGAACCTGGTGACTGGTGATCACATCCCCACCCCACAGGATCTGCCCCAGAGAAAGTCCTCGCTCGTCACCA GCAAGCTTGCGGGCTATGGCATTGCTGGTTCTACCAACGTGACAGGTGATCAAGTTAAGAAGCTGGACGTCCTCTCCAACGACCTGGTTA TGAACATGTTAAAGTCATCCTTTGCCACGTGTGTTCTCGTGTCAGAAGAAGATAAACACGCCATCATAGTGGAACCGGAGAAAAGGGGTA AATATGTGGTCTGTTTTGATCCCCTTGATGGATCTTCCAACATCGATTGCCTTGTGTCCGTTGGAACCATTTTTGGCATCTATAGAAAGA AATCAACTGATGAGCCTTCTGAGAAGGATGCTCTGCAACCAGGCCGGAACCTGGTGGCAGCCGGCTACGCACTGTATGGCAGTGCCACCA TGCTGGTCCTTGCCATGGACTGTGGGGTCAACTGCTTCATGCTGGACCCGGCCATCGGGGAGTTCATTTTGGTGGACAAGGATGTGAAGA TAAAAAAGAAAGGTAAAATCTACAGCCTTAACGAGGGCTACGCCAGGGACTTTGACCCTGCCGTCACTGAGTACATCCAGAGGAAGAAGT TCCCCCCAGATAATTCAGCTCCTTATGGGGCCCGGTATGTGGGCTCCATGGTGGCTGATGTTCATCGCACTCTGGTCTACGGAGGGATAT TTCTGTACCCCGCTAACAAGAAGAGCCCCAATGGAAAGCTGAGACTGCTGTACGAATGCAACCCCATGGCCTACGTCATGGAGAAGGCTG GGGGAATGGCCACCACTGGGAAGGAGGCCGTGTTAGACGTCATTCCCACAGACATTCACCAGAGGGCGCCGGTGATCTTGGGATCCCCCG ACGACGTGCTCGAGTTCCTGAAGGTGTATGAGAAGCACTCTGCCCAGTGAGCACCTGCCCTGCCTGCATCCGGAGAATTGCCTCTACCTG GACCTTTTGTCTCACACAGCAGTACCCTGACCTGCTGTGCACCTTACATTCCTAGAGAGCAGAAATAAAAAGCATGACTATTTCCACCAT CAAATGCTGTAGAATGCTTGGCACTCCCTAACCAAATGCTGTCTCCATAATGCCACTGGTGTTAAGATATATTTTGAGTGGATGGAGGAG >26664_26664_3_ENSA-FBP1_ENSA_chr1_150598117_ENST00000361532_FBP1_chr9_97382773_ENST00000375326_length(amino acids)=394AA_BP=103 MAGGLGCDVCYWFVEDTQEKEGILPERAEEAKLKAKYPSLGQKPGGSDFLMKRLQKGQKYFDSGDYNMAKAKMKNKQLPSAGPDKNLVTG DHIPTPQDLPQRKSSLVTSKLAGYGIAGSTNVTGDQVKKLDVLSNDLVMNMLKSSFATCVLVSEEDKHAIIVEPEKRGKYVVCFDPLDGS SNIDCLVSVGTIFGIYRKKSTDEPSEKDALQPGRNLVAAGYALYGSATMLVLAMDCGVNCFMLDPAIGEFILVDKDVKIKKKGKIYSLNE GYARDFDPAVTEYIQRKKFPPDNSAPYGARYVGSMVADVHRTLVYGGIFLYPANKKSPNGKLRLLYECNPMAYVMEKAGGMATTGKEAVL -------------------------------------------------------------- >26664_26664_4_ENSA-FBP1_ENSA_chr1_150598117_ENST00000361532_FBP1_chr9_97382773_ENST00000415431_length(transcript)=1460nt_BP=373nt GGCGGGTGGTTGGAAAGACGGTGAATGGAAAGGGGATGGCTGGTGGTCTTGGGTGTGATGTGTGTTATTGGTTTGTAGAGGACACGCAGG AGAAAGAAGGTATTCTGCCTGAGAGAGCTGAAGAGGCAAAGCTAAAGGCCAAATACCCAAGCCTAGGACAAAAGCCTGGAGGCTCCGACT TCCTCATGAAGAGACTCCAGAAAGGGCAAAAGTACTTTGACTCAGGAGACTACAACATGGCCAAAGCCAAGATGAAGAATAAGCAGCTGC CAAGTGCAGGACCAGACAAGAACCTGGTGACTGGTGATCACATCCCCACCCCACAGGATCTGCCCCAGAGAAAGTCCTCGCTCGTCACCA GCAAGCTTGCGGGCTATGGCATTGCTGGTTCTACCAACGTGACAGGTGATCAAGTTAAGAAGCTGGACGTCCTCTCCAACGACCTGGTTA TGAACATGTTAAAGTCATCCTTTGCCACGTGTGTTCTCGTGTCAGAAGAAGATAAACACGCCATCATAGTGGAACCGGAGAAAAGGGGTA AATATGTGGTCTGTTTTGATCCCCTTGATGGATCTTCCAACATCGATTGCCTTGTGTCCGTTGGAACCATTTTTGGCATCTATAGAAAGA AATCAACTGATGAGCCTTCTGAGAAGGATGCTCTGCAACCAGGCCGGAACCTGGTGGCAGCCGGCTACGCACTGTATGGCAGTGCCACCA TGCTGGTCCTTGCCATGGACTGTGGGGTCAACTGCTTCATGCTGGACCCGGCCATCGGGGAGTTCATTTTGGTGGACAAGGATGTGAAGA TAAAAAAGAAAGGTAAAATCTACAGCCTTAACGAGGGCTACGCCAGGGACTTTGACCCTGCCGTCACTGAGTACATCCAGAGGAAGAAGT