
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:EP300-HAUS8 (FusionGDB2 ID:26751) |
Fusion Gene Summary for EP300-HAUS8 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: EP300-HAUS8 | Fusion gene ID: 26751 | Hgene | Tgene | Gene symbol | EP300 | HAUS8 | Gene ID | 2033 | 93323 |
| Gene name | E1A binding protein p300 | HAUS augmin like complex subunit 8 | |
| Synonyms | KAT3B|MKHK2|RSTS2|p300 | DGT4|HICE1|NY-SAR-48 | |
| Cytomap | 22q13.2 | 19p13.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | histone acetyltransferase p300E1A-associated protein p300E1A-binding protein, 300kDhistone butyryltransferase p300histone crotonyltransferase p300p300 HATprotein 2-hydroxyisobutyryltransferase p300protein propionyltransferase p300 | HAUS augmin-like complex subunit 8HEC1/NDC80 interacting, centrosome associated 1HEC1/NDC80-interacting centrosome-associated protein 1Hec1-interacting and centrosome-associated 1sarcoma antigen NY-SAR-48 | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | Q09472 | Q9BT25 | |
| Ensembl transtripts involved in fusion gene | ENST00000263253, | ENST00000253669, ENST00000448593, ENST00000593360, | |
| Fusion gene scores | * DoF score | 20 X 17 X 12=4080 | 7 X 8 X 6=336 |
| # samples | 29 | 7 | |
| ** MAII score | log2(29/4080*10)=-3.81444434684392 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/336*10)=-2.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: EP300 [Title/Abstract] AND HAUS8 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | EP300(41513825)-HAUS8(17166812), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | EP300 | GO:0000122 | negative regulation of transcription by RNA polymerase II | 10733570 |
| Hgene | EP300 | GO:0001666 | response to hypoxia | 9887100|15261140 |
| Hgene | EP300 | GO:0006110 | regulation of glycolytic process | 29775581 |
| Hgene | EP300 | GO:0006355 | regulation of transcription, DNA-templated | 15261140 |
| Hgene | EP300 | GO:0006473 | protein acetylation | 21030595|24939902 |
| Hgene | EP300 | GO:0006475 | internal protein amino acid acetylation | 18722353 |
| Hgene | EP300 | GO:0010742 | macrophage derived foam cell differentiation | 26504087 |
| Hgene | EP300 | GO:0010976 | positive regulation of neuron projection development | 27256286 |
| Hgene | EP300 | GO:0016573 | histone acetylation | 25818647|27256286 |
| Hgene | EP300 | GO:0018076 | N-terminal peptidyl-lysine acetylation | 12435739 |
| Hgene | EP300 | GO:0018393 | internal peptidyl-lysine acetylation | 17403783 |
| Hgene | EP300 | GO:0018394 | peptidyl-lysine acetylation | 23811396|23962722 |
| Hgene | EP300 | GO:0031333 | negative regulation of protein complex assembly | 23962722 |
| Hgene | EP300 | GO:0034644 | cellular response to UV | 24939902 |
| Hgene | EP300 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 17403783 |
| Hgene | EP300 | GO:0043627 | response to estrogen | 11581164 |
| Hgene | EP300 | GO:0043923 | positive regulation by host of viral transcription | 16687403 |
| Hgene | EP300 | GO:0043969 | histone H2B acetylation | 23415232 |
| Hgene | EP300 | GO:0045721 | negative regulation of gluconeogenesis | 30193097 |
| Hgene | EP300 | GO:0045815 | positive regulation of gene expression, epigenetic | 25818647 |
| Hgene | EP300 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 12586840|18722353|23811396 |
| Hgene | EP300 | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 10518217|25818647 |
| Hgene | EP300 | GO:0060765 | regulation of androgen receptor signaling pathway | 18487222 |
| Hgene | EP300 | GO:0061921 | peptidyl-lysine propionylation | 17267393 |
| Hgene | EP300 | GO:0090043 | regulation of tubulin deacetylation | 18722353 |
| Hgene | EP300 | GO:0140066 | peptidyl-lysine crotonylation | 25818647 |
| Hgene | EP300 | GO:0140067 | peptidyl-lysine butyrylation | 17267393|29775581 |
| Hgene | EP300 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling | 23811396 |
| Hgene | EP300 | GO:1905636 | positive regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 23811396 |
 Fusion gene breakpoints across EP300 (5'-gene) Fusion gene breakpoints across EP300 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
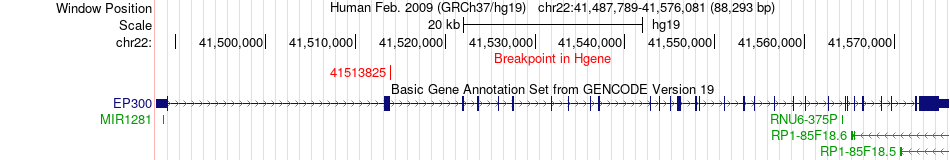 |
 Fusion gene breakpoints across HAUS8 (3'-gene) Fusion gene breakpoints across HAUS8 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
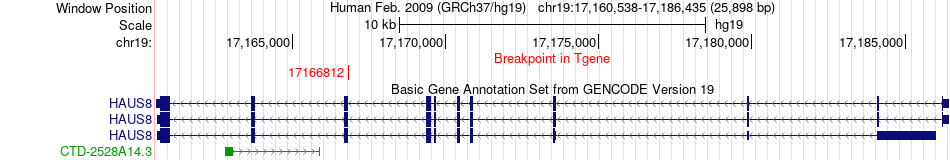 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BLCA | TCGA-4Z-AA84-01A | EP300 | chr22 | 41513825 | - | HAUS8 | chr19 | 17166812 | - |
| ChimerDB4 | BLCA | TCGA-4Z-AA84-01A | EP300 | chr22 | 41513825 | + | HAUS8 | chr19 | 17166812 | - |
| ChimerDB4 | BLCA | TCGA-4Z-AA84 | EP300 | chr22 | 41513825 | + | HAUS8 | chr19 | 17166812 | - |
Top |
Fusion Gene ORF analysis for EP300-HAUS8 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000263253 | ENST00000253669 | EP300 | chr22 | 41513825 | + | HAUS8 | chr19 | 17166812 | - |
| In-frame | ENST00000263253 | ENST00000448593 | EP300 | chr22 | 41513825 | + | HAUS8 | chr19 | 17166812 | - |
| In-frame | ENST00000263253 | ENST00000593360 | EP300 | chr22 | 41513825 | + | HAUS8 | chr19 | 17166812 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000263253 | EP300 | chr22 | 41513825 | + | ENST00000253669 | HAUS8 | chr19 | 17166812 | - | 2680 | 1948 | 1219 | 2535 | 438 |
| ENST00000263253 | EP300 | chr22 | 41513825 | + | ENST00000448593 | HAUS8 | chr19 | 17166812 | - | 2648 | 1948 | 1219 | 2535 | 438 |
| ENST00000263253 | EP300 | chr22 | 41513825 | + | ENST00000593360 | HAUS8 | chr19 | 17166812 | - | 2643 | 1948 | 1219 | 2535 | 438 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000263253 | ENST00000253669 | EP300 | chr22 | 41513825 | + | HAUS8 | chr19 | 17166812 | - | 0.007326284 | 0.9926737 |
| ENST00000263253 | ENST00000448593 | EP300 | chr22 | 41513825 | + | HAUS8 | chr19 | 17166812 | - | 0.007775414 | 0.9922246 |
| ENST00000263253 | ENST00000593360 | EP300 | chr22 | 41513825 | + | HAUS8 | chr19 | 17166812 | - | 0.007857205 | 0.9921428 |
Top |
Fusion Genomic Features for EP300-HAUS8 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
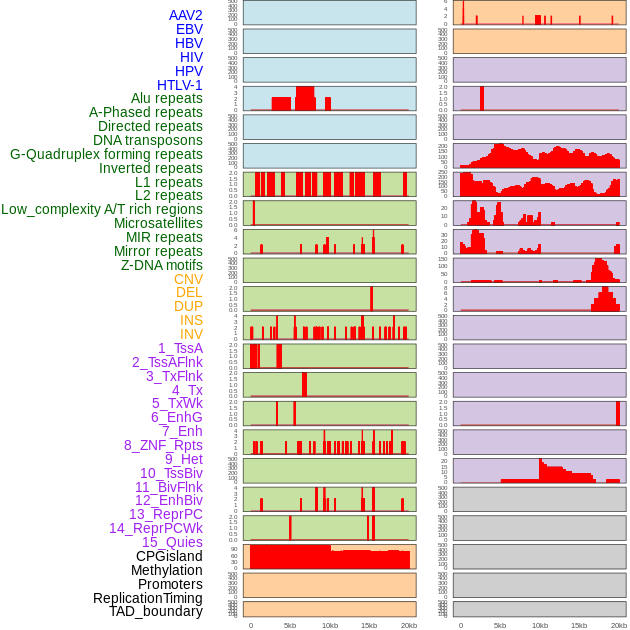 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for EP300-HAUS8 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr22:41513825/chr19:17166812) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| EP300 | HAUS8 |
| FUNCTION: Functions as histone acetyltransferase and regulates transcription via chromatin remodeling (PubMed:23415232, PubMed:23934153, PubMed:8945521). Acetylates all four core histones in nucleosomes. Histone acetylation gives an epigenetic tag for transcriptional activation (PubMed:23415232, PubMed:23934153, PubMed:8945521). Mediates cAMP-gene regulation by binding specifically to phosphorylated CREB protein. Mediates acetylation of histone H3 at 'Lys-122' (H3K122ac), a modification that localizes at the surface of the histone octamer and stimulates transcription, possibly by promoting nucleosome instability. Mediates acetylation of histone H3 at 'Lys-27' (H3K27ac) (PubMed:23911289). Also functions as acetyltransferase for non-histone targets, such as ALX1, HDAC1, PRMT1 or SIRT2 (PubMed:12929931, PubMed:16762839, PubMed:18722353). Acetylates 'Lys-131' of ALX1 and acts as its coactivator (PubMed:12929931). Acetylates SIRT2 and is proposed to indirectly increase the transcriptional activity of p53/TP53 through acetylation and subsequent attenuation of SIRT2 deacetylase function (PubMed:18722353). Following DNA damage, forms a stress-responsive p53/TP53 coactivator complex with JMY which mediates p53/TP53 acetylation, thereby increasing p53/TP53-dependent transcription and apoptosis (PubMed:11511361, PubMed:15448695). Promotes chromatin acetylation in heat shock responsive HSP genes during the heat shock response (HSR), thereby stimulating HSR transcription (PubMed:18451878). Acetylates HDAC1 leading to its inactivation and modulation of transcription (PubMed:16762839). Acetylates 'Lys-247' of EGR2 (By similarity). Acts as a TFAP2A-mediated transcriptional coactivator in presence of CITED2 (PubMed:12586840). Plays a role as a coactivator of NEUROD1-dependent transcription of the secretin and p21 genes and controls terminal differentiation of cells in the intestinal epithelium. Promotes cardiac myocyte enlargement. Can also mediate transcriptional repression. Acetylates FOXO1 and enhances its transcriptional activity (PubMed:15890677). Acetylates BCL6 wich disrupts its ability to recruit histone deacetylases and hinders its transcriptional repressor activity (PubMed:12402037). Participates in CLOCK or NPAS2-regulated rhythmic gene transcription; exhibits a circadian association with CLOCK or NPAS2, correlating with increase in PER1/2 mRNA and histone H3 acetylation on the PER1/2 promoter (PubMed:14645221). Acetylates MTA1 at 'Lys-626' which is essential for its transcriptional coactivator activity (PubMed:16617102). Acetylates XBP1 isoform 2; acetylation increases protein stability of XBP1 isoform 2 and enhances its transcriptional activity (PubMed:20955178). Acetylates PCNA; acetylation promotes removal of chromatin-bound PCNA and its degradation during nucleotide excision repair (NER) (PubMed:24939902). Acetylates MEF2D (PubMed:21030595). Acetylates and stabilizes ZBTB7B protein by antagonizing ubiquitin conjugation and degradation, this mechanism may be involved in CD4/CD8 lineage differentiation (PubMed:20810990). Acetylates GABPB1, impairing GABPB1 heterotetramerization and activity (By similarity). Acetylates PCK1 and promotes PCK1 anaplerotic activity (PubMed:30193097). Acetylates RXRA and RXRG (PubMed:17761950). In addition to protein acetyltransferase, can use different acyl-CoA substrates, such as (2E)-butenoyl-CoA (crotonyl-CoA), butanoyl-CoA (butyryl-CoA), 2-hydroxyisobutanoyl-CoA (2-hydroxyisobutyryl-CoA), lactoyl-CoA or propanoyl-CoA (propionyl-CoA), and is able to mediate protein crotonylation, butyrylation, 2-hydroxyisobutyrylation, lactylation or propionylation, respectively (PubMed:17267393, PubMed:25818647, PubMed:29775581, PubMed:31645732). Acts as a histone crotonyltransferase; crotonylation marks active promoters and enhancers and confers resistance to transcriptional repressors (PubMed:25818647). Histone crotonyltransferase activity is dependent on the concentration of (2E)-butenoyl-CoA (crotonyl-CoA) substrate and such activity is weak when (2E)-butenoyl-CoA (crotonyl-CoA) concentration is low (PubMed:25818647). Also acts as a histone butyryltransferase; butyrylation marks active promoters (PubMed:17267393). Catalyzes histone lactylation in macrophages by using lactoyl-CoA directly derived from endogenous or exogenous lactate, leading to stimulates gene transcription (PubMed:31645732). Acts as a protein-lysine 2-hydroxyisobutyryltransferase; regulates glycolysis by mediating 2-hydroxyisobutyrylation of glycolytic enzymes (PubMed:29775581). Functions as a transcriptional coactivator for SMAD4 in the TGF-beta signaling pathway (PubMed:25514493). {ECO:0000250|UniProtKB:B2RWS6, ECO:0000269|PubMed:10733570, ECO:0000269|PubMed:11430825, ECO:0000269|PubMed:11511361, ECO:0000269|PubMed:11701890, ECO:0000269|PubMed:12402037, ECO:0000269|PubMed:12586840, ECO:0000269|PubMed:12929931, ECO:0000269|PubMed:14645221, ECO:0000269|PubMed:15186775, ECO:0000269|PubMed:15448695, ECO:0000269|PubMed:15890677, ECO:0000269|PubMed:16617102, ECO:0000269|PubMed:16762839, ECO:0000269|PubMed:17267393, ECO:0000269|PubMed:17761950, ECO:0000269|PubMed:18451878, ECO:0000269|PubMed:18722353, ECO:0000269|PubMed:18995842, ECO:0000269|PubMed:20810990, ECO:0000269|PubMed:21030595, ECO:0000269|PubMed:23415232, ECO:0000269|PubMed:23911289, ECO:0000269|PubMed:23934153, ECO:0000269|PubMed:24939902, ECO:0000269|PubMed:25514493, ECO:0000269|PubMed:25818647, ECO:0000269|PubMed:29775581, ECO:0000269|PubMed:30193097, ECO:0000269|PubMed:31645732, ECO:0000269|PubMed:8945521, ECO:0000305|PubMed:20955178}.; FUNCTION: (Microbial infection) In case of HIV-1 infection, it is recruited by the viral protein Tat. Regulates Tat's transactivating activity and may help inducing chromatin remodeling of proviral genes. Binds to and may be involved in the transforming capacity of the adenovirus E1A protein. {ECO:0000269|PubMed:10545121, ECO:0000269|PubMed:11080476}. | FUNCTION: Contributes to mitotic spindle assembly, maintenance of centrosome integrity and completion of cytokinesis as part of the HAUS augmin-like complex. {ECO:0000269|PubMed:18362163, ECO:0000269|PubMed:19369198, ECO:0000269|PubMed:19427217}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 11_17 | 243 | 2415.0 | Motif | Nuclear localization signal |
| Tgene | HAUS8 | chr22:41513825 | chr19:17166812 | ENST00000593360 | 6 | 10 | 156_208 | 154 | 350.0 | Coiled coil | Ontology_term=ECO:0000255 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 1519_1526 | 243 | 2415.0 | Compositional bias | Note=Poly-Glu |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 2066_2069 | 243 | 2415.0 | Compositional bias | Note=Poly-Gln |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 2190_2195 | 243 | 2415.0 | Compositional bias | Note=Poly-Gln |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 797_800 | 243 | 2415.0 | Compositional bias | Note=Poly-Ser |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 1067_1139 | 243 | 2415.0 | Domain | Bromo |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 1287_1663 | 243 | 2415.0 | Domain | CBP/p300-type HAT |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 566_645 | 243 | 2415.0 | Domain | KIX |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 1017_1029 | 243 | 2415.0 | Region | Note=CRD1%3B mediates transcriptional repression |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 1398_1400 | 243 | 2415.0 | Region | Acetyl-CoA binding |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 1410_1411 | 243 | 2415.0 | Region | Acetyl-CoA binding |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 1572_1818 | 243 | 2415.0 | Region | Note=Binding region for E1A adenovirus |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 1664_1707 | 243 | 2415.0 | Zinc finger | ZZ-type |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 1728_1809 | 243 | 2415.0 | Zinc finger | TAZ-type 2 |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 331_417 | 243 | 2415.0 | Zinc finger | TAZ-type 1 |
| Tgene | HAUS8 | chr22:41513825 | chr19:17166812 | ENST00000253669 | 7 | 11 | 156_208 | 215 | 411.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | HAUS8 | chr22:41513825 | chr19:17166812 | ENST00000448593 | 7 | 11 | 156_208 | 214 | 410.0 | Coiled coil | Ontology_term=ECO:0000255 |
Top |
Fusion Gene Sequence for EP300-HAUS8 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >26751_26751_1_EP300-HAUS8_EP300_chr22_41513825_ENST00000263253_HAUS8_chr19_17166812_ENST00000253669_length(transcript)=2680nt_BP=1948nt CTTGTTTGTGTGCTAGGCTGGGGGGGAGAGAGGGCGAGAGAGAGCGGGCGAGAGTGGGCAAGCAGGACGCCGGGCTGAGTGCTAACTGCG GGAGCCAGAGAGTGCGGAGGGGAGTCGGGTCGGAGAGAGGCGGCAGGGGCCGAGACAGTGGCAGGGGGCCCGGGGCGCACGGGCTGAGGC GACCCCCAGCCCCCTCCCGTCCGCACACACCCCCACCGCGGTCCAGCAGCCGGGCCGGCGTCGACGCCTAGGGGGGACCATTACATAACC CCGCGCCCCGCGGCGTCTTCTCCCGCCGCCGCGGGCGGCCCCGAACGGAGCCCCGGGGGGCGGGCGCTCCCAGCACCTGGCCGCCGGCGG TGGGGGCCGTAGCAGCGGCCGTATTTATTTATTTTCCGCGGGAAAGGAAGGCGAAGGAGGGGAGCGCGGCGCGAGGAGGGGCCGCCTGCG CCGCCGCCGGAGCGGGGCCTCCTCGGTGGGCTCCGCGTCGGCGCGGGCGTGCGGGCGGCGCTGCTCGGCCCGGCCCCCTCGGCCCTCTGG TCCGGCCAGCTCCGCTCCCGGCGTCCTTGCCGCGCCTCCGCCGGCCGCCGCGCGATGTGAGGCGGCGGCGCCAGCCTGGCTCTCGGCTCG GGCGAGTTCTCTGCGGCCATTAGGGGCCGGTGCGGCGGCGGCGGCGCGGAGCGCGGCGGCAGGAGGAGGGTTCGGAGGGTGGGGGCGCAG GCCCGGGAGGGGGCACCGGGAGGAGGTGAGTGTCTCTTGTCGCCTCCTCCTCTCCCCCCTTTTCGCCCCCGCCTCCTTGTGGCGATGAGA AGGAGGAGGACAGCGCCGAGGAGGAAGAGGTTGATGGCGGCGGCGGAGCTCCGAGAGACCTCGGCTGGGCAGGGGCCGGCCGTGGCGGGC CGGGGACTGCGCCTCTAGAGCCGCGAGTTCTCGGGAATTCGCCGCAGCGGACGCGCTCGGCGAATTTGTGCTCTTGTGCCCTCCTCCGGG CTTGGGCCCAGGCCCGGCCCCTCGCACTTGCCCTTACCTTTTCTATCGAGTCCGCATCCCTCTCCAGCCACTGCGACCCGGCGAAGAGAA AAAGGAACTTCCCCCACCCCCTCGGGTGCCGTCGGAGCCCCCCAGCCCACCCCTGGGTGCGGCGCGGGGACCCCGGGCCGAAGAAGAGAT TTCCTGAGGATTCTGGTTTTCCTCGCTTGTATCTCCGAAAGAATTAAAAATGGCCGAGAATGTGGTGGAACCGGGGCCGCCTTCAGCCAA GCGGCCTAAACTCTCATCTCCGGCCCTCTCGGCGTCCGCCAGCGATGGCACAGATTTTGGCTCTCTATTTGACTTGGAGCACGACTTACC AGATGAATTAATCAACTCTACAGAATTGGGACTAACCAATGGTGGTGATATTAATCAGCTTCAGACAAGTCTTGGCATGGTACAAGATGC AGCTTCTAAACATAAACAGCTGTCAGAATTGCTGCGATCTGGTAGTTCCCCTAACCTCAATATGGGAGTTGGTGGCCCAGGTCAAGTCAT GGCCAGCCAGGCCCAACAGAGCAGTCCTGGATTAGGTTTGATAAATAGCATGGTCAAAAGCCCAATGACACAGGCAGGCTTGACTTCTCC CAACATGGGGATGGGCACTAGTGGACCAAATCAGGGTCCTACGCAGTCAACAGGTATGATGAACAGTCCAGTAAATCAGCCTGCCATGGG AATGAACACAGGGATGAATGCGGGCATGAATCCTGGAATGTTGGCTGCAGGCAATGGACAAGGGATAATGCCTAATCAAGTCATGAACGG TTCAATTGGAGCAGGCCGAGGGCGACAGAATATGCAGTACCCAAACCCAGGCATGGGAAGTGCTGGCAACTTACTGACTGAGCCTCTTCA GCAGGGCTCTCCCCAGATGGGAGGACAAACAGGATTGAGAGGCCCCCAGCCTCTTAAGATCGAGATGCTCAGCCCCTTCGAGGCAGTGGC CACACGCTTCAAGGAGCAATACAGGACATTCGCCACGGCCCTGGACACTACCAGGCACGAGCTGCCCGTGAGGTCCATCCACCTGGAGGG AGATGGGCAGCAGCTCTTAGACGCCCTGCAGCATGAACTGGTGACCACTCAGCGCCTCCTGGGAGAACTTGATGTTGGTGATTCGGAAGA AAATGTGCAGGTGCTGGACTTACTGAGCGAACTCAAGGACGTGACGGCGAAAAAGGACCTTGAGCTCCGAAGGAGCTTTGCCCAGGTGCT GGAACTCTCCGCAGAGGCAAGCAAAGAGGCAGCCTTGGCAAACCAGGAAGTCTGGGAAGAGACCCAGGGCATGGCGCCCCCCAGCCGGTG GTATTTCAATCAAGACAGTGCCTGCAGAGAATCTGGGGGAGCACCCAAGAACACGCCCCTGTCTGAGGACGACAACCCGGGTGCCTCGTC AGCCCCCGCTCAGGCCACGTTCATCAGCCCAAGCGAAGATTTTTCTTCAAGCAGCCAGGCAGAAGTCCCGCCCTCTCTCTCTCGTTCAGG GAGGGACTTGTCATGACTCATGGTTACATTCAGGATACTTGAGCACTTTATATACTACCGTAGCACTGTAGCTATTTTTTATGTGATAAT >26751_26751_1_EP300-HAUS8_EP300_chr22_41513825_ENST00000263253_HAUS8_chr19_17166812_ENST00000253669_length(amino acids)=438AA_BP=241 MAENVVEPGPPSAKRPKLSSPALSASASDGTDFGSLFDLEHDLPDELINSTELGLTNGGDINQLQTSLGMVQDAASKHKQLSELLRSGSS PNLNMGVGGPGQVMASQAQQSSPGLGLINSMVKSPMTQAGLTSPNMGMGTSGPNQGPTQSTGMMNSPVNQPAMGMNTGMNAGMNPGMLAA GNGQGIMPNQVMNGSIGAGRGRQNMQYPNPGMGSAGNLLTEPLQQGSPQMGGQTGLRGPQPLKIEMLSPFEAVATRFKEQYRTFATALDT TRHELPVRSIHLEGDGQQLLDALQHELVTTQRLLGELDVGDSEENVQVLDLLSELKDVTAKKDLELRRSFAQVLELSAEASKEAALANQE -------------------------------------------------------------- >26751_26751_2_EP300-HAUS8_EP300_chr22_41513825_ENST00000263253_HAUS8_chr19_17166812_ENST00000448593_length(transcript)=2648nt_BP=1948nt CTTGTTTGTGTGCTAGGCTGGGGGGGAGAGAGGGCGAGAGAGAGCGGGCGAGAGTGGGCAAGCAGGACGCCGGGCTGAGTGCTAACTGCG GGAGCCAGAGAGTGCGGAGGGGAGTCGGGTCGGAGAGAGGCGGCAGGGGCCGAGACAGTGGCAGGGGGCCCGGGGCGCACGGGCTGAGGC GACCCCCAGCCCCCTCCCGTCCGCACACACCCCCACCGCGGTCCAGCAGCCGGGCCGGCGTCGACGCCTAGGGGGGACCATTACATAACC CCGCGCCCCGCGGCGTCTTCTCCCGCCGCCGCGGGCGGCCCCGAACGGAGCCCCGGGGGGCGGGCGCTCCCAGCACCTGGCCGCCGGCGG TGGGGGCCGTAGCAGCGGCCGTATTTATTTATTTTCCGCGGGAAAGGAAGGCGAAGGAGGGGAGCGCGGCGCGAGGAGGGGCCGCCTGCG CCGCCGCCGGAGCGGGGCCTCCTCGGTGGGCTCCGCGTCGGCGCGGGCGTGCGGGCGGCGCTGCTCGGCCCGGCCCCCTCGGCCCTCTGG TCCGGCCAGCTCCGCTCCCGGCGTCCTTGCCGCGCCTCCGCCGGCCGCCGCGCGATGTGAGGCGGCGGCGCCAGCCTGGCTCTCGGCTCG GGCGAGTTCTCTGCGGCCATTAGGGGCCGGTGCGGCGGCGGCGGCGCGGAGCGCGGCGGCAGGAGGAGGGTTCGGAGGGTGGGGGCGCAG GCCCGGGAGGGGGCACCGGGAGGAGGTGAGTGTCTCTTGTCGCCTCCTCCTCTCCCCCCTTTTCGCCCCCGCCTCCTTGTGGCGATGAGA AGGAGGAGGACAGCGCCGAGGAGGAAGAGGTTGATGGCGGCGGCGGAGCTCCGAGAGACCTCGGCTGGGCAGGGGCCGGCCGTGGCGGGC CGGGGACTGCGCCTCTAGAGCCGCGAGTTCTCGGGAATTCGCCGCAGCGGACGCGCTCGGCGAATTTGTGCTCTTGTGCCCTCCTCCGGG CTTGGGCCCAGGCCCGGCCCCTCGCACTTGCCCTTACCTTTTCTATCGAGTCCGCATCCCTCTCCAGCCACTGCGACCCGGCGAAGAGAA AAAGGAACTTCCCCCACCCCCTCGGGTGCCGTCGGAGCCCCCCAGCCCACCCCTGGGTGCGGCGCGGGGACCCCGGGCCGAAGAAGAGAT TTCCTGAGGATTCTGGTTTTCCTCGCTTGTATCTCCGAAAGAATTAAAAATGGCCGAGAATGTGGTGGAACCGGGGCCGCCTTCAGCCAA GCGGCCTAAACTCTCATCTCCGGCCCTCTCGGCGTCCGCCAGCGATGGCACAGATTTTGGCTCTCTATTTGACTTGGAGCACGACTTACC AGATGAATTAATCAACTCTACAGAATTGGGACTAACCAATGGTGGTGATATTAATCAGCTTCAGACAAGTCTTGGCATGGTACAAGATGC AGCTTCTAAACATAAACAGCTGTCAGAATTGCTGCGATCTGGTAGTTCCCCTAACCTCAATATGGGAGTTGGTGGCCCAGGTCAAGTCAT GGCCAGCCAGGCCCAACAGAGCAGTCCTGGATTAGGTTTGATAAATAGCATGGTCAAAAGCCCAATGACACAGGCAGGCTTGACTTCTCC CAACATGGGGATGGGCACTAGTGGACCAAATCAGGGTCCTACGCAGTCAACAGGTATGATGAACAGTCCAGTAAATCAGCCTGCCATGGG AATGAACACAGGGATGAATGCGGGCATGAATCCTGGAATGTTGGCTGCAGGCAATGGACAAGGGATAATGCCTAATCAAGTCATGAACGG TTCAATTGGAGCAGGCCGAGGGCGACAGAATATGCAGTACCCAAACCCAGGCATGGGAAGTGCTGGCAACTTACTGACTGAGCCTCTTCA GCAGGGCTCTCCCCAGATGGGAGGACAAACAGGATTGAGAGGCCCCCAGCCTCTTAAGATCGAGATGCTCAGCCCCTTCGAGGCAGTGGC CACACGCTTCAAGGAGCAATACAGGACATTCGCCACGGCCCTGGACACTACCAGGCACGAGCTGCCCGTGAGGTCCATCCACCTGGAGGG AGATGGGCAGCAGCTCTTAGACGCCCTGCAGCATGAACTGGTGACCACTCAGCGCCTCCTGGGAGAACTTGATGTTGGTGATTCGGAAGA AAATGTGCAGGTGCTGGACTTACTGAGCGAACTCAAGGACGTGACGGCGAAAAAGGACCTTGAGCTCCGAAGGAGCTTTGCCCAGGTGCT GGAACTCTCCGCAGAGGCAAGCAAAGAGGCAGCCTTGGCAAACCAGGAAGTCTGGGAAGAGACCCAGGGCATGGCGCCCCCCAGCCGGTG GTATTTCAATCAAGACAGTGCCTGCAGAGAATCTGGGGGAGCACCCAAGAACACGCCCCTGTCTGAGGACGACAACCCGGGTGCCTCGTC AGCCCCCGCTCAGGCCACGTTCATCAGCCCAAGCGAAGATTTTTCTTCAAGCAGCCAGGCAGAAGTCCCGCCCTCTCTCTCTCGTTCAGG GAGGGACTTGTCATGACTCATGGTTACATTCAGGATACTTGAGCACTTTATATACTACCGTAGCACTGTAGCTATTTTTTATGTGATAAT >26751_26751_2_EP300-HAUS8_EP300_chr22_41513825_ENST00000263253_HAUS8_chr19_17166812_ENST00000448593_length(amino acids)=438AA_BP=241 MAENVVEPGPPSAKRPKLSSPALSASASDGTDFGSLFDLEHDLPDELINSTELGLTNGGDINQLQTSLGMVQDAASKHKQLSELLRSGSS PNLNMGVGGPGQVMASQAQQSSPGLGLINSMVKSPMTQAGLTSPNMGMGTSGPNQGPTQSTGMMNSPVNQPAMGMNTGMNAGMNPGMLAA GNGQGIMPNQVMNGSIGAGRGRQNMQYPNPGMGSAGNLLTEPLQQGSPQMGGQTGLRGPQPLKIEMLSPFEAVATRFKEQYRTFATALDT TRHELPVRSIHLEGDGQQLLDALQHELVTTQRLLGELDVGDSEENVQVLDLLSELKDVTAKKDLELRRSFAQVLELSAEASKEAALANQE -------------------------------------------------------------- >26751_26751_3_EP300-HAUS8_EP300_chr22_41513825_ENST00000263253_HAUS8_chr19_17166812_ENST00000593360_length(transcript)=2643nt_BP=1948nt CTTGTTTGTGTGCTAGGCTGGGGGGGAGAGAGGGCGAGAGAGAGCGGGCGAGAGTGGGCAAGCAGGACGCCGGGCTGAGTGCTAACTGCG GGAGCCAGAGAGTGCGGAGGGGAGTCGGGTCGGAGAGAGGCGGCAGGGGCCGAGACAGTGGCAGGGGGCCCGGGGCGCACGGGCTGAGGC GACCCCCAGCCCCCTCCCGTCCGCACACACCCCCACCGCGGTCCAGCAGCCGGGCCGGCGTCGACGCCTAGGGGGGACCATTACATAACC CCGCGCCCCGCGGCGTCTTCTCCCGCCGCCGCGGGCGGCCCCGAACGGAGCCCCGGGGGGCGGGCGCTCCCAGCACCTGGCCGCCGGCGG TGGGGGCCGTAGCAGCGGCCGTATTTATTTATTTTCCGCGGGAAAGGAAGGCGAAGGAGGGGAGCGCGGCGCGAGGAGGGGCCGCCTGCG CCGCCGCCGGAGCGGGGCCTCCTCGGTGGGCTCCGCGTCGGCGCGGGCGTGCGGGCGGCGCTGCTCGGCCCGGCCCCCTCGGCCCTCTGG TCCGGCCAGCTCCGCTCCCGGCGTCCTTGCCGCGCCTCCGCCGGCCGCCGCGCGATGTGAGGCGGCGGCGCCAGCCTGGCTCTCGGCTCG GGCGAGTTCTCTGCGGCCATTAGGGGCCGGTGCGGCGGCGGCGGCGCGGAGCGCGGCGGCAGGAGGAGGGTTCGGAGGGTGGGGGCGCAG GCCCGGGAGGGGGCACCGGGAGGAGGTGAGTGTCTCTTGTCGCCTCCTCCTCTCCCCCCTTTTCGCCCCCGCCTCCTTGTGGCGATGAGA AGGAGGAGGACAGCGCCGAGGAGGAAGAGGTTGATGGCGGCGGCGGAGCTCCGAGAGACCTCGGCTGGGCAGGGGCCGGCCGTGGCGGGC CGGGGACTGCGCCTCTAGAGCCGCGAGTTCTCGGGAATTCGCCGCAGCGGACGCGCTCGGCGAATTTGTGCTCTTGTGCCCTCCTCCGGG CTTGGGCCCAGGCCCGGCCCCTCGCACTTGCCCTTACCTTTTCTATCGAGTCCGCATCCCTCTCCAGCCACTGCGACCCGGCGAAGAGAA AAAGGAACTTCCCCCACCCCCTCGGGTGCCGTCGGAGCCCCCCAGCCCACCCCTGGGTGCGGCGCGGGGACCCCGGGCCGAAGAAGAGAT TTCCTGAGGATTCTGGTTTTCCTCGCTTGTATCTCCGAAAGAATTAAAAATGGCCGAGAATGTGGTGGAACCGGGGCCGCCTTCAGCCAA GCGGCCTAAACTCTCATCTCCGGCCCTCTCGGCGTCCGCCAGCGATGGCACAGATTTTGGCTCTCTATTTGACTTGGAGCACGACTTACC AGATGAATTAATCAACTCTACAGAATTGGGACTAACCAATGGTGGTGATATTAATCAGCTTCAGACAAGTCTTGGCATGGTACAAGATGC AGCTTCTAAACATAAACAGCTGTCAGAATTGCTGCGATCTGGTAGTTCCCCTAACCTCAATATGGGAGTTGGTGGCCCAGGTCAAGTCAT GGCCAGCCAGGCCCAACAGAGCAGTCCTGGATTAGGTTTGATAAATAGCATGGTCAAAAGCCCAATGACACAGGCAGGCTTGACTTCTCC CAACATGGGGATGGGCACTAGTGGACCAAATCAGGGTCCTACGCAGTCAACAGGTATGATGAACAGTCCAGTAAATCAGCCTGCCATGGG AATGAACACAGGGATGAATGCGGGCATGAATCCTGGAATGTTGGCTGCAGGCAATGGACAAGGGATAATGCCTAATCAAGTCATGAACGG TTCAATTGGAGCAGGCCGAGGGCGACAGAATATGCAGTACCCAAACCCAGGCATGGGAAGTGCTGGCAACTTACTGACTGAGCCTCTTCA GCAGGGCTCTCCCCAGATGGGAGGACAAACAGGATTGAGAGGCCCCCAGCCTCTTAAGATCGAGATGCTCAGCCCCTTCGAGGCAGTGGC CACACGCTTCAAGGAGCAATACAGGACATTCGCCACGGCCCTGGACACTACCAGGCACGAGCTGCCCGTGAGGTCCATCCACCTGGAGGG AGATGGGCAGCAGCTCTTAGACGCCCTGCAGCATGAACTGGTGACCACTCAGCGCCTCCTGGGAGAACTTGATGTTGGTGATTCGGAAGA AAATGTGCAGGTGCTGGACTTACTGAGCGAACTCAAGGACGTGACGGCGAAAAAGGACCTTGAGCTCCGAAGGAGCTTTGCCCAGGTGCT GGAACTCTCCGCAGAGGCAAGCAAAGAGGCAGCCTTGGCAAACCAGGAAGTCTGGGAAGAGACCCAGGGCATGGCGCCCCCCAGCCGGTG GTATTTCAATCAAGACAGTGCCTGCAGAGAATCTGGGGGAGCACCCAAGAACACGCCCCTGTCTGAGGACGACAACCCGGGTGCCTCGTC AGCCCCCGCTCAGGCCACGTTCATCAGCCCAAGCGAAGATTTTTCTTCAAGCAGCCAGGCAGAAGTCCCGCCCTCTCTCTCTCGTTCAGG GAGGGACTTGTCATGACTCATGGTTACATTCAGGATACTTGAGCACTTTATATACTACCGTAGCACTGTAGCTATTTTTTATGTGATAAT >26751_26751_3_EP300-HAUS8_EP300_chr22_41513825_ENST00000263253_HAUS8_chr19_17166812_ENST00000593360_length(amino acids)=438AA_BP=241 MAENVVEPGPPSAKRPKLSSPALSASASDGTDFGSLFDLEHDLPDELINSTELGLTNGGDINQLQTSLGMVQDAASKHKQLSELLRSGSS PNLNMGVGGPGQVMASQAQQSSPGLGLINSMVKSPMTQAGLTSPNMGMGTSGPNQGPTQSTGMMNSPVNQPAMGMNTGMNAGMNPGMLAA GNGQGIMPNQVMNGSIGAGRGRQNMQYPNPGMGSAGNLLTEPLQQGSPQMGGQTGLRGPQPLKIEMLSPFEAVATRFKEQYRTFATALDT TRHELPVRSIHLEGDGQQLLDALQHELVTTQRLLGELDVGDSEENVQVLDLLSELKDVTAKKDLELRRSFAQVLELSAEASKEAALANQE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for EP300-HAUS8 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 2_139 | 243.0 | 2415.0 | ALX1 |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 2_149 | 243.0 | 2415.0 | RORA |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 1397_1399 | 243.0 | 2415.0 | histone |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 2003_2212 | 243.0 | 2415.0 | HTLV-1 Tax |
| Hgene | EP300 | chr22:41513825 | chr19:17166812 | ENST00000263253 | + | 2 | 31 | 2041_2240 | 243.0 | 2415.0 | NCOA2 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for EP300-HAUS8 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for EP300-HAUS8 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
