
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ERBB3-EGFR (FusionGDB2 ID:27211) |
Fusion Gene Summary for ERBB3-EGFR |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ERBB3-EGFR | Fusion gene ID: 27211 | Hgene | Tgene | Gene symbol | ERBB3 | EGFR | Gene ID | 2065 | 1956 |
| Gene name | erb-b2 receptor tyrosine kinase 3 | epidermal growth factor receptor | |
| Synonyms | ErbB-3|FERLK|HER3|LCCS2|MDA-BF-1|c-erbB-3|c-erbB3|erbB3-S|p180-ErbB3|p45-sErbB3|p85-sErbB3 | ERBB|ERBB1|HER1|NISBD2|PIG61|mENA | |
| Cytomap | 12q13.2 | 7p11.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | receptor tyrosine-protein kinase erbB-3human epidermal growth factor receptor 3proto-oncogene-like protein c-ErbB-3tyrosine kinase-type cell surface receptor HER3v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 | epidermal growth factor receptoravian erythroblastic leukemia viral (v-erb-b) oncogene homologcell growth inhibiting protein 40cell proliferation-inducing protein 61epidermal growth factor receptor tyrosine kinase domainerb-b2 receptor tyrosine kinas | |
| Modification date | 20200327 | 20200329 | |
| UniProtAcc | P21860 | P00533 | |
| Ensembl transtripts involved in fusion gene | ENST00000267101, ENST00000415288, ENST00000450146, ENST00000553131, ENST00000549832, ENST00000411731, | ENST00000275493, ENST00000342916, ENST00000344576, ENST00000420316, ENST00000442591, ENST00000454757, ENST00000463948, ENST00000455089, | |
| Fusion gene scores | * DoF score | 16 X 18 X 11=3168 | 17 X 20 X 8=2720 |
| # samples | 19 | 22 | |
| ** MAII score | log2(19/3168*10)=-4.05950101174866 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(22/2720*10)=-3.62803122261304 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ERBB3 [Title/Abstract] AND EGFR [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ERBB3(56492359)-EGFR(55266410), # samples:5 EGFR(55087058)-ERBB3(56492542), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ERBB3 | GO:0007162 | negative regulation of cell adhesion | 7556068 |
| Hgene | ERBB3 | GO:0007165 | signal transduction | 10572067 |
| Hgene | ERBB3 | GO:0009968 | negative regulation of signal transduction | 11389077 |
| Hgene | ERBB3 | GO:0014065 | phosphatidylinositol 3-kinase signaling | 7556068 |
| Hgene | ERBB3 | GO:0042127 | regulation of cell proliferation | 11389077 |
| Hgene | ERBB3 | GO:0051048 | negative regulation of secretion | 10559227 |
| Tgene | EGFR | GO:0001934 | positive regulation of protein phosphorylation | 20551055 |
| Tgene | EGFR | GO:0007165 | signal transduction | 10572067 |
| Tgene | EGFR | GO:0007166 | cell surface receptor signaling pathway | 7736574 |
| Tgene | EGFR | GO:0007173 | epidermal growth factor receptor signaling pathway | 7736574|12435727 |
| Tgene | EGFR | GO:0008283 | cell proliferation | 17115032 |
| Tgene | EGFR | GO:0008284 | positive regulation of cell proliferation | 7736574 |
| Tgene | EGFR | GO:0010750 | positive regulation of nitric oxide mediated signal transduction | 12828935 |
| Tgene | EGFR | GO:0018108 | peptidyl-tyrosine phosphorylation | 22732145 |
| Tgene | EGFR | GO:0030307 | positive regulation of cell growth | 15467833 |
| Tgene | EGFR | GO:0042177 | negative regulation of protein catabolic process | 17115032 |
| Tgene | EGFR | GO:0042327 | positive regulation of phosphorylation | 15082764 |
| Tgene | EGFR | GO:0043406 | positive regulation of MAP kinase activity | 10572067 |
| Tgene | EGFR | GO:0045739 | positive regulation of DNA repair | 17115032 |
| Tgene | EGFR | GO:0045740 | positive regulation of DNA replication | 17115032 |
| Tgene | EGFR | GO:0045944 | positive regulation of transcription by RNA polymerase II | 20551055 |
| Tgene | EGFR | GO:0050679 | positive regulation of epithelial cell proliferation | 10572067 |
| Tgene | EGFR | GO:0050999 | regulation of nitric-oxide synthase activity | 12828935 |
| Tgene | EGFR | GO:0070141 | response to UV-A | 18483258 |
| Tgene | EGFR | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 20551055 |
| Tgene | EGFR | GO:0071392 | cellular response to estradiol stimulus | 20551055 |
| Tgene | EGFR | GO:1900020 | positive regulation of protein kinase C activity | 22732145 |
| Tgene | EGFR | GO:1903078 | positive regulation of protein localization to plasma membrane | 22732145 |
 Fusion gene breakpoints across ERBB3 (5'-gene) Fusion gene breakpoints across ERBB3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
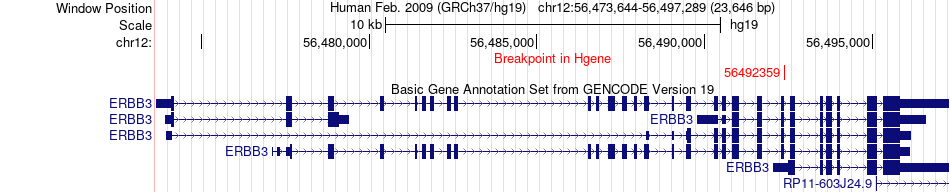 |
 Fusion gene breakpoints across EGFR (3'-gene) Fusion gene breakpoints across EGFR (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
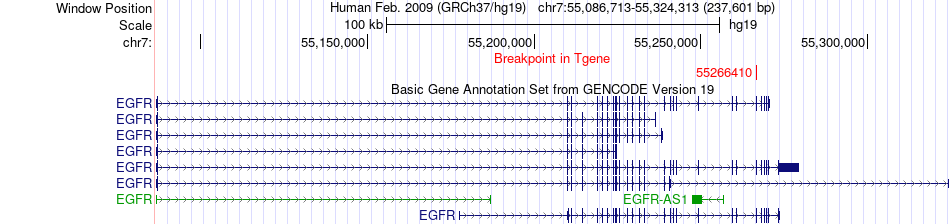 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-LN-A4A8 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| ChimerDB4 | ESCA | TCGA-Q9-A6FU | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| ChimerDB4 | ESCA | TCGA-VR-A8EY | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| ChimerDB4 | LGG | TCGA-DU-7292 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| ChimerDB4 | LGG | TCGA-HT-8104 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