TCCCCCCAGATAATTCAGCTCCTTATGGGGCCCGGTATGTGGGCTCCATGGTGGCTGATGTTCATCGCACTCTGGTCTACGGAGGGATAT TTCTGTACCCCGCTAACAAGAAGAGCCCCAATGGAAAGCTGAGACTGCTGTACGAATGCAACCCCATGGCCTACGTCATGGAGAAGGCTG GGGGAATGGCCACCACTGGGAAGGAGGCCGTGTTAGACGTCATTCCCACAGACATTCACCAGAGGGCGCCGGTGATCTTGGGATCCCCCG ACGACGTGCTCGAGTTCCTGAAGGTGTATGAGAAGCACTCTGCCCAGTGAGCACCTGCCCTGCCTGCATCCGGAGAATTGCCTCTACCTG GACCTTTTGTCTCACACAGCAGTACCCTGACCTGCTGTGCACCTTACATTCCTAGAGAGCAGAAATAAAAAGCATGACTATTTCCACCAT CAAATGCTGTAGAATGCTTGGCACTCCCTAACCAAATGCTGTCTCCATAATGCCACTGGTGTTAAGATATATTTTGAGTGGATGGAGGAG >26664_26664_4_ENSA-FBP1_ENSA_chr1_150598117_ENST00000361532_FBP1_chr9_97382773_ENST00000415431_length(amino acids)=394AA_BP=103 MAGGLGCDVCYWFVEDTQEKEGILPERAEEAKLKAKYPSLGQKPGGSDFLMKRLQKGQKYFDSGDYNMAKAKMKNKQLPSAGPDKNLVTG DHIPTPQDLPQRKSSLVTSKLAGYGIAGSTNVTGDQVKKLDVLSNDLVMNMLKSSFATCVLVSEEDKHAIIVEPEKRGKYVVCFDPLDGS SNIDCLVSVGTIFGIYRKKSTDEPSEKDALQPGRNLVAAGYALYGSATMLVLAMDCGVNCFMLDPAIGEFILVDKDVKIKKKGKIYSLNE GYARDFDPAVTEYIQRKKFPPDNSAPYGARYVGSMVADVHRTLVYGGIFLYPANKKSPNGKLRLLYECNPMAYVMEKAGGMATTGKEAVL -------------------------------------------------------------- >26664_26664_5_ENSA-FBP1_ENSA_chr1_150598117_ENST00000361631_FBP1_chr9_97382773_ENST00000375326_length(transcript)=1504nt_BP=409nt GAAAGACGGTGAATGGAAAGGGGATGGCTGGTGGTCTTGGGTGTGATGTGTGTTATTGGTTTGTAGAGGACACGCAGGAGAAAGAAGGTA TTCTGCCTGAGAGAGCTGAAGAGGCAAAGCTAAAGGCCAAATACCCAAGCCTAGGACAAAAGCCTGGAGGCTCCGACTTCCTCATGAAGA GACTCCAGAAAGGGGATTATAAATCATTACATTGGAGTGTGCTTCTCTGTGCGGATGAAATGCAAAAGTACTTTGACTCAGGAGACTACA ACATGGCCAAAGCCAAGATGAAGAATAAGCAGCTGCCAAGTGCAGGACCAGACAAGAACCTGGTGACTGGTGATCACATCCCCACCCCAC AGGATCTGCCCCAGAGAAAGTCCTCGCTCGTCACCAGCAAGCTTGCGGGCTATGGCATTGCTGGTTCTACCAACGTGACAGGTGATCAAG TTAAGAAGCTGGACGTCCTCTCCAACGACCTGGTTATGAACATGTTAAAGTCATCCTTTGCCACGTGTGTTCTCGTGTCAGAAGAAGATA AACACGCCATCATAGTGGAACCGGAGAAAAGGGGTAAATATGTGGTCTGTTTTGATCCCCTTGATGGATCTTCCAACATCGATTGCCTTG TGTCCGTTGGAACCATTTTTGGCATCTATAGAAAGAAATCAACTGATGAGCCTTCTGAGAAGGATGCTCTGCAACCAGGCCGGAACCTGG TGGCAGCCGGCTACGCACTGTATGGCAGTGCCACCATGCTGGTCCTTGCCATGGACTGTGGGGTCAACTGCTTCATGCTGGACCCGGCCA TCGGGGAGTTCATTTTGGTGGACAAGGATGTGAAGATAAAAAAGAAAGGTAAAATCTACAGCCTTAACGAGGGCTACGCCAGGGACTTTG ACCCTGCCGTCACTGAGTACATCCAGAGGAAGAAGTTCCCCCCAGATAATTCAGCTCCTTATGGGGCCCGGTATGTGGGCTCCATGGTGG CTGATGTTCATCGCACTCTGGTCTACGGAGGGATATTTCTGTACCCCGCTAACAAGAAGAGCCCCAATGGAAAGCTGAGACTGCTGTACG AATGCAACCCCATGGCCTACGTCATGGAGAAGGCTGGGGGAATGGCCACCACTGGGAAGGAGGCCGTGTTAGACGTCATTCCCACAGACA TTCACCAGAGGGCGCCGGTGATCTTGGGATCCCCCGACGACGTGCTCGAGTTCCTGAAGGTGTATGAGAAGCACTCTGCCCAGTGAGCAC CTGCCCTGCCTGCATCCGGAGAATTGCCTCTACCTGGACCTTTTGTCTCACACAGCAGTACCCTGACCTGCTGTGCACCTTACATTCCTA GAGAGCAGAAATAAAAAGCATGACTATTTCCACCATCAAATGCTGTAGAATGCTTGGCACTCCCTAACCAAATGCTGTCTCCATAATGCC >26664_26664_5_ENSA-FBP1_ENSA_chr1_150598117_ENST00000361631_FBP1_chr9_97382773_ENST00000375326_length(amino acids)=410AA_BP=119 MAGGLGCDVCYWFVEDTQEKEGILPERAEEAKLKAKYPSLGQKPGGSDFLMKRLQKGDYKSLHWSVLLCADEMQKYFDSGDYNMAKAKMK NKQLPSAGPDKNLVTGDHIPTPQDLPQRKSSLVTSKLAGYGIAGSTNVTGDQVKKLDVLSNDLVMNMLKSSFATCVLVSEEDKHAIIVEP EKRGKYVVCFDPLDGSSNIDCLVSVGTIFGIYRKKSTDEPSEKDALQPGRNLVAAGYALYGSATMLVLAMDCGVNCFMLDPAIGEFILVD KDVKIKKKGKIYSLNEGYARDFDPAVTEYIQRKKFPPDNSAPYGARYVGSMVADVHRTLVYGGIFLYPANKKSPNGKLRLLYECNPMAYV -------------------------------------------------------------- >26664_26664_6_ENSA-FBP1_ENSA_chr1_150598117_ENST00000361631_FBP1_chr9_97382773_ENST00000415431_length(transcript)=1496nt_BP=409nt GAAAGACGGTGAATGGAAAGGGGATGGCTGGTGGTCTTGGGTGTGATGTGTGTTATTGGTTTGTAGAGGACACGCAGGAGAAAGAAGGTA TTCTGCCTGAGAGAGCTGAAGAGGCAAAGCTAAAGGCCAAATACCCAAGCCTAGGACAAAAGCCTGGAGGCTCCGACTTCCTCATGAAGA GACTCCAGAAAGGGGATTATAAATCATTACATTGGAGTGTGCTTCTCTGTGCGGATGAAATGCAAAAGTACTTTGACTCAGGAGACTACA ACATGGCCAAAGCCAAGATGAAGAATAAGCAGCTGCCAAGTGCAGGACCAGACAAGAACCTGGTGACTGGTGATCACATCCCCACCCCAC AGGATCTGCCCCAGAGAAAGTCCTCGCTCGTCACCAGCAAGCTTGCGGGCTATGGCATTGCTGGTTCTACCAACGTGACAGGTGATCAAG TTAAGAAGCTGGACGTCCTCTCCAACGACCTGGTTATGAACATGTTAAAGTCATCCTTTGCCACGTGTGTTCTCGTGTCAGAAGAAGATA AACACGCCATCATAGTGGAACCGGAGAAAAGGGGTAAATATGTGGTCTGTTTTGATCCCCTTGATGGATCTTCCAACATCGATTGCCTTG TGTCCGTTGGAACCATTTTTGGCATCTATAGAAAGAAATCAACTGATGAGCCTTCTGAGAAGGATGCTCTGCAACCAGGCCGGAACCTGG TGGCAGCCGGCTACGCACTGTATGGCAGTGCCACCATGCTGGTCCTTGCCATGGACTGTGGGGTCAACTGCTTCATGCTGGACCCGGCCA TCGGGGAGTTCATTTTGGTGGACAAGGATGTGAAGATAAAAAAGAAAGGTAAAATCTACAGCCTTAACGAGGGCTACGCCAGGGACTTTG ACCCTGCCGTCACTGAGTACATCCAGAGGAAGAAGTTCCCCCCAGATAATTCAGCTCCTTATGGGGCCCGGTATGTGGGCTCCATGGTGG CTGATGTTCATCGCACTCTGGTCTACGGAGGGATATTTCTGTACCCCGCTAACAAGAAGAGCCCCAATGGAAAGCTGAGACTGCTGTACG AATGCAACCCCATGGCCTACGTCATGGAGAAGGCTGGGGGAATGGCCACCACTGGGAAGGAGGCCGTGTTAGACGTCATTCCCACAGACA TTCACCAGAGGGCGCCGGTGATCTTGGGATCCCCCGACGACGTGCTCGAGTTCCTGAAGGTGTATGAGAAGCACTCTGCCCAGTGAGCAC CTGCCCTGCCTGCATCCGGAGAATTGCCTCTACCTGGACCTTTTGTCTCACACAGCAGTACCCTGACCTGCTGTGCACCTTACATTCCTA GAGAGCAGAAATAAAAAGCATGACTATTTCCACCATCAAATGCTGTAGAATGCTTGGCACTCCCTAACCAAATGCTGTCTCCATAATGCC >26664_26664_6_ENSA-FBP1_ENSA_chr1_150598117_ENST00000361631_FBP1_chr9_97382773_ENST00000415431_length(amino acids)=410AA_BP=119 MAGGLGCDVCYWFVEDTQEKEGILPERAEEAKLKAKYPSLGQKPGGSDFLMKRLQKGDYKSLHWSVLLCADEMQKYFDSGDYNMAKAKMK NKQLPSAGPDKNLVTGDHIPTPQDLPQRKSSLVTSKLAGYGIAGSTNVTGDQVKKLDVLSNDLVMNMLKSSFATCVLVSEEDKHAIIVEP EKRGKYVVCFDPLDGSSNIDCLVSVGTIFGIYRKKSTDEPSEKDALQPGRNLVAAGYALYGSATMLVLAMDCGVNCFMLDPAIGEFILVD KDVKIKKKGKIYSLNEGYARDFDPAVTEYIQRKKFPPDNSAPYGARYVGSMVADVHRTLVYGGIFLYPANKKSPNGKLRLLYECNPMAYV -------------------------------------------------------------- >26664_26664_7_ENSA-FBP1_ENSA_chr1_150598117_ENST00000369009_FBP1_chr9_97382773_ENST00000375326_length(transcript)=1640nt_BP=545nt