
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ERLIN1-DOCK1 (FusionGDB2 ID:27420) |
Fusion Gene Summary for ERLIN1-DOCK1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ERLIN1-DOCK1 | Fusion gene ID: 27420 | Hgene | Tgene | Gene symbol | ERLIN1 | DOCK1 | Gene ID | 10613 | 1793 |
| Gene name | ER lipid raft associated 1 | dedicator of cytokinesis 1 | |
| Synonyms | C10orf69|Erlin-1|KE04|KEO4|SPFH1|SPG62 | DOCK180|ced5 | |
| Cytomap | 10q24.31 | 10q26.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | erlin-1Band_7 23-211 Keo4 (Interim) similar to C.elegans protein C42C1.9SPFH domain family, member 1SPFH domain-containing protein 1endoplasmic reticulum lipid raft-associated protein 1stomatin-prohibitin-flotillin-HflC/K domain-containing protein 1 | dedicator of cytokinesis protein 1180 kDa protein downstream of CRKDOwnstream of CrK | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | O75477 | Q5JSL3 | |
| Ensembl transtripts involved in fusion gene | ENST00000407654, ENST00000421367, | ENST00000484400, ENST00000280333, | |
| Fusion gene scores | * DoF score | 6 X 6 X 5=180 | 16 X 15 X 8=1920 |
| # samples | 6 | 16 | |
| ** MAII score | log2(6/180*10)=-1.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(16/1920*10)=-3.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ERLIN1 [Title/Abstract] AND DOCK1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ERLIN1(101933965)-DOCK1(129201319), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ERLIN1 | GO:0030433 | ubiquitin-dependent ERAD pathway | 19240031 |
 Fusion gene breakpoints across ERLIN1 (5'-gene) Fusion gene breakpoints across ERLIN1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
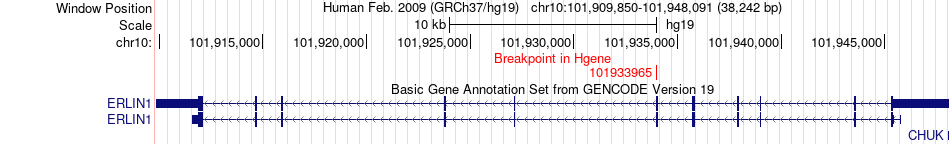 |
 Fusion gene breakpoints across DOCK1 (3'-gene) Fusion gene breakpoints across DOCK1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
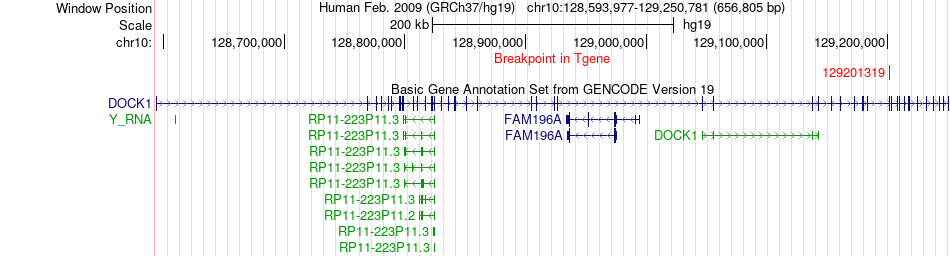 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | DLBC | TCGA-VB-A8QN-01A | ERLIN1 | chr10 | 101933965 | - | DOCK1 | chr10 | 129201319 | + |
| ChimerDB4 | DLBC | TCGA-VB-A8QN | ERLIN1 | chr10 | 101933965 | - | DOCK1 | chr10 | 129201319 | + |
Top |
Fusion Gene ORF analysis for ERLIN1-DOCK1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000407654 | ENST00000484400 | ERLIN1 | chr10 | 101933965 | - | DOCK1 | chr10 | 129201319 | + |
| 5CDS-intron | ENST00000421367 | ENST00000484400 | ERLIN1 | chr10 | 101933965 | - | DOCK1 | chr10 | 129201319 | + |
| In-frame | ENST00000407654 | ENST00000280333 | ERLIN1 | chr10 | 101933965 | - | DOCK1 | chr10 | 129201319 | + |
| In-frame | ENST00000421367 | ENST00000280333 | ERLIN1 | chr10 | 101933965 | - | DOCK1 | chr10 | 129201319 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000421367 | ERLIN1 | chr10 | 101933965 | - | ENST00000280333 | DOCK1 | chr10 | 129201319 | + | 6036 | 3212 | 2708 | 4945 | 745 |
| ENST00000407654 | ERLIN1 | chr10 | 101933965 | - | ENST00000280333 | DOCK1 | chr10 | 129201319 | + | 3411 | 587 | 2 | 2320 | 772 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000421367 | ENST00000280333 | ERLIN1 | chr10 | 101933965 | - | DOCK1 | chr10 | 129201319 | + | 0.000377695 | 0.9996222 |
| ENST00000407654 | ENST00000280333 | ERLIN1 | chr10 | 101933965 | - | DOCK1 | chr10 | 129201319 | + | 0.000715491 | 0.99928457 |
Top |
Fusion Genomic Features for ERLIN1-DOCK1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ERLIN1 | chr10 | 101933964 | - | DOCK1 | chr10 | 129201318 | + | 7.05E-06 | 0.99999297 |
| ERLIN1 | chr10 | 101933964 | - | DOCK1 | chr10 | 129201318 | + | 7.05E-06 | 0.99999297 |
| ERLIN1 | chr10 | 101933964 | - | DOCK1 | chr10 | 129201318 | + | 7.05E-06 | 0.99999297 |
| ERLIN1 | chr10 | 101933964 | - | DOCK1 | chr10 | 129201318 | + | 7.05E-06 | 0.99999297 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
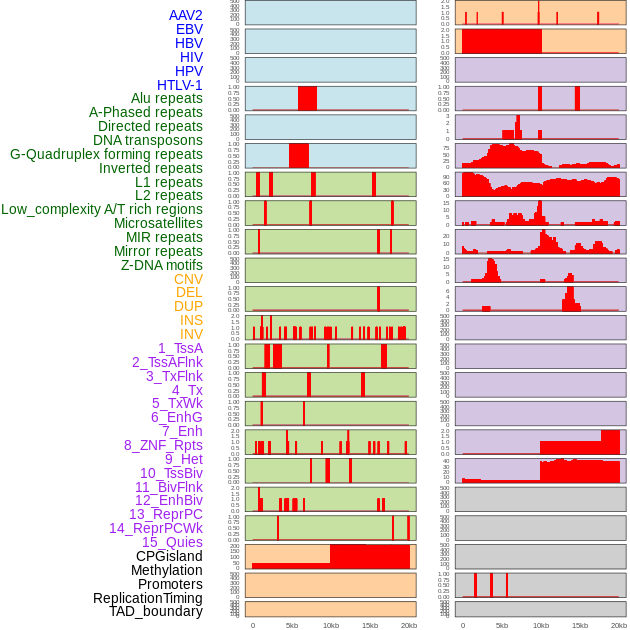 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
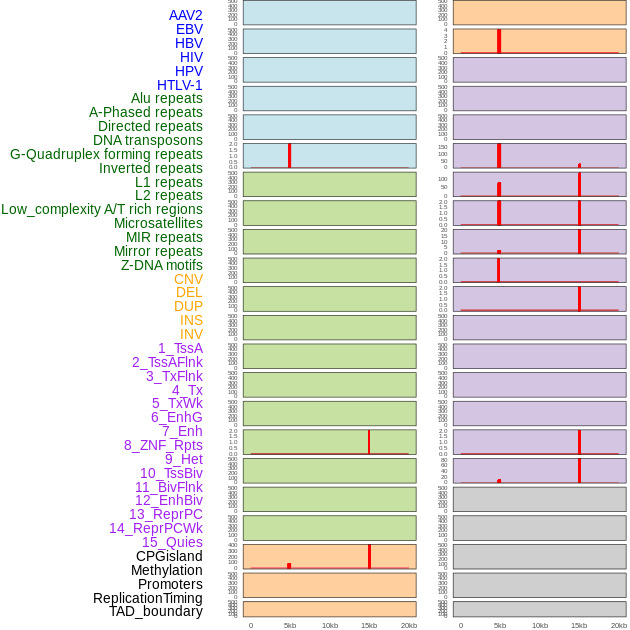 |
Top |
Fusion Protein Features for ERLIN1-DOCK1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:101933965/chr10:129201319) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ERLIN1 | DOCK1 |
| FUNCTION: Component of the ERLIN1/ERLIN2 complex which mediates the endoplasmic reticulum-associated degradation (ERAD) of inositol 1,4,5-trisphosphate receptors (IP3Rs). Involved in regulation of cellular cholesterol homeostasis by regulation the SREBP signaling pathway. Binds cholesterol and may promote ER retention of the SCAP-SREBF complex (PubMed:24217618). {ECO:0000269|PubMed:19240031, ECO:0000269|PubMed:24217618}.; FUNCTION: (Microbial infection) Required early in hepatitis C virus (HCV) infection to initiate RNA replication, and later in the infection to support infectious virus production. {ECO:0000269|PubMed:31810281}. | FUNCTION: Guanine nucleotide-exchange factor (GEF) that activates CDC42 by exchanging bound GDP for free GTP. Required for marginal zone (MZ) B-cell development, is associated with early bone marrow B-cell development, MZ B-cell formation, MZ B-cell number and marginal metallophilic macrophages morphology. Facilitates filopodia formation through the activation of CDC42. {ECO:0000250|UniProtKB:A2AF47}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ERLIN1 | chr10:101933965 | chr10:129201319 | ENST00000407654 | - | 7 | 12 | 1_7 | 168 | 349.0 | Topological domain | Cytoplasmic |
| Hgene | ERLIN1 | chr10:101933965 | chr10:129201319 | ENST00000421367 | - | 6 | 11 | 1_7 | 168 | 349.0 | Topological domain | Cytoplasmic |
| Hgene | ERLIN1 | chr10:101933965 | chr10:129201319 | ENST00000407654 | - | 7 | 12 | 8_28 | 168 | 349.0 | Transmembrane | Helical |
| Hgene | ERLIN1 | chr10:101933965 | chr10:129201319 | ENST00000421367 | - | 6 | 11 | 8_28 | 168 | 349.0 | Transmembrane | Helical |
| Tgene | DOCK1 | chr10:101933965 | chr10:129201319 | ENST00000280333 | 37 | 52 | 1687_1695 | 1288 | 1866.0 | Region | Phosphoinositide-binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ERLIN1 | chr10:101933965 | chr10:129201319 | ENST00000407654 | - | 7 | 12 | 29_348 | 168 | 349.0 | Topological domain | Lumenal |
| Hgene | ERLIN1 | chr10:101933965 | chr10:129201319 | ENST00000421367 | - | 6 | 11 | 29_348 | 168 | 349.0 | Topological domain | Lumenal |
| Tgene | DOCK1 | chr10:101933965 | chr10:129201319 | ENST00000280333 | 37 | 52 | 1207_1617 | 1288 | 1866.0 | Domain | DOCKER | |
| Tgene | DOCK1 | chr10:101933965 | chr10:129201319 | ENST00000280333 | 37 | 52 | 425_609 | 1288 | 1866.0 | Domain | C2 DOCK-type | |