Top |
Fusion Gene ORF analysis for ERBB3-EGFR |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000267101 | ENST00000275493 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000267101 | ENST00000342916 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000267101 | ENST00000344576 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000267101 | ENST00000420316 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000267101 | ENST00000442591 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000267101 | ENST00000454757 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000267101 | ENST00000463948 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000415288 | ENST00000275493 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000415288 | ENST00000342916 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000415288 | ENST00000344576 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000415288 | ENST00000420316 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000415288 | ENST00000442591 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000415288 | ENST00000454757 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000415288 | ENST00000463948 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000450146 | ENST00000275493 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000450146 | ENST00000342916 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000450146 | ENST00000344576 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000450146 | ENST00000420316 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000450146 | ENST00000442591 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000450146 | ENST00000454757 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000450146 | ENST00000463948 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000553131 | ENST00000275493 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000553131 | ENST00000342916 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000553131 | ENST00000344576 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000553131 | ENST00000420316 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000553131 | ENST00000442591 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000553131 | ENST00000454757 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5CDS-intron | ENST00000553131 | ENST00000463948 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5UTR-3CDS | ENST00000549832 | ENST00000455089 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5UTR-intron | ENST00000549832 | ENST00000275493 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5UTR-intron | ENST00000549832 | ENST00000342916 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5UTR-intron | ENST00000549832 | ENST00000344576 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5UTR-intron | ENST00000549832 | ENST00000420316 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5UTR-intron | ENST00000549832 | ENST00000442591 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5UTR-intron | ENST00000549832 | ENST00000454757 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| 5UTR-intron | ENST00000549832 | ENST00000463948 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| In-frame | ENST00000267101 | ENST00000455089 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| In-frame | ENST00000415288 | ENST00000455089 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| In-frame | ENST00000450146 | ENST00000455089 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| In-frame | ENST00000553131 | ENST00000455089 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| intron-3CDS | ENST00000411731 | ENST00000455089 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| intron-intron | ENST00000411731 | ENST00000275493 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| intron-intron | ENST00000411731 | ENST00000342916 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| intron-intron | ENST00000411731 | ENST00000344576 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| intron-intron | ENST00000411731 | ENST00000420316 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| intron-intron | ENST00000411731 | ENST00000442591 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| intron-intron | ENST00000411731 | ENST00000454757 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
| intron-intron | ENST00000411731 | ENST00000463948 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000267101 | ERBB3 | chr12 | 56492359 | + | ENST00000455089 | EGFR | chr7 | 55266410 | + | 4153 | 3132 | 440 | 3841 | 1133 |
| ENST00000450146 | ERBB3 | chr12 | 56492359 | + | ENST00000455089 | EGFR | chr7 | 55266410 | + | 2088 | 1067 | 106 | 1776 | 556 |
| ENST00000415288 | ERBB3 | chr12 | 56492359 | + | ENST00000455089 | EGFR | chr7 | 55266410 | + | 3756 | 2735 | 85 | 3444 | 1119 |
| ENST00000553131 | ERBB3 | chr12 | 56492359 | + | ENST00000455089 | EGFR | chr7 | 55266410 | + | 2170 | 1149 | 482 | 1858 | 458 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000267101 | ENST00000455089 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + | 0.001773078 | 0.99822694 |
| ENST00000450146 | ENST00000455089 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + | 0.002794822 | 0.9972052 |
| ENST00000415288 | ENST00000455089 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + | 0.000902222 | 0.9990978 |
| ENST00000553131 | ENST00000455089 | ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266410 | + | 0.004832051 | 0.995168 |
Top |
Fusion Genomic Features for ERBB3-EGFR |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266409 | + | 0.007797451 | 0.9922025 |
| ERBB3 | chr12 | 56492359 | + | EGFR | chr7 | 55266409 | + | 0.007797451 | 0.9922025 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
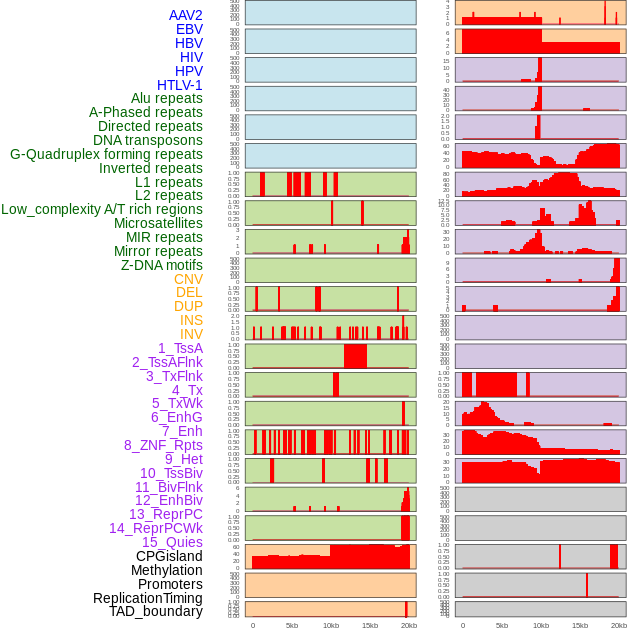 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for ERBB3-EGFR |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:56492359/chr7:55266410) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ERBB3 | EGFR |
| FUNCTION: Tyrosine-protein kinase that plays an essential role as cell surface receptor for neuregulins. Binds to neuregulin-1 (NRG1) and is activated by it; ligand-binding increases phosphorylation on tyrosine residues and promotes its association with the p85 subunit of phosphatidylinositol 3-kinase (PubMed:20682778). May also be activated by CSPG5 (PubMed:15358134). Involved in the regulation of myeloid cell differentiation (PubMed:27416908). {ECO:0000269|PubMed:15358134, ECO:0000269|PubMed:20682778, ECO:0000269|PubMed:27416908}. | FUNCTION: Receptor tyrosine kinase binding ligands of the EGF family and activating several signaling cascades to convert extracellular cues into appropriate cellular responses (PubMed:2790960, PubMed:10805725, PubMed:27153536). Known ligands include EGF, TGFA/TGF-alpha, AREG, epigen/EPGN, BTC/betacellulin, epiregulin/EREG and HBEGF/heparin-binding EGF (PubMed:2790960, PubMed:7679104, PubMed:8144591, PubMed:9419975, PubMed:15611079, PubMed:12297049, PubMed:27153536, PubMed:20837704, PubMed:17909029). Ligand binding triggers receptor homo- and/or heterodimerization and autophosphorylation on key cytoplasmic residues. The phosphorylated receptor recruits adapter proteins like GRB2 which in turn activates complex downstream signaling cascades. Activates at least 4 major downstream signaling cascades including the RAS-RAF-MEK-ERK, PI3 kinase-AKT, PLCgamma-PKC and STATs modules (PubMed:27153536). May also activate the NF-kappa-B signaling cascade (PubMed:11116146). Also directly phosphorylates other proteins like RGS16, activating its GTPase activity and probably coupling the EGF receptor signaling to the G protein-coupled receptor signaling (PubMed:11602604). Also phosphorylates MUC1 and increases its interaction with SRC and CTNNB1/beta-catenin (PubMed:11483589). Positively regulates cell migration via interaction with CCDC88A/GIV which retains EGFR at the cell membrane following ligand stimulation, promoting EGFR signaling which triggers cell migration (PubMed:20462955). Plays a role in enhancing learning and memory performance (By similarity). {ECO:0000250|UniProtKB:Q01279, ECO:0000269|PubMed:10805725, ECO:0000269|PubMed:11116146, ECO:0000269|PubMed:11483589, ECO:0000269|PubMed:11602604, ECO:0000269|PubMed:12297049, ECO:0000269|PubMed:12297050, ECO:0000269|PubMed:12620237, ECO:0000269|PubMed:12873986, ECO:0000269|PubMed:15374980, ECO:0000269|PubMed:15590694, ECO:0000269|PubMed:15611079, ECO:0000269|PubMed:17115032, ECO:0000269|PubMed:17909029, ECO:0000269|PubMed:19560417, ECO:0000269|PubMed:20462955, ECO:0000269|PubMed:20837704, ECO:0000269|PubMed:21258366, ECO:0000269|PubMed:27153536, ECO:0000269|PubMed:2790960, ECO:0000269|PubMed:7679104, ECO:0000269|PubMed:8144591, ECO:0000269|PubMed:9419975}.; FUNCTION: Isoform 2 may act as an antagonist of EGF action.; FUNCTION: (Microbial infection) Acts as a receptor for hepatitis C virus (HCV) in hepatocytes and facilitates its cell entry. Mediates HCV entry by promoting the formation of the CD81-CLDN1 receptor complexes that are essential for HCV entry and by enhancing membrane fusion of cells expressing HCV envelope glycoproteins. {ECO:0000269|PubMed:21516087}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000267101 | + | 22 | 28 | 715_723 | 897 | 1343.0 | Nucleotide binding | Note=ATP |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000267101 | + | 22 | 28 | 788_790 | 897 | 1343.0 | Nucleotide binding | Note=ATP |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000267101 | + | 22 | 28 | 834_839 | 897 | 1343.0 | Nucleotide binding | Note=ATP |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000415288 | + | 23 | 29 | 715_723 | 838 | 1284.0 | Nucleotide binding | Note=ATP |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000415288 | + | 23 | 29 | 788_790 | 838 | 1284.0 | Nucleotide binding | Note=ATP |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000415288 | + | 23 | 29 | 834_839 | 838 | 1284.0 | Nucleotide binding | Note=ATP |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000267101 | + | 22 | 28 | 20_643 | 897 | 1343.0 | Topological domain | Extracellular |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000415288 | + | 23 | 29 | 20_643 | 838 | 1284.0 | Topological domain | Extracellular |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000267101 | + | 22 | 28 | 644_664 | 897 | 1343.0 | Transmembrane | Helical |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000415288 | + | 23 | 29 | 644_664 | 838 | 1284.0 | Transmembrane | Helical |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000342916 | 0 | 16 | 712_979 | 0 | 629.0 | Domain | Protein kinase | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000344576 | 0 | 16 | 712_979 | 0 | 706.0 | Domain | Protein kinase | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000420316 | 0 | 10 | 712_979 | 0 | 406.0 | Domain | Protein kinase | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000342916 | 0 | 16 | 718_726 | 0 | 629.0 | Nucleotide binding | ATP | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000342916 | 0 | 16 | 790_791 | 0 | 629.0 | Nucleotide binding | ATP | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000344576 | 0 | 16 | 718_726 | 0 | 706.0 | Nucleotide binding | ATP | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000344576 | 0 | 16 | 790_791 | 0 | 706.0 | Nucleotide binding | ATP | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000420316 | 0 | 10 | 718_726 | 0 | 406.0 | Nucleotide binding | ATP | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000420316 | 0 | 10 | 790_791 | 0 | 406.0 | Nucleotide binding | ATP | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000342916 | 0 | 16 | 688_704 | 0 | 629.0 | Region | Note=Important for dimerization%2C phosphorylation and activation | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000344576 | 0 | 16 | 688_704 | 0 | 706.0 | Region | Note=Important for dimerization%2C phosphorylation and activation | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000420316 | 0 | 10 | 688_704 | 0 | 406.0 | Region | Note=Important for dimerization%2C phosphorylation and activation | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000342916 | 0 | 16 | 390_600 | 0 | 629.0 | Repeat | Note=Approximate | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000342916 | 0 | 16 | 75_300 | 0 | 629.0 | Repeat | Note=Approximate | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000344576 | 0 | 16 | 390_600 | 0 | 706.0 | Repeat | Note=Approximate | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000344576 | 0 | 16 | 75_300 | 0 | 706.0 | Repeat | Note=Approximate | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000420316 | 0 | 10 | 390_600 | 0 | 406.0 | Repeat | Note=Approximate | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000420316 | 0 | 10 | 75_300 | 0 | 406.0 | Repeat | Note=Approximate | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000342916 | 0 | 16 | 25_645 | 0 | 629.0 | Topological domain | Extracellular | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000342916 | 0 | 16 | 669_1210 | 0 | 629.0 | Topological domain | Cytoplasmic | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000344576 | 0 | 16 | 25_645 | 0 | 706.0 | Topological domain | Extracellular | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000344576 | 0 | 16 | 669_1210 | 0 | 706.0 | Topological domain | Cytoplasmic | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000420316 | 0 | 10 | 25_645 | 0 | 406.0 | Topological domain | Extracellular | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000420316 | 0 | 10 | 669_1210 | 0 | 406.0 | Topological domain | Cytoplasmic | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000342916 | 0 | 16 | 646_668 | 0 | 629.0 | Transmembrane | Helical | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000344576 | 0 | 16 | 646_668 | 0 | 706.0 | Transmembrane | Helical | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000420316 | 0 | 10 | 646_668 | 0 | 406.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000267101 | + | 22 | 28 | 709_966 | 897 | 1343.0 | Domain | Protein kinase |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000411731 | + | 1 | 3 | 709_966 | 0 | 184.0 | Domain | Protein kinase |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000415288 | + | 23 | 29 | 709_966 | 838 | 1284.0 | Domain | Protein kinase |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000450146 | + | 9 | 15 | 709_966 | 254 | 700.0 | Domain | Protein kinase |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000411731 | + | 1 | 3 | 715_723 | 0 | 184.0 | Nucleotide binding | Note=ATP |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000411731 | + | 1 | 3 | 788_790 | 0 | 184.0 | Nucleotide binding | Note=ATP |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000411731 | + | 1 | 3 | 834_839 | 0 | 184.0 | Nucleotide binding | Note=ATP |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000450146 | + | 9 | 15 | 715_723 | 254 | 700.0 | Nucleotide binding | Note=ATP |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000450146 | + | 9 | 15 | 788_790 | 254 | 700.0 | Nucleotide binding | Note=ATP |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000450146 | + | 9 | 15 | 834_839 | 254 | 700.0 | Nucleotide binding | Note=ATP |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000267101 | + | 22 | 28 | 665_1342 | 897 | 1343.0 | Topological domain | Cytoplasmic |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000411731 | + | 1 | 3 | 20_643 | 0 | 184.0 | Topological domain | Extracellular |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000411731 | + | 1 | 3 | 665_1342 | 0 | 184.0 | Topological domain | Cytoplasmic |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000415288 | + | 23 | 29 | 665_1342 | 838 | 1284.0 | Topological domain | Cytoplasmic |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000450146 | + | 9 | 15 | 20_643 | 254 | 700.0 | Topological domain | Extracellular |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000450146 | + | 9 | 15 | 665_1342 | 254 | 700.0 | Topological domain | Cytoplasmic |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000411731 | + | 1 | 3 | 644_664 | 0 | 184.0 | Transmembrane | Helical |
| Hgene | ERBB3 | chr12:56492359 | chr7:55266410 | ENST00000450146 | + | 9 | 15 | 644_664 | 254 | 700.0 | Transmembrane | Helical |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000275493 | 21 | 28 | 712_979 | 900 | 1211.0 | Domain | Protein kinase | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000275493 | 21 | 28 | 718_726 | 900 | 1211.0 | Nucleotide binding | ATP | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000275493 | 21 | 28 | 790_791 | 900 | 1211.0 | Nucleotide binding | ATP | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000275493 | 21 | 28 | 688_704 | 900 | 1211.0 | Region | Note=Important for dimerization%2C phosphorylation and activation | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000275493 | 21 | 28 | 390_600 | 900 | 1211.0 | Repeat | Note=Approximate | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000275493 | 21 | 28 | 75_300 | 900 | 1211.0 | Repeat | Note=Approximate | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000275493 | 21 | 28 | 25_645 | 900 | 1211.0 | Topological domain | Extracellular | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000275493 | 21 | 28 | 669_1210 | 900 | 1211.0 | Topological domain | Cytoplasmic | |
| Tgene | EGFR | chr12:56492359 | chr7:55266410 | ENST00000275493 | 21 | 28 | 646_668 | 900 | 1211.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for ERBB3-EGFR |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >27211_27211_1_ERBB3-EGFR_ERBB3_chr12_56492359_ENST00000267101_EGFR_chr7_55266410_ENST00000455089_length(transcript)=4153nt_BP=3132nt ATTACCCTTCCCTGGATCTGGGGGCTTTCGGAATCTCGACCTCCCCTTGGCCTATCTCCTGCAGAAAAATTAGGGTGAGCCCCATCCTCG ATCTGCTCCGCCAAGTTGCGGGACCGCGGGGCGTGGCACGCTCGGGGCAGGCGGTCCGAGGCTCCGCAATCCCTACTCCAGCCTCGCGCG GGAGGGGGCGCGGCCGTGACTCACCCCCTTCCCTCTGCGTTCCTCCCTCCCTCTCTCTCTCTCTCTCACACACACACACCCCTCCCCTGC CATCCCTCCCCGGACTCCGGCTCCGGCTCCGATTGCAATTTGCAACCTCCGCTGCCGTCGCCGCAGCAGCCACCAATTCGCCAGCGGTTC AGGTGGCTCTTGCCTCGATGTCCTAGCCTAGGGGCCCCCGGGCCGGACTTGGCTGGGCTCCCTTCACCCTCTGCGGAGTCATGAGGGCGA ACGACGCTCTGCAGGTGCTGGGCTTGCTTTTCAGCCTGGCCCGGGGCTCCGAGGTGGGCAACTCTCAGGCAGTGTGTCCTGGGACTCTGA ATGGCCTGAGTGTGACCGGCGATGCTGAGAACCAATACCAGACACTGTACAAGCTCTACGAGAGGTGTGAGGTGGTGATGGGGAACCTTG AGATTGTGCTCACGGGACACAATGCCGACCTCTCCTTCCTGCAGTGGATTCGAGAAGTGACAGGCTATGTCCTCGTGGCCATGAATGAAT TCTCTACTCTACCATTGCCCAACCTCCGCGTGGTGCGAGGGACCCAGGTCTACGATGGGAAGTTTGCCATCTTCGTCATGTTGAACTATA ACACCAACTCCAGCCACGCTCTGCGCCAGCTCCGCTTGACTCAGCTCACCGAGATTCTGTCAGGGGGTGTTTATATTGAGAAGAACGATA AGCTTTGTCACATGGACACAATTGACTGGAGGGACATCGTGAGGGACCGAGATGCTGAGATAGTGGTGAAGGACAATGGCAGAAGCTGTC CCCCCTGTCATGAGGTTTGCAAGGGGCGATGCTGGGGTCCTGGATCAGAAGACTGCCAGACATTGACCAAGACCATCTGTGCTCCTCAGT GTAATGGTCACTGCTTTGGGCCCAACCCCAACCAGTGCTGCCATGATGAGTGTGCCGGGGGCTGCTCAGGCCCTCAGGACACAGACTGCT TTGCCTGCCGGCACTTCAATGACAGTGGAGCCTGTGTACCTCGCTGTCCACAGCCTCTTGTCTACAACAAGCTAACTTTCCAGCTGGAAC CCAATCCCCACACCAAGTATCAGTATGGAGGAGTTTGTGTAGCCAGCTGTCCCCATAACTTTGTGGTGGATCAAACATCCTGTGTCAGGG CCTGTCCTCCTGACAAGATGGAAGTAGATAAAAATGGGCTCAAGATGTGTGAGCCTTGTGGGGGACTATGTCCCAAAGCCTGTGAGGGAA CAGGCTCTGGGAGCCGCTTCCAGACTGTGGACTCGAGCAACATTGATGGATTTGTGAACTGCACCAAGATCCTGGGCAACCTGGACTTTC TGATCACCGGCCTCAATGGAGACCCCTGGCACAAGATCCCTGCCCTGGACCCAGAGAAGCTCAATGTCTTCCGGACAGTACGGGAGATCA CAGGTTACCTGAACATCCAGTCCTGGCCGCCCCACATGCACAACTTCAGTGTTTTTTCCAATTTGACAACCATTGGAGGCAGAAGCCTCT ACAACCGGGGCTTCTCATTGTTGATCATGAAGAACTTGAATGTCACATCTCTGGGCTTCCGATCCCTGAAGGAAATTAGTGCTGGGCGTA TCTATATAAGTGCCAATAGGCAGCTCTGCTACCACCACTCTTTGAACTGGACCAAGGTGCTTCGGGGGCCTACGGAAGAGCGACTAGACA TCAAGCATAATCGGCCGCGCAGAGACTGCGTGGCAGAGGGCAAAGTGTGTGACCCACTGTGCTCCTCTGGGGGATGCTGGGGCCCAGGCC CTGGTCAGTGCTTGTCCTGTCGAAATTATAGCCGAGGAGGTGTCTGTGTGACCCACTGCAACTTTCTGAATGGGGAGCCTCGAGAATTTG CCCATGAGGCCGAATGCTTCTCCTGCCACCCGGAATGCCAACCCATGGAGGGCACTGCCACATGCAATGGCTCGGGCTCTGATACTTGTG CTCAATGTGCCCATTTTCGAGATGGGCCCCACTGTGTGAGCAGCTGCCCCCATGGAGTCCTAGGTGCCAAGGGCCCAATCTACAAGTACC CAGATGTTCAGAATGAATGTCGGCCCTGCCATGAGAACTGCACCCAGGGGTGTAAAGGACCAGAGCTTCAAGACTGTTTAGGACAAACAC TGGTGCTGATCGGCAAAACCCATCTGACAATGGCTTTGACAGTGATAGCAGGATTGGTAGTGATTTTCATGATGCTGGGCGGCACTTTTC TCTACTGGCGTGGGCGCCGGATTCAGAATAAAAGGGCTATGAGGCGATACTTGGAACGGGGTGAGAGCATAGAGCCTCTGGACCCCAGTG AGAAGGCTAACAAAGTCTTGGCCAGAATCTTCAAAGAGACAGAGCTAAGGAAGCTTAAAGTGCTTGGCTCGGGTGTCTTTGGAACTGTGC ACAAAGGAGTGTGGATCCCTGAGGGTGAATCAATCAAGATTCCAGTCTGCATTAAAGTCATTGAGGACAAGAGTGGACGGCAGAGTTTTC AAGCTGTGACAGATCATATGCTGGCCATTGGCAGCCTGGACCATGCCCACATTGTAAGGCTGCTGGGACTATGCCCAGGGTCATCTCTGC AGCTTGTCACTCAATATTTGCCTCTGGGTTCTCTGCTGGATCATGTGAGACAACACCGGGGGGCACTGGGGCCACAGCTGCTGCTCAACT GGGGAGTACAAATTGCCAAGGGAATGTACTACCTTGAGGAACATGGTATGGTGCATAGAAACCTGGCTGCCCGAAACGTGCTACTCAAGT CACCCAGTCAGGTTCAGGTGGCAGATTTTGGTGTGGCTGACCTGCTGCCTCCTGATGATAAGCAGCTGCTATACAGTGAGGCCAAGACTC CAATTAAGTGGATGGCCCTTGAGAGTATCCACTTTGGGAAATACACACACCAGAGTGATGTCTGGAGCTATGGGGTGACTGTTTGGGAGT TGATGACCTTTGGATCCAAGCCATATGACGGAATCCCTGCCAGCGAGATCTCCTCCATCCTGGAGAAAGGAGAACGCCTCCCTCAGCCAC CCATATGTACCATCGATGTCTACATGATCATGGTCAAGTGCTGGATGATAGACGCAGATAGTCGCCCAAAGTTCCGTGAGTTGATCATCG AATTCTCCAAAATGGCCCGAGACCCCCAGCGCTACCTTGTCATTCAGGGGGATGAAAGAATGCATTTGCCAAGTCCTACAGACTCCAACT TCTACCGTGCCCTGATGGATGAAGAAGACATGGACGACGTGGTGGATGCCGACGAGTACCTCATCCCACAGCAGGGCTTCTTCAGCAGCC CCTCCACGTCACGGACTCCCCTCCTGAGCTCTCTGAGTGCAACCAGCAACAATTCCACCGTGGCTTGCATTGATAGAAATGGGCTGCAAA GCTGTCCCATCAAGGAAGACAGCTTCTTGCAGCGATACAGCTCAGACCCCACAGGCGCCTTGACTGAGGACAGCATAGACGACACCTTCC TCCCAGTGCCTGGTGAGTGGCTTGTCTGGAAACAGTCCTGCTCCTCAACCTCCTCGACCCACTCAGCAGCAGCCAGTCTCCAGTGTCCAA GCCAGGTGCTCCCTCCAGCATCTCCAGAGGGGGAAACAGTGGCAGATTTGCAGACACAGTGAAGGGCGTAAGGAGCAGATAAACACATGA CCGAGCCTGCACAAGCTCTTTGTTGTGTCTGGTTGTTTGCTGTACCTCTGTTGTAAGAATGAATCTGCAAAATTTCTAGCTTATGAAGCA AATCACGGACATACACATCTGTGTGTGTGAGTGTTCATGATGTGTGTACATCTGTGTATGTGTGTGTGTGTATGTGTGTGTTTGTGACAG ATTTGATCCCTGTTCTCTCTGCTGGCTCTATCTTGACCTGTGAAACGTATATTTAACTAATTAAATATTAGTTAATATTAATAAATTTTA >27211_27211_1_ERBB3-EGFR_ERBB3_chr12_56492359_ENST00000267101_EGFR_chr7_55266410_ENST00000455089_length(amino acids)=1133AA_BP=47 MRANDALQVLGLLFSLARGSEVGNSQAVCPGTLNGLSVTGDAENQYQTLYKLYERCEVVMGNLEIVLTGHNADLSFLQWIREVTGYVLVA MNEFSTLPLPNLRVVRGTQVYDGKFAIFVMLNYNTNSSHALRQLRLTQLTEILSGGVYIEKNDKLCHMDTIDWRDIVRDRDAEIVVKDNG RSCPPCHEVCKGRCWGPGSEDCQTLTKTICAPQCNGHCFGPNPNQCCHDECAGGCSGPQDTDCFACRHFNDSGACVPRCPQPLVYNKLTF QLEPNPHTKYQYGGVCVASCPHNFVVDQTSCVRACPPDKMEVDKNGLKMCEPCGGLCPKACEGTGSGSRFQTVDSSNIDGFVNCTKILGN LDFLITGLNGDPWHKIPALDPEKLNVFRTVREITGYLNIQSWPPHMHNFSVFSNLTTIGGRSLYNRGFSLLIMKNLNVTSLGFRSLKEIS AGRIYISANRQLCYHHSLNWTKVLRGPTEERLDIKHNRPRRDCVAEGKVCDPLCSSGGCWGPGPGQCLSCRNYSRGGVCVTHCNFLNGEP REFAHEAECFSCHPECQPMEGTATCNGSGSDTCAQCAHFRDGPHCVSSCPHGVLGAKGPIYKYPDVQNECRPCHENCTQGCKGPELQDCL GQTLVLIGKTHLTMALTVIAGLVVIFMMLGGTFLYWRGRRIQNKRAMRRYLERGESIEPLDPSEKANKVLARIFKETELRKLKVLGSGVF GTVHKGVWIPEGESIKIPVCIKVIEDKSGRQSFQAVTDHMLAIGSLDHAHIVRLLGLCPGSSLQLVTQYLPLGSLLDHVRQHRGALGPQL LLNWGVQIAKGMYYLEEHGMVHRNLAARNVLLKSPSQVQVADFGVADLLPPDDKQLLYSEAKTPIKWMALESIHFGKYTHQSDVWSYGVT VWELMTFGSKPYDGIPASEISSILEKGERLPQPPICTIDVYMIMVKCWMIDADSRPKFRELIIEFSKMARDPQRYLVIQGDERMHLPSPT DSNFYRALMDEEDMDDVVDADEYLIPQQGFFSSPSTSRTPLLSSLSATSNNSTVACIDRNGLQSCPIKEDSFLQRYSSDPTGALTEDSID -------------------------------------------------------------- >27211_27211_2_ERBB3-EGFR_ERBB3_chr12_56492359_ENST00000415288_EGFR_chr7_55266410_ENST00000455089_length(transcript)=3756nt_BP=2735nt ATTTATTCCAGGACCTAGAGAGAGGCAGGATGAGGGTAGAGAATTAACCCCTTAGTACCCAAAAGAAGCACAGAGGGGTCCCAGGTTGAA AAAGGAAATCTTTTCACCTTCCCATTCATGGAATGTGTGTCCTGGGACTCTGAATGGCCTGAGTGTGACCGGCGATGCTGAGAACCAATA CCAGACACTGTACAAGCTCTACGAGAGGTGTGAGGTGGTGATGGGGAACCTTGAGATTGTGCTCACGGGACACAATGCCGACCTCTCCTT CCTGCAGTGGATTCGAGAAGTGACAGGCTATGTCCTCGTGGCCATGAATGAATTCTCTACTCTACCATTGCCCAACCTCCGCGTGGTGCG AGGGACCCAGGTCTACGATGGGAAGTTTGCCATCTTCGTCATGTTGAACTATAACACCAACTCCAGCCACGCTCTGCGCCAGCTCCGCTT GACTCAGCTCACCGAGATTCTGTCAGGGGGTGTTTATATTGAGAAGAACGATAAGCTTTGTCACATGGACACAATTGACTGGAGGGACAT CGTGAGGGACCGAGATGCTGAGATAGTGGTGAAGGACAATGGCAGAAGCTGTCCCCCCTGTCATGAGGTTTGCAAGGGGCGATGCTGGGG TCCTGGATCAGAAGACTGCCAGACATTGACCAAGACCATCTGTGCTCCTCAGTGTAATGGTCACTGCTTTGGGCCCAACCCCAACCAGTG CTGCCATGATGAGTGTGCCGGGGGCTGCTCAGGCCCTCAGGACACAGACTGCTTTGCCTGCCGGCACTTCAATGACAGTGGAGCCTGTGT ACCTCGCTGTCCACAGCCTCTTGTCTACAACAAGCTAACTTTCCAGCTGGAACCCAATCCCCACACCAAGTATCAGTATGGAGGAGTTTG TGTAGCCAGCTGTCCCCATAACTTTGTGGTGGATCAAACATCCTGTGTCAGGGCCTGTCCTCCTGACAAGATGGAAGTAGATAAAAATGG GCTCAAGATGTGTGAGCCTTGTGGGGGACTATGTCCCAAAGCCTGTGAGGGAACAGGCTCTGGGAGCCGCTTCCAGACTGTGGACTCGAG CAACATTGATGGATTTGTGAACTGCACCAAGATCCTGGGCAACCTGGACTTTCTGATCACCGGCCTCAATGGAGACCCCTGGCACAAGAT CCCTGCCCTGGACCCAGAGAAGCTCAATGTCTTCCGGACAGTACGGGAGATCACAGGTTACCTGAACATCCAGTCCTGGCCGCCCCACAT GCACAACTTCAGTGTTTTTTCCAATTTGACAACCATTGGAGGCAGAAGCCTCTACAACCGGGGCTTCTCATTGTTGATCATGAAGAACTT GAATGTCACATCTCTGGGCTTCCGATCCCTGAAGGAAATTAGTGCTGGGCGTATCTATATAAGTGCCAATAGGCAGCTCTGCTACCACCA CTCTTTGAACTGGACCAAGGTGCTTCGGGGGCCTACGGAAGAGCGACTAGACATCAAGCATAATCGGCCGCGCAGAGACTGCGTGGCAGA GGGCAAAGTGTGTGACCCACTGTGCTCCTCTGGGGGATGCTGGGGCCCAGGCCCTGGTCAGTGCTTGTCCTGTCGAAATTATAGCCGAGG AGGTGTCTGTGTGACCCACTGCAACTTTCTGAATGGGGAGCCTCGAGAATTTGCCCATGAGGCCGAATGCTTCTCCTGCCACCCGGAATG CCAACCCATGGAGGGCACTGCCACATGCAATGGCTCGGGCTCTGATACTTGTGCTCAATGTGCCCATTTTCGAGATGGGCCCCACTGTGT GAGCAGCTGCCCCCATGGAGTCCTAGGTGCCAAGGGCCCAATCTACAAGTACCCAGATGTTCAGAATGAATGTCGGCCCTGCCATGAGAA CTGCACCCAGGGGTGTAAAGGACCAGAGCTTCAAGACTGTTTAGGACAAACACTGGTGCTGATCGGCAAAACCCATCTGACAATGGCTTT GACAGTGATAGCAGGATTGGTAGTGATTTTCATGATGCTGGGCGGCACTTTTCTCTACTGGCGTGGGCGCCGGATTCAGAATAAAAGGGC TATGAGGCGATACTTGGAACGGGGTGAGAGCATAGAGCCTCTGGACCCCAGTGAGAAGGCTAACAAAGTCTTGGCCAGAATCTTCAAAGA GACAGAGCTAAGGAAGCTTAAAGTGCTTGGCTCGGGTGTCTTTGGAACTGTGCACAAAGGAGTGTGGATCCCTGAGGGTGAATCAATCAA GATTCCAGTCTGCATTAAAGTCATTGAGGACAAGAGTGGACGGCAGAGTTTTCAAGCTGTGACAGATCATATGCTGGCCATTGGCAGCCT GGACCATGCCCACATTGTAAGGCTGCTGGGACTATGCCCAGGGTCATCTCTGCAGCTTGTCACTCAATATTTGCCTCTGGGTTCTCTGCT GGATCATGTGAGACAACACCGGGGGGCACTGGGGCCACAGCTGCTGCTCAACTGGGGAGTACAAATTGCCAAGGGAATGTACTACCTTGA GGAACATGGTATGGTGCATAGAAACCTGGCTGCCCGAAACGTGCTACTCAAGTCACCCAGTCAGGTTCAGGTGGCAGATTTTGGTGTGGC TGACCTGCTGCCTCCTGATGATAAGCAGCTGCTATACAGTGAGGCCAAGACTCCAATTAAGTGGATGGCCCTTGAGAGTATCCACTTTGG GAAATACACACACCAGAGTGATGTCTGGAGCTATGGGGTGACTGTTTGGGAGTTGATGACCTTTGGATCCAAGCCATATGACGGAATCCC TGCCAGCGAGATCTCCTCCATCCTGGAGAAAGGAGAACGCCTCCCTCAGCCACCCATATGTACCATCGATGTCTACATGATCATGGTCAA GTGCTGGATGATAGACGCAGATAGTCGCCCAAAGTTCCGTGAGTTGATCATCGAATTCTCCAAAATGGCCCGAGACCCCCAGCGCTACCT TGTCATTCAGGGGGATGAAAGAATGCATTTGCCAAGTCCTACAGACTCCAACTTCTACCGTGCCCTGATGGATGAAGAAGACATGGACGA CGTGGTGGATGCCGACGAGTACCTCATCCCACAGCAGGGCTTCTTCAGCAGCCCCTCCACGTCACGGACTCCCCTCCTGAGCTCTCTGAG TGCAACCAGCAACAATTCCACCGTGGCTTGCATTGATAGAAATGGGCTGCAAAGCTGTCCCATCAAGGAAGACAGCTTCTTGCAGCGATA CAGCTCAGACCCCACAGGCGCCTTGACTGAGGACAGCATAGACGACACCTTCCTCCCAGTGCCTGGTGAGTGGCTTGTCTGGAAACAGTC CTGCTCCTCAACCTCCTCGACCCACTCAGCAGCAGCCAGTCTCCAGTGTCCAAGCCAGGTGCTCCCTCCAGCATCTCCAGAGGGGGAAAC AGTGGCAGATTTGCAGACACAGTGAAGGGCGTAAGGAGCAGATAAACACATGACCGAGCCTGCACAAGCTCTTTGTTGTGTCTGGTTGTT TGCTGTACCTCTGTTGTAAGAATGAATCTGCAAAATTTCTAGCTTATGAAGCAAATCACGGACATACACATCTGTGTGTGTGAGTGTTCA TGATGTGTGTACATCTGTGTATGTGTGTGTGTGTATGTGTGTGTTTGTGACAGATTTGATCCCTGTTCTCTCTGCTGGCTCTATCTTGAC >27211_27211_2_ERBB3-EGFR_ERBB3_chr12_56492359_ENST00000415288_EGFR_chr7_55266410_ENST00000455089_length(amino acids)=1119AA_BP=33 MKKEIFSPSHSWNVCPGTLNGLSVTGDAENQYQTLYKLYERCEVVMGNLEIVLTGHNADLSFLQWIREVTGYVLVAMNEFSTLPLPNLRV VRGTQVYDGKFAIFVMLNYNTNSSHALRQLRLTQLTEILSGGVYIEKNDKLCHMDTIDWRDIVRDRDAEIVVKDNGRSCPPCHEVCKGRC WGPGSEDCQTLTKTICAPQCNGHCFGPNPNQCCHDECAGGCSGPQDTDCFACRHFNDSGACVPRCPQPLVYNKLTFQLEPNPHTKYQYGG VCVASCPHNFVVDQTSCVRACPPDKMEVDKNGLKMCEPCGGLCPKACEGTGSGSRFQTVDSSNIDGFVNCTKILGNLDFLITGLNGDPWH KIPALDPEKLNVFRTVREITGYLNIQSWPPHMHNFSVFSNLTTIGGRSLYNRGFSLLIMKNLNVTSLGFRSLKEISAGRIYISANRQLCY HHSLNWTKVLRGPTEERLDIKHNRPRRDCVAEGKVCDPLCSSGGCWGPGPGQCLSCRNYSRGGVCVTHCNFLNGEPREFAHEAECFSCHP ECQPMEGTATCNGSGSDTCAQCAHFRDGPHCVSSCPHGVLGAKGPIYKYPDVQNECRPCHENCTQGCKGPELQDCLGQTLVLIGKTHLTM ALTVIAGLVVIFMMLGGTFLYWRGRRIQNKRAMRRYLERGESIEPLDPSEKANKVLARIFKETELRKLKVLGSGVFGTVHKGVWIPEGES IKIPVCIKVIEDKSGRQSFQAVTDHMLAIGSLDHAHIVRLLGLCPGSSLQLVTQYLPLGSLLDHVRQHRGALGPQLLLNWGVQIAKGMYY LEEHGMVHRNLAARNVLLKSPSQVQVADFGVADLLPPDDKQLLYSEAKTPIKWMALESIHFGKYTHQSDVWSYGVTVWELMTFGSKPYDG IPASEISSILEKGERLPQPPICTIDVYMIMVKCWMIDADSRPKFRELIIEFSKMARDPQRYLVIQGDERMHLPSPTDSNFYRALMDEEDM DDVVDADEYLIPQQGFFSSPSTSRTPLLSSLSATSNNSTVACIDRNGLQSCPIKEDSFLQRYSSDPTGALTEDSIDDTFLPVPGEWLVWK -------------------------------------------------------------- >27211_27211_3_ERBB3-EGFR_ERBB3_chr12_56492359_ENST00000450146_EGFR_chr7_55266410_ENST00000455089_length(transcript)=2088nt_BP=1067nt AATTTGCAACCTCCGCTGCCGTCGCCGCAGCAGCCACCAATTCGCCAGCGGTTCAGGTGGCTCTTGCCTCGATGTCCTAGCCTAGGGGCC CCCGGGCCGGACTTGGCTGGGCTCCCTTCACCCTCTGCGGAGTCATGAGGGCGAACGACGCTCTGCAGGTGCCAAGGGCCCAATCTACAA GTACCCAGATGTTCAGAATGAATGTCGGCCCTGCCATGAGAACTGCACCCAGGGGTGTAAAGGACCAGAGCTTCAAGACTGTTTAGGACA AACACTGGTGCTGATCGGCAAAACCCATCTGACAATGGCTTTGACAGTGATAGCAGGATTGGTAGTGATTTTCATGATGCTGGGCGGCAC TTTTCTCTACTGGCGTGGGCGCCGGATTCAGAATAAAAGGGCTATGAGGCGATACTTGGAACGGGGTGAGAGCATAGAGCCTCTGGACCC CAGTGAGAAGGCTAACAAAGTCTTGGCCAGAATCTTCAAAGAGACAGAGCTAAGGAAGCTTAAAGTGCTTGGCTCGGGTGTCTTTGGAAC TGTGCACAAAGGAGTGTGGATCCCTGAGGGTGAATCAATCAAGATTCCAGTCTGCATTAAAGTCATTGAGGACAAGAGTGGACGGCAGAG TTTTCAAGCTGTGACAGATCATATGCTGGCCATTGGCAGCCTGGACCATGCCCACATTGTAAGGCTGCTGGGACTATGCCCAGGGTCATC TCTGCAGCTTGTCACTCAATATTTGCCTCTGGGTTCTCTGCTGGATCATGTGAGACAACACCGGGGGGCACTGGGGCCACAGCTGCTGCT CAACTGGGGAGTACAAATTGCCAAGGGAATGTACTACCTTGAGGAACATGGTATGGTGCATAGAAACCTGGCTGCCCGAAACGTGCTACT CAAGTCACCCAGTCAGGTTCAGGTGGCAGATTTTGGTGTGGCTGACCTGCTGCCTCCTGATGATAAGCAGCTGCTATACAGTGAGGCCAA GACTCCAATTAAGTGGATGGCCCTTGAGAGTATCCACTTTGGGAAATACACACACCAGAGTGATGTCTGGAGCTATGGGGTGACTGTTTG GGAGTTGATGACCTTTGGATCCAAGCCATATGACGGAATCCCTGCCAGCGAGATCTCCTCCATCCTGGAGAAAGGAGAACGCCTCCCTCA GCCACCCATATGTACCATCGATGTCTACATGATCATGGTCAAGTGCTGGATGATAGACGCAGATAGTCGCCCAAAGTTCCGTGAGTTGAT CATCGAATTCTCCAAAATGGCCCGAGACCCCCAGCGCTACCTTGTCATTCAGGGGGATGAAAGAATGCATTTGCCAAGTCCTACAGACTC CAACTTCTACCGTGCCCTGATGGATGAAGAAGACATGGACGACGTGGTGGATGCCGACGAGTACCTCATCCCACAGCAGGGCTTCTTCAG CAGCCCCTCCACGTCACGGACTCCCCTCCTGAGCTCTCTGAGTGCAACCAGCAACAATTCCACCGTGGCTTGCATTGATAGAAATGGGCT GCAAAGCTGTCCCATCAAGGAAGACAGCTTCTTGCAGCGATACAGCTCAGACCCCACAGGCGCCTTGACTGAGGACAGCATAGACGACAC CTTCCTCCCAGTGCCTGGTGAGTGGCTTGTCTGGAAACAGTCCTGCTCCTCAACCTCCTCGACCCACTCAGCAGCAGCCAGTCTCCAGTG TCCAAGCCAGGTGCTCCCTCCAGCATCTCCAGAGGGGGAAACAGTGGCAGATTTGCAGACACAGTGAAGGGCGTAAGGAGCAGATAAACA CATGACCGAGCCTGCACAAGCTCTTTGTTGTGTCTGGTTGTTTGCTGTACCTCTGTTGTAAGAATGAATCTGCAAAATTTCTAGCTTATG AAGCAAATCACGGACATACACATCTGTGTGTGTGAGTGTTCATGATGTGTGTACATCTGTGTATGTGTGTGTGTGTATGTGTGTGTTTGT GACAGATTTGATCCCTGTTCTCTCTGCTGGCTCTATCTTGACCTGTGAAACGTATATTTAACTAATTAAATATTAGTTAATATTAATAAA >27211_27211_3_ERBB3-EGFR_ERBB3_chr12_56492359_ENST00000450146_EGFR_chr7_55266410_ENST00000455089_length(amino acids)=556AA_BP=122 MGSLHPLRSHEGERRSAGAKGPIYKYPDVQNECRPCHENCTQGCKGPELQDCLGQTLVLIGKTHLTMALTVIAGLVVIFMMLGGTFLYWR GRRIQNKRAMRRYLERGESIEPLDPSEKANKVLARIFKETELRKLKVLGSGVFGTVHKGVWIPEGESIKIPVCIKVIEDKSGRQSFQAVT DHMLAIGSLDHAHIVRLLGLCPGSSLQLVTQYLPLGSLLDHVRQHRGALGPQLLLNWGVQIAKGMYYLEEHGMVHRNLAARNVLLKSPSQ VQVADFGVADLLPPDDKQLLYSEAKTPIKWMALESIHFGKYTHQSDVWSYGVTVWELMTFGSKPYDGIPASEISSILEKGERLPQPPICT IDVYMIMVKCWMIDADSRPKFRELIIEFSKMARDPQRYLVIQGDERMHLPSPTDSNFYRALMDEEDMDDVVDADEYLIPQQGFFSSPSTS RTPLLSSLSATSNNSTVACIDRNGLQSCPIKEDSFLQRYSSDPTGALTEDSIDDTFLPVPGEWLVWKQSCSSTSSTHSAAASLQCPSQVL -------------------------------------------------------------- >27211_27211_4_ERBB3-EGFR_ERBB3_chr12_56492359_ENST00000553131_EGFR_chr7_55266410_ENST00000455089_length(transcript)=2170nt_BP=1149nt AGAACACTGGTCAGAGAAATGGGAGGCATGCATTCTAGTCCTGATTTTGCCATTAATTTGCCACATGACTTTGAAGAAGTTACTTATCTT CTCTGTGCCTCGGTTTATGCATCTATACAGAGGAAATAACATTTGTCCTTCCAGGATGGCTGTAAGGGTAAAGGGGGATGATGTATGTGA AAGTGCTTTGGAAAGCACAGAGCACTGTATAAAAGGTACTCAAGGTGGTAATAGTACTACCAACTCTCCCTAGCTGTCCCCTTCCCCACT TTGTGCTCCTCCATCAAAGGGAAAACCCAACCCCTTTGATTCCTGATCTCATGAGCACAAATAACTTCCTCAGTTCTCAGGGTCTGTACC TCAATATGCCTATAATCCATTCCAGGACTAACGGTGCTTCCTCTTCCTGCCCTTTCAGCTGTGCTGCTTTTGGCATTCACCTATGAGGAG CGGGTTGGAGTGGGACATGGGAATGGCCTTTCCTGAGTAACTCCTTCCCATTTGCTCCTCAGAGCATAGAGCCTCTGGACCCCAGTGAGA AGGCTAACAAAGTCTTGGCCAGAATCTTCAAAGAGACAGAGCTAAGGAAGCTTAAAGTGCTTGGCTCGGGTGTCTTTGGAACTGTGCACA AAGGAGTGTGGATCCCTGAGGGTGAATCAATCAAGATTCCAGTCTGCATTAAAGTCATTGAGGACAAGAGTGGACGGCAGAGTTTTCAAG CTGTGACAGATCATATGCTGGCCATTGGCAGCCTGGACCATGCCCACATTGTAAGGCTGCTGGGACTATGCCCAGGGTCATCTCTGCAGC TTGTCACTCAATATTTGCCTCTGGGTTCTCTGCTGGATCATGTGAGACAACACCGGGGGGCACTGGGGCCACAGCTGCTGCTCAACTGGG GAGTACAAATTGCCAAGGGAATGTACTACCTTGAGGAACATGGTATGGTGCATAGAAACCTGGCTGCCCGAAACGTGCTACTCAAGTCAC CCAGTCAGGTTCAGGTGGCAGATTTTGGTGTGGCTGACCTGCTGCCTCCTGATGATAAGCAGCTGCTATACAGTGAGGCCAAGACTCCAA TTAAGTGGATGGCCCTTGAGAGTATCCACTTTGGGAAATACACACACCAGAGTGATGTCTGGAGCTATGGGGTGACTGTTTGGGAGTTGA TGACCTTTGGATCCAAGCCATATGACGGAATCCCTGCCAGCGAGATCTCCTCCATCCTGGAGAAAGGAGAACGCCTCCCTCAGCCACCCA TATGTACCATCGATGTCTACATGATCATGGTCAAGTGCTGGATGATAGACGCAGATAGTCGCCCAAAGTTCCGTGAGTTGATCATCGAAT TCTCCAAAATGGCCCGAGACCCCCAGCGCTACCTTGTCATTCAGGGGGATGAAAGAATGCATTTGCCAAGTCCTACAGACTCCAACTTCT ACCGTGCCCTGATGGATGAAGAAGACATGGACGACGTGGTGGATGCCGACGAGTACCTCATCCCACAGCAGGGCTTCTTCAGCAGCCCCT CCACGTCACGGACTCCCCTCCTGAGCTCTCTGAGTGCAACCAGCAACAATTCCACCGTGGCTTGCATTGATAGAAATGGGCTGCAAAGCT GTCCCATCAAGGAAGACAGCTTCTTGCAGCGATACAGCTCAGACCCCACAGGCGCCTTGACTGAGGACAGCATAGACGACACCTTCCTCC CAGTGCCTGGTGAGTGGCTTGTCTGGAAACAGTCCTGCTCCTCAACCTCCTCGACCCACTCAGCAGCAGCCAGTCTCCAGTGTCCAAGCC AGGTGCTCCCTCCAGCATCTCCAGAGGGGGAAACAGTGGCAGATTTGCAGACACAGTGAAGGGCGTAAGGAGCAGATAAACACATGACCG AGCCTGCACAAGCTCTTTGTTGTGTCTGGTTGTTTGCTGTACCTCTGTTGTAAGAATGAATCTGCAAAATTTCTAGCTTATGAAGCAAAT CACGGACATACACATCTGTGTGTGTGAGTGTTCATGATGTGTGTACATCTGTGTATGTGTGTGTGTGTATGTGTGTGTTTGTGACAGATT TGATCCCTGTTCTCTCTGCTGGCTCTATCTTGACCTGTGAAACGTATATTTAACTAATTAAATATTAGTTAATATTAATAAATTTTAAGC >27211_27211_4_ERBB3-EGFR_ERBB3_chr12_56492359_ENST00000553131_EGFR_chr7_55266410_ENST00000455089_length(amino acids)=458AA_BP=24 MSNSFPFAPQSIEPLDPSEKANKVLARIFKETELRKLKVLGSGVFGTVHKGVWIPEGESIKIPVCIKVIEDKSGRQSFQAVTDHMLAIGS LDHAHIVRLLGLCPGSSLQLVTQYLPLGSLLDHVRQHRGALGPQLLLNWGVQIAKGMYYLEEHGMVHRNLAARNVLLKSPSQVQVADFGV ADLLPPDDKQLLYSEAKTPIKWMALESIHFGKYTHQSDVWSYGVTVWELMTFGSKPYDGIPASEISSILEKGERLPQPPICTIDVYMIMV KCWMIDADSRPKFRELIIEFSKMARDPQRYLVIQGDERMHLPSPTDSNFYRALMDEEDMDDVVDADEYLIPQQGFFSSPSTSRTPLLSSL SATSNNSTVACIDRNGLQSCPIKEDSFLQRYSSDPTGALTEDSIDDTFLPVPGEWLVWKQSCSSTSSTHSAAASLQCPSQVLPPASPEGE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ERBB3-EGFR |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ERBB3-EGFR |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ERBB3-EGFR |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