ATTTTGACTGAGCAACCCTAGTGACAGGAGCCGAAGCAGCAGCGCAGGTTGTCCCCGTTTCCCCTCCCCCTTCCCTTCTCCGGTTGCCTT CCCGGGCCCCTTACACTCCACAGTCCCGGTCCCGCCATGTCCCAGAAACAAGAAGAAGAGAACCCTGCGGAGGAGACCGGCGAGGAGAAG CAGGACACGCAGGAGAAAGAAGGTATTCTGCCTGAGAGAGCTGAAGAGGCAAAGCTAAAGGCCAAATACCCAAGCCTAGGACAAAAGCCT GGAGGCTCCGACTTCCTCATGAAGAGACTCCAGAAAGGGGTATGGGGCATAGTCTCTTACCCTCTTTCTTTGGAGCTAAAGGAGGTTCTT CGAATGAAGTCTGTAGAGCAAAAGTACTTTGACTCAGGAGACTACAACATGGCCAAAGCCAAGATGAAGAATAAGCAGCTGCCAAGTGCA GGACCAGACAAGAACCTGGTGACTGGTGATCACATCCCCACCCCACAGGATCTGCCCCAGAGAAAGTCCTCGCTCGTCACCAGCAAGCTT GCGGGCTATGGCATTGCTGGTTCTACCAACGTGACAGGTGATCAAGTTAAGAAGCTGGACGTCCTCTCCAACGACCTGGTTATGAACATG TTAAAGTCATCCTTTGCCACGTGTGTTCTCGTGTCAGAAGAAGATAAACACGCCATCATAGTGGAACCGGAGAAAAGGGGTAAATATGTG GTCTGTTTTGATCCCCTTGATGGATCTTCCAACATCGATTGCCTTGTGTCCGTTGGAACCATTTTTGGCATCTATAGAAAGAAATCAACT GATGAGCCTTCTGAGAAGGATGCTCTGCAACCAGGCCGGAACCTGGTGGCAGCCGGCTACGCACTGTATGGCAGTGCCACCATGCTGGTC CTTGCCATGGACTGTGGGGTCAACTGCTTCATGCTGGACCCGGCCATCGGGGAGTTCATTTTGGTGGACAAGGATGTGAAGATAAAAAAG AAAGGTAAAATCTACAGCCTTAACGAGGGCTACGCCAGGGACTTTGACCCTGCCGTCACTGAGTACATCCAGAGGAAGAAGTTCCCCCCA GATAATTCAGCTCCTTATGGGGCCCGGTATGTGGGCTCCATGGTGGCTGATGTTCATCGCACTCTGGTCTACGGAGGGATATTTCTGTAC CCCGCTAACAAGAAGAGCCCCAATGGAAAGCTGAGACTGCTGTACGAATGCAACCCCATGGCCTACGTCATGGAGAAGGCTGGGGGAATG GCCACCACTGGGAAGGAGGCCGTGTTAGACGTCATTCCCACAGACATTCACCAGAGGGCGCCGGTGATCTTGGGATCCCCCGACGACGTG CTCGAGTTCCTGAAGGTGTATGAGAAGCACTCTGCCCAGTGAGCACCTGCCCTGCCTGCATCCGGAGAATTGCCTCTACCTGGACCTTTT GTCTCACACAGCAGTACCCTGACCTGCTGTGCACCTTACATTCCTAGAGAGCAGAAATAAAAAGCATGACTATTTCCACCATCAAATGCT GTAGAATGCTTGGCACTCCCTAACCAAATGCTGTCTCCATAATGCCACTGGTGTTAAGATATATTTTGAGTGGATGGAGGAGAAATAAAC >26664_26664_7_ENSA-FBP1_ENSA_chr1_150598117_ENST00000369009_FBP1_chr9_97382773_ENST00000375326_length(amino acids)=447AA_BP=156 MSPFPLPLPFSGCLPGPLTLHSPGPAMSQKQEEENPAEETGEEKQDTQEKEGILPERAEEAKLKAKYPSLGQKPGGSDFLMKRLQKGVWG IVSYPLSLELKEVLRMKSVEQKYFDSGDYNMAKAKMKNKQLPSAGPDKNLVTGDHIPTPQDLPQRKSSLVTSKLAGYGIAGSTNVTGDQV KKLDVLSNDLVMNMLKSSFATCVLVSEEDKHAIIVEPEKRGKYVVCFDPLDGSSNIDCLVSVGTIFGIYRKKSTDEPSEKDALQPGRNLV AAGYALYGSATMLVLAMDCGVNCFMLDPAIGEFILVDKDVKIKKKGKIYSLNEGYARDFDPAVTEYIQRKKFPPDNSAPYGARYVGSMVA -------------------------------------------------------------- >26664_26664_8_ENSA-FBP1_ENSA_chr1_150598117_ENST00000369009_FBP1_chr9_97382773_ENST00000415431_length(transcript)=1632nt_BP=545nt ATTTTGACTGAGCAACCCTAGTGACAGGAGCCGAAGCAGCAGCGCAGGTTGTCCCCGTTTCCCCTCCCCCTTCCCTTCTCCGGTTGCCTT CCCGGGCCCCTTACACTCCACAGTCCCGGTCCCGCCATGTCCCAGAAACAAGAAGAAGAGAACCCTGCGGAGGAGACCGGCGAGGAGAAG CAGGACACGCAGGAGAAAGAAGGTATTCTGCCTGAGAGAGCTGAAGAGGCAAAGCTAAAGGCCAAATACCCAAGCCTAGGACAAAAGCCT GGAGGCTCCGACTTCCTCATGAAGAGACTCCAGAAAGGGGTATGGGGCATAGTCTCTTACCCTCTTTCTTTGGAGCTAAAGGAGGTTCTT CGAATGAAGTCTGTAGAGCAAAAGTACTTTGACTCAGGAGACTACAACATGGCCAAAGCCAAGATGAAGAATAAGCAGCTGCCAAGTGCA GGACCAGACAAGAACCTGGTGACTGGTGATCACATCCCCACCCCACAGGATCTGCCCCAGAGAAAGTCCTCGCTCGTCACCAGCAAGCTT GCGGGCTATGGCATTGCTGGTTCTACCAACGTGACAGGTGATCAAGTTAAGAAGCTGGACGTCCTCTCCAACGACCTGGTTATGAACATG TTAAAGTCATCCTTTGCCACGTGTGTTCTCGTGTCAGAAGAAGATAAACACGCCATCATAGTGGAACCGGAGAAAAGGGGTAAATATGTG GTCTGTTTTGATCCCCTTGATGGATCTTCCAACATCGATTGCCTTGTGTCCGTTGGAACCATTTTTGGCATCTATAGAAAGAAATCAACT GATGAGCCTTCTGAGAAGGATGCTCTGCAACCAGGCCGGAACCTGGTGGCAGCCGGCTACGCACTGTATGGCAGTGCCACCATGCTGGTC CTTGCCATGGACTGTGGGGTCAACTGCTTCATGCTGGACCCGGCCATCGGGGAGTTCATTTTGGTGGACAAGGATGTGAAGATAAAAAAG AAAGGTAAAATCTACAGCCTTAACGAGGGCTACGCCAGGGACTTTGACCCTGCCGTCACTGAGTACATCCAGAGGAAGAAGTTCCCCCCA GATAATTCAGCTCCTTATGGGGCCCGGTATGTGGGCTCCATGGTGGCTGATGTTCATCGCACTCTGGTCTACGGAGGGATATTTCTGTAC CCCGCTAACAAGAAGAGCCCCAATGGAAAGCTGAGACTGCTGTACGAATGCAACCCCATGGCCTACGTCATGGAGAAGGCTGGGGGAATG GCCACCACTGGGAAGGAGGCCGTGTTAGACGTCATTCCCACAGACATTCACCAGAGGGCGCCGGTGATCTTGGGATCCCCCGACGACGTG CTCGAGTTCCTGAAGGTGTATGAGAAGCACTCTGCCCAGTGAGCACCTGCCCTGCCTGCATCCGGAGAATTGCCTCTACCTGGACCTTTT GTCTCACACAGCAGTACCCTGACCTGCTGTGCACCTTACATTCCTAGAGAGCAGAAATAAAAAGCATGACTATTTCCACCATCAAATGCT GTAGAATGCTTGGCACTCCCTAACCAAATGCTGTCTCCATAATGCCACTGGTGTTAAGATATATTTTGAGTGGATGGAGGAGAAATAAAC >26664_26664_8_ENSA-FBP1_ENSA_chr1_150598117_ENST00000369009_FBP1_chr9_97382773_ENST00000415431_length(amino acids)=447AA_BP=156 MSPFPLPLPFSGCLPGPLTLHSPGPAMSQKQEEENPAEETGEEKQDTQEKEGILPERAEEAKLKAKYPSLGQKPGGSDFLMKRLQKGVWG IVSYPLSLELKEVLRMKSVEQKYFDSGDYNMAKAKMKNKQLPSAGPDKNLVTGDHIPTPQDLPQRKSSLVTSKLAGYGIAGSTNVTGDQV KKLDVLSNDLVMNMLKSSFATCVLVSEEDKHAIIVEPEKRGKYVVCFDPLDGSSNIDCLVSVGTIFGIYRKKSTDEPSEKDALQPGRNLV AAGYALYGSATMLVLAMDCGVNCFMLDPAIGEFILVDKDVKIKKKGKIYSLNEGYARDFDPAVTEYIQRKKFPPDNSAPYGARYVGSMVA -------------------------------------------------------------- >26664_26664_9_ENSA-FBP1_ENSA_chr1_150598117_ENST00000369014_FBP1_chr9_97382773_ENST00000375326_length(transcript)=1571nt_BP=476nt ATTTTGACTGAGCAACCCTAGTGACAGGAGCCGAAGCAGCAGCGCAGGTTGTCCCCGTTTCCCCTCCCCCTTCCCTTCTCCGGTTGCCTT CCCGGGCCCCTTACACTCCACAGTCCCGGTCCCGCCATGTCCCAGAAACAAGAAGAAGAGAACCCTGCGGAGGAGACCGGCGAGGAGAAG CAGGACACGCAGGAGAAAGAAGGTATTCTGCCTGAGAGAGCTGAAGAGGCAAAGCTAAAGGCCAAATACCCAAGCCTAGGACAAAAGCCT GGAGGCTCCGACTTCCTCATGAAGAGACTCCAGAAAGGGCAAAAGTACTTTGACTCAGGAGACTACAACATGGCCAAAGCCAAGATGAAG AATAAGCAGCTGCCAAGTGCAGGACCAGACAAGAACCTGGTGACTGGTGATCACATCCCCACCCCACAGGATCTGCCCCAGAGAAAGTCC TCGCTCGTCACCAGCAAGCTTGCGGGCTATGGCATTGCTGGTTCTACCAACGTGACAGGTGATCAAGTTAAGAAGCTGGACGTCCTCTCC AACGACCTGGTTATGAACATGTTAAAGTCATCCTTTGCCACGTGTGTTCTCGTGTCAGAAGAAGATAAACACGCCATCATAGTGGAACCG GAGAAAAGGGGTAAATATGTGGTCTGTTTTGATCCCCTTGATGGATCTTCCAACATCGATTGCCTTGTGTCCGTTGGAACCATTTTTGGC ATCTATAGAAAGAAATCAACTGATGAGCCTTCTGAGAAGGATGCTCTGCAACCAGGCCGGAACCTGGTGGCAGCCGGCTACGCACTGTAT GGCAGTGCCACCATGCTGGTCCTTGCCATGGACTGTGGGGTCAACTGCTTCATGCTGGACCCGGCCATCGGGGAGTTCATTTTGGTGGAC AAGGATGTGAAGATAAAAAAGAAAGGTAAAATCTACAGCCTTAACGAGGGCTACGCCAGGGACTTTGACCCTGCCGTCACTGAGTACATC CAGAGGAAGAAGTTCCCCCCAGATAATTCAGCTCCTTATGGGGCCCGGTATGTGGGCTCCATGGTGGCTGATGTTCATCGCACTCTGGTC TACGGAGGGATATTTCTGTACCCCGCTAACAAGAAGAGCCCCAATGGAAAGCTGAGACTGCTGTACGAATGCAACCCCATGGCCTACGTC ATGGAGAAGGCTGGGGGAATGGCCACCACTGGGAAGGAGGCCGTGTTAGACGTCATTCCCACAGACATTCACCAGAGGGCGCCGGTGATC TTGGGATCCCCCGACGACGTGCTCGAGTTCCTGAAGGTGTATGAGAAGCACTCTGCCCAGTGAGCACCTGCCCTGCCTGCATCCGGAGAA TTGCCTCTACCTGGACCTTTTGTCTCACACAGCAGTACCCTGACCTGCTGTGCACCTTACATTCCTAGAGAGCAGAAATAAAAAGCATGA CTATTTCCACCATCAAATGCTGTAGAATGCTTGGCACTCCCTAACCAAATGCTGTCTCCATAATGCCACTGGTGTTAAGATATATTTTGA >26664_26664_9_ENSA-FBP1_ENSA_chr1_150598117_ENST00000369014_FBP1_chr9_97382773_ENST00000375326_length(amino acids)=424AA_BP=133 MSPFPLPLPFSGCLPGPLTLHSPGPAMSQKQEEENPAEETGEEKQDTQEKEGILPERAEEAKLKAKYPSLGQKPGGSDFLMKRLQKGQKY FDSGDYNMAKAKMKNKQLPSAGPDKNLVTGDHIPTPQDLPQRKSSLVTSKLAGYGIAGSTNVTGDQVKKLDVLSNDLVMNMLKSSFATCV LVSEEDKHAIIVEPEKRGKYVVCFDPLDGSSNIDCLVSVGTIFGIYRKKSTDEPSEKDALQPGRNLVAAGYALYGSATMLVLAMDCGVNC FMLDPAIGEFILVDKDVKIKKKGKIYSLNEGYARDFDPAVTEYIQRKKFPPDNSAPYGARYVGSMVADVHRTLVYGGIFLYPANKKSPNG -------------------------------------------------------------- >26664_26664_10_ENSA-FBP1_ENSA_chr1_150598117_ENST00000369014_FBP1_chr9_97382773_ENST00000415431_length(transcript)=1563nt_BP=476nt ATTTTGACTGAGCAACCCTAGTGACAGGAGCCGAAGCAGCAGCGCAGGTTGTCCCCGTTTCCCCTCCCCCTTCCCTTCTCCGGTTGCCTT CCCGGGCCCCTTACACTCCACAGTCCCGGTCCCGCCATGTCCCAGAAACAAGAAGAAGAGAACCCTGCGGAGGAGACCGGCGAGGAGAAG CAGGACACGCAGGAGAAAGAAGGTATTCTGCCTGAGAGAGCTGAAGAGGCAAAGCTAAAGGCCAAATACCCAAGCCTAGGACAAAAGCCT GGAGGCTCCGACTTCCTCATGAAGAGACTCCAGAAAGGGCAAAAGTACTTTGACTCAGGAGACTACAACATGGCCAAAGCCAAGATGAAG AATAAGCAGCTGCCAAGTGCAGGACCAGACAAGAACCTGGTGACTGGTGATCACATCCCCACCCCACAGGATCTGCCCCAGAGAAAGTCC TCGCTCGTCACCAGCAAGCTTGCGGGCTATGGCATTGCTGGTTCTACCAACGTGACAGGTGATCAAGTTAAGAAGCTGGACGTCCTCTCC AACGACCTGGTTATGAACATGTTAAAGTCATCCTTTGCCACGTGTGTTCTCGTGTCAGAAGAAGATAAACACGCCATCATAGTGGAACCG GAGAAAAGGGGTAAATATGTGGTCTGTTTTGATCCCCTTGATGGATCTTCCAACATCGATTGCCTTGTGTCCGTTGGAACCATTTTTGGC ATCTATAGAAAGAAATCAACTGATGAGCCTTCTGAGAAGGATGCTCTGCAACCAGGCCGGAACCTGGTGGCAGCCGGCTACGCACTGTAT GGCAGTGCCACCATGCTGGTCCTTGCCATGGACTGTGGGGTCAACTGCTTCATGCTGGACCCGGCCATCGGGGAGTTCATTTTGGTGGAC AAGGATGTGAAGATAAAAAAGAAAGGTAAAATCTACAGCCTTAACGAGGGCTACGCCAGGGACTTTGACCCTGCCGTCACTGAGTACATC CAGAGGAAGAAGTTCCCCCCAGATAATTCAGCTCCTTATGGGGCCCGGTATGTGGGCTCCATGGTGGCTGATGTTCATCGCACTCTGGTC TACGGAGGGATATTTCTGTACCCCGCTAACAAGAAGAGCCCCAATGGAAAGCTGAGACTGCTGTACGAATGCAACCCCATGGCCTACGTC ATGGAGAAGGCTGGGGGAATGGCCACCACTGGGAAGGAGGCCGTGTTAGACGTCATTCCCACAGACATTCACCAGAGGGCGCCGGTGATC TTGGGATCCCCCGACGACGTGCTCGAGTTCCTGAAGGTGTATGAGAAGCACTCTGCCCAGTGAGCACCTGCCCTGCCTGCATCCGGAGAA TTGCCTCTACCTGGACCTTTTGTCTCACACAGCAGTACCCTGACCTGCTGTGCACCTTACATTCCTAGAGAGCAGAAATAAAAAGCATGA CTATTTCCACCATCAAATGCTGTAGAATGCTTGGCACTCCCTAACCAAATGCTGTCTCCATAATGCCACTGGTGTTAAGATATATTTTGA >26664_26664_10_ENSA-FBP1_ENSA_chr1_150598117_ENST00000369014_FBP1_chr9_97382773_ENST00000415431_length(amino acids)=424AA_BP=133 MSPFPLPLPFSGCLPGPLTLHSPGPAMSQKQEEENPAEETGEEKQDTQEKEGILPERAEEAKLKAKYPSLGQKPGGSDFLMKRLQKGQKY FDSGDYNMAKAKMKNKQLPSAGPDKNLVTGDHIPTPQDLPQRKSSLVTSKLAGYGIAGSTNVTGDQVKKLDVLSNDLVMNMLKSSFATCV LVSEEDKHAIIVEPEKRGKYVVCFDPLDGSSNIDCLVSVGTIFGIYRKKSTDEPSEKDALQPGRNLVAAGYALYGSATMLVLAMDCGVNC FMLDPAIGEFILVDKDVKIKKKGKIYSLNEGYARDFDPAVTEYIQRKKFPPDNSAPYGARYVGSMVADVHRTLVYGGIFLYPANKKSPNG -------------------------------------------------------------- >26664_26664_11_ENSA-FBP1_ENSA_chr1_150598117_ENST00000369016_FBP1_chr9_97382773_ENST00000375326_length(transcript)=1589nt_BP=494nt CCGAAGCAGCAGCGCAGGTTGTCCCCGTTTCCCCTCCCCCTTCCCTTCTCCGGTTGCCTTCCCGGGCCCCTTACACTCCACAGTCCCGGT CCCGCCATGTCCCAGAAACAAGAAGAAGAGAACCCTGCGGAGGAGACCGGCGAGGAGAAGCAGGACACGCAGGAGAAAGAAGGTATTCTG CCTGAGAGAGCTGAAGAGGCAAAGCTAAAGGCCAAATACCCAAGCCTAGGACAAAAGCCTGGAGGCTCCGACTTCCTCATGAAGAGACTC CAGAAAGGGGATTATAAATCATTACATTGGAGTGTGCTTCTCTGTGCGGATGAAATGCAAAAGTACTTTGACTCAGGAGACTACAACATG GCCAAAGCCAAGATGAAGAATAAGCAGCTGCCAAGTGCAGGACCAGACAAGAACCTGGTGACTGGTGATCACATCCCCACCCCACAGGAT CTGCCCCAGAGAAAGTCCTCGCTCGTCACCAGCAAGCTTGCGGGCTATGGCATTGCTGGTTCTACCAACGTGACAGGTGATCAAGTTAAG AAGCTGGACGTCCTCTCCAACGACCTGGTTATGAACATGTTAAAGTCATCCTTTGCCACGTGTGTTCTCGTGTCAGAAGAAGATAAACAC GCCATCATAGTGGAACCGGAGAAAAGGGGTAAATATGTGGTCTGTTTTGATCCCCTTGATGGATCTTCCAACATCGATTGCCTTGTGTCC GTTGGAACCATTTTTGGCATCTATAGAAAGAAATCAACTGATGAGCCTTCTGAGAAGGATGCTCTGCAACCAGGCCGGAACCTGGTGGCA GCCGGCTACGCACTGTATGGCAGTGCCACCATGCTGGTCCTTGCCATGGACTGTGGGGTCAACTGCTTCATGCTGGACCCGGCCATCGGG GAGTTCATTTTGGTGGACAAGGATGTGAAGATAAAAAAGAAAGGTAAAATCTACAGCCTTAACGAGGGCTACGCCAGGGACTTTGACCCT GCCGTCACTGAGTACATCCAGAGGAAGAAGTTCCCCCCAGATAATTCAGCTCCTTATGGGGCCCGGTATGTGGGCTCCATGGTGGCTGAT GTTCATCGCACTCTGGTCTACGGAGGGATATTTCTGTACCCCGCTAACAAGAAGAGCCCCAATGGAAAGCTGAGACTGCTGTACGAATGC AACCCCATGGCCTACGTCATGGAGAAGGCTGGGGGAATGGCCACCACTGGGAAGGAGGCCGTGTTAGACGTCATTCCCACAGACATTCAC CAGAGGGCGCCGGTGATCTTGGGATCCCCCGACGACGTGCTCGAGTTCCTGAAGGTGTATGAGAAGCACTCTGCCCAGTGAGCACCTGCC CTGCCTGCATCCGGAGAATTGCCTCTACCTGGACCTTTTGTCTCACACAGCAGTACCCTGACCTGCTGTGCACCTTACATTCCTAGAGAG CAGAAATAAAAAGCATGACTATTTCCACCATCAAATGCTGTAGAATGCTTGGCACTCCCTAACCAAATGCTGTCTCCATAATGCCACTGG >26664_26664_11_ENSA-FBP1_ENSA_chr1_150598117_ENST00000369016_FBP1_chr9_97382773_ENST00000375326_length(amino acids)=440AA_BP=149 MSPFPLPLPFSGCLPGPLTLHSPGPAMSQKQEEENPAEETGEEKQDTQEKEGILPERAEEAKLKAKYPSLGQKPGGSDFLMKRLQKGDYK SLHWSVLLCADEMQKYFDSGDYNMAKAKMKNKQLPSAGPDKNLVTGDHIPTPQDLPQRKSSLVTSKLAGYGIAGSTNVTGDQVKKLDVLS NDLVMNMLKSSFATCVLVSEEDKHAIIVEPEKRGKYVVCFDPLDGSSNIDCLVSVGTIFGIYRKKSTDEPSEKDALQPGRNLVAAGYALY GSATMLVLAMDCGVNCFMLDPAIGEFILVDKDVKIKKKGKIYSLNEGYARDFDPAVTEYIQRKKFPPDNSAPYGARYVGSMVADVHRTLV -------------------------------------------------------------- >26664_26664_12_ENSA-FBP1_ENSA_chr1_150598117_ENST00000369016_FBP1_chr9_97382773_ENST00000415431_length(transcript)=1581nt_BP=494nt CCGAAGCAGCAGCGCAGGTTGTCCCCGTTTCCCCTCCCCCTTCCCTTCTCCGGTTGCCTTCCCGGGCCCCTTACACTCCACAGTCCCGGT CCCGCCATGTCCCAGAAACAAGAAGAAGAGAACCCTGCGGAGGAGACCGGCGAGGAGAAGCAGGACACGCAGGAGAAAGAAGGTATTCTG CCTGAGAGAGCTGAAGAGGCAAAGCTAAAGGCCAAATACCCAAGCCTAGGACAAAAGCCTGGAGGCTCCGACTTCCTCATGAAGAGACTC CAGAAAGGGGATTATAAATCATTACATTGGAGTGTGCTTCTCTGTGCGGATGAAATGCAAAAGTACTTTGACTCAGGAGACTACAACATG GCCAAAGCCAAGATGAAGAATAAGCAGCTGCCAAGTGCAGGACCAGACAAGAACCTGGTGACTGGTGATCACATCCCCACCCCACAGGAT CTGCCCCAGAGAAAGTCCTCGCTCGTCACCAGCAAGCTTGCGGGCTATGGCATTGCTGGTTCTACCAACGTGACAGGTGATCAAGTTAAG AAGCTGGACGTCCTCTCCAACGACCTGGTTATGAACATGTTAAAGTCATCCTTTGCCACGTGTGTTCTCGTGTCAGAAGAAGATAAACAC GCCATCATAGTGGAACCGGAGAAAAGGGGTAAATATGTGGTCTGTTTTGATCCCCTTGATGGATCTTCCAACATCGATTGCCTTGTGTCC GTTGGAACCATTTTTGGCATCTATAGAAAGAAATCAACTGATGAGCCTTCTGAGAAGGATGCTCTGCAACCAGGCCGGAACCTGGTGGCA GCCGGCTACGCACTGTATGGCAGTGCCACCATGCTGGTCCTTGCCATGGACTGTGGGGTCAACTGCTTCATGCTGGACCCGGCCATCGGG GAGTTCATTTTGGTGGACAAGGATGTGAAGATAAAAAAGAAAGGTAAAATCTACAGCCTTAACGAGGGCTACGCCAGGGACTTTGACCCT GCCGTCACTGAGTACATCCAGAGGAAGAAGTTCCCCCCAGATAATTCAGCTCCTTATGGGGCCCGGTATGTGGGCTCCATGGTGGCTGAT GTTCATCGCACTCTGGTCTACGGAGGGATATTTCTGTACCCCGCTAACAAGAAGAGCCCCAATGGAAAGCTGAGACTGCTGTACGAATGC AACCCCATGGCCTACGTCATGGAGAAGGCTGGGGGAATGGCCACCACTGGGAAGGAGGCCGTGTTAGACGTCATTCCCACAGACATTCAC CAGAGGGCGCCGGTGATCTTGGGATCCCCCGACGACGTGCTCGAGTTCCTGAAGGTGTATGAGAAGCACTCTGCCCAGTGAGCACCTGCC CTGCCTGCATCCGGAGAATTGCCTCTACCTGGACCTTTTGTCTCACACAGCAGTACCCTGACCTGCTGTGCACCTTACATTCCTAGAGAG CAGAAATAAAAAGCATGACTATTTCCACCATCAAATGCTGTAGAATGCTTGGCACTCCCTAACCAAATGCTGTCTCCATAATGCCACTGG >26664_26664_12_ENSA-FBP1_ENSA_chr1_150598117_ENST00000369016_FBP1_chr9_97382773_ENST00000415431_length(amino acids)=440AA_BP=149 MSPFPLPLPFSGCLPGPLTLHSPGPAMSQKQEEENPAEETGEEKQDTQEKEGILPERAEEAKLKAKYPSLGQKPGGSDFLMKRLQKGDYK SLHWSVLLCADEMQKYFDSGDYNMAKAKMKNKQLPSAGPDKNLVTGDHIPTPQDLPQRKSSLVTSKLAGYGIAGSTNVTGDQVKKLDVLS NDLVMNMLKSSFATCVLVSEEDKHAIIVEPEKRGKYVVCFDPLDGSSNIDCLVSVGTIFGIYRKKSTDEPSEKDALQPGRNLVAAGYALY GSATMLVLAMDCGVNCFMLDPAIGEFILVDKDVKIKKKGKIYSLNEGYARDFDPAVTEYIQRKKFPPDNSAPYGARYVGSMVADVHRTLV -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ENSA-FBP1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ENSA-FBP1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ENSA-FBP1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