| Tgene | DOCK1 | chr10:101933965 | chr10:129201319 | ENST00000280333 | 37 | 52 | 9_70 | 1288 | 1866.0 | Domain | SH3 |
Top |
Fusion Gene Sequence for ERLIN1-DOCK1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >27420_27420_1_ERLIN1-DOCK1_ERLIN1_chr10_101933965_ENST00000407654_DOCK1_chr10_129201319_ENST00000280333_length(transcript)=3411nt_BP=587nt GGCTGTTGGCGGCGGTTGGCTCGGCGCGGGAGTCGGCTGCACGTGCGGGCGGGGGCGATGCGTCACTGATCGGAGGAACGAGAATGAATA TGACTCAAGCCCGGGTTCTGGTGGCTGCAGTGGTGGGGTTGGTGGCTGTCCTGCTCTACGCCTCCATCCACAAGATTGAGGAGGGCCATC TGGCTGTGTACTACAGGGGAGGAGCTTTACTAACTAGCCCCAGTGGACCAGGCTATCATATCATGTTGCCTTTCATTACTACGTTCAGAT CTGTGCAGACAACACTACAAACTGATGAAGTTAAAAATGTGCCTTGTGGAACAAGTGGTGGGGTCATGATCTATATTGACCGAATAGAAG TGGTTAATATGTTGGCTCCTTATGCAGTGTTTGATATCGTGAGGAACTATACTGCAGATTATGACAAGACCTTAATCTTCAATAAAATCC ACCATGAGCTGAACCAGTTCTGCAGTGCCCACACACTTCAGGAAGTTTACATTGAATTGTTTGATCAAATAGATGAAAACCTGAAGCAAG CTCTGCAGAAAGACTTAAACCTCATGGCCCCAGGTCTCACTATACAGATGTGGGAGGAGGCCATTGCCTTGGGCAAGGAGCTAGCCGAGC AGTATGAGAACGAAATGTTTGATTATGAGCAACTCAGCGAATTGCTGAAAAAACAGGCTCAGTTTTATGAAAACATCGTCAAAGTGATCA GGCCCAAGCCTGACTATTTTGCTGTTGGCTACTACGGACAAGGGTTCCCCACATTCCTGCGGGGAAAAGTTTTCATTTACCGAGGGAAAG AGTATGAGCGCCGGGAAGATTTTGAGGCTCGGCTCTTAACTCAGTTTCCAAACGCCGAGAAAATGAAGACAACATCTCCACCAGGCGACG ATATTAAAAACTCTCCTGGCCAGTATATTCAGTGCTTCACAGTGAAGCCCAAACTCGATCTGCCTCCTAAGTTTCACAGGCCAGTGTCAG AGCAGATTGTAAGTTTTTACAGGGTGAACGAGGTCCAGCGATTTGAATATTCTCGGCCAATCCGGAAGGGAGAGAAAAACCCAGACAATG AATTTGCGAATATGTGGATCGAGAGAACCATATATACAACTGCATATAAATTACCTGGAATTTTAAGGTGGTTTGAGGTCAAGTCTGTTT TCATGGTGGAAATCAGCCCCCTGGAGAATGCCATTGAGACCATGCAGCTGACGAACGACAAGATCAACAGCATGGTGCAGCAGCACCTGG ATGACCCCAGCCTGCCCATCAACCCGCTCTCCATGCTCCTGAACGGCATCGTGGACCCAGCTGTCATGGGGGGCTTCGCAAACTACGAAA AGGCCTTCTTTACAGACCGGTACCTGCAGGAGCACCCTGAGGCCCATGAAAAGATCGAGAAGCTCAAGGACCTGATTGCTTGGCAGATTC CTTTTCTGGCCGAAGGGATCAGAATCCATGGAGACAAAGTCACGGAGGCACTGAGGCCGTTCCACGAGAGGATGGAGGCCTGTTTCAAAC AGCTGAAGGAAAAGGTGGAGAAAGAGTACGGCGTCCGAATCATGCCCTCAAGTCTGGATGATAGAAGAGGCAGCCGCCCCCGGTCCATGG TGCGGTCCTTCACGATGCCTTCCTCATCCCGCCCTCTGTCTGTGGCCTCTGTCTCTTCCCTCTCATCGGACAGCACCCCTTCCAGACCAG GCTCCGACGGGTTTGCCCTGGAGCCTCTCCTGCCAAAGAAAATGCACTCCAGGTCCCAGGACAAGCTGGACAAGGATGACCTGGAGAAGG AGAAGAAGGACAAGAAGAAGGAAAAAAGGAACAGCAAACATCAAGAGATATTTGAGAAAGAATTTAAACCCACCGACATTTCCCTGCAGC AGTCTGAGGCTGTGATCCTTTCGGAAACGATAAGTCCCCTGCGGCCCCAGAGACCGAAGAGCCAGGTGATGAACGTCATTGGAAGCGAAA GGCGCTTCTCGGTGTCCCCCTCGTCACCGTCCTCCCAGCAAACACCCCCTCCAGTTACACCAAGGGCCAAGCTCAGCTTCAGCATGCAGT CGAGCTTGGAGCTGAACGGCATGACGGGGGCGGACGTGGCCGATGTCCCACCCCCTCTGCCTCTCAAAGGCAGCGTGGCAGATTACGGGA ATTTGATGGAAAACCAGGACTTGCTGGGCTCGCCAACACCTCCACCTCCCCCTCCACACCAGAGGCATCTGCCACCTCCACTGCCCAGCA AAACTCCGCCTCCTCCCCCTCCAAAGACAACTCGCAAGCAGGCATCGGTGGACTCCGGGATCGTGCAGTGACGTCGCAAGCCTCTCTGGA AAGAGTGTGCTGCCCCTCCCCATCTCCATGCCCTCTCCTTCTGTGTCCCCTGAGTCTGCTGTTTACCTCATTGGGCCTGTGATGTTAACA TTTCGTGCGACTGCTTTTTCTTCAAAGGAGTTCAGTTCTCACCATGGAGTGAGTGGCCTTTAGCGTCATGGAGCAAGGTGGGTCTGGGAG GTAGATATGGGTCCGGGATGTGCTATCGTAGTTATCAGAGTTGGGGGCCTCTGAGTGTGTCTGGCTCTGAGAGAGTCTGAGTCTTGCCCA AACATTCTTTCTTTTTGTGCCAAATGACTTGCATTTGCAAAGAGCTCAATTGCTCTGAGCTCAGCCAAGTAGGAGAGGCTAGGCCATCAC TCTTAGGAAGCTGTGTAGTGATGATGTATAAGAATCCTCCTCACTGTCATGGGATGTTGTATCCAGCCCCTCCTTGTTCCAGCCGGTGGT GTGACTTCGTTGGTTGAGGTGTGTCTCCAACCTACATCAGACCATGAAGTTCAACCCCTCCAGGGAAGCTCCTGATTTCCCCTGCATAAT TGAAAATAGGATATTTCTCAGCTATTGAACAGTTACTAATTTATGGGGTGGAAACAGCATTAAGAATACTGAATCAAATGGAAAAACAAA TGAATACAGGAAGATAAGTGTTCGTTCTTTTCTGAAAAAGAGTATGTGTACCACAAGAGCTGGTTTTAATTGGGTGAATTGTTTTTGTCC TCATTCTGTACAGAAATTTGTATATATGATGGTTCTTAGAACTTGTTTTAATTTTTGTGGTCCTTCCGTTTATTATAATAGGCGTCCACC AATGATTATCCATATGTGTTCTTAATTTTTAACTGCTGGAAGTGTTAAAACACACACACACACACACACACATTTTTTTTTTGAGAACTC CAAAGTCCTGAAAATTTTGGTGGACAATGATTTTTAAAAAACTAACTTTGTATAACCTAATATTTGTATTCTCTCATCTATATTTTTTTA >27420_27420_1_ERLIN1-DOCK1_ERLIN1_chr10_101933965_ENST00000407654_DOCK1_chr10_129201319_ENST00000280333_length(amino acids)=772AA_BP=194 LLAAVGSARESAARAGGGDASLIGGTRMNMTQARVLVAAVVGLVAVLLYASIHKIEEGHLAVYYRGGALLTSPSGPGYHIMLPFITTFRS VQTTLQTDEVKNVPCGTSGGVMIYIDRIEVVNMLAPYAVFDIVRNYTADYDKTLIFNKIHHELNQFCSAHTLQEVYIELFDQIDENLKQA LQKDLNLMAPGLTIQMWEEAIALGKELAEQYENEMFDYEQLSELLKKQAQFYENIVKVIRPKPDYFAVGYYGQGFPTFLRGKVFIYRGKE YERREDFEARLLTQFPNAEKMKTTSPPGDDIKNSPGQYIQCFTVKPKLDLPPKFHRPVSEQIVSFYRVNEVQRFEYSRPIRKGEKNPDNE FANMWIERTIYTTAYKLPGILRWFEVKSVFMVEISPLENAIETMQLTNDKINSMVQQHLDDPSLPINPLSMLLNGIVDPAVMGGFANYEK AFFTDRYLQEHPEAHEKIEKLKDLIAWQIPFLAEGIRIHGDKVTEALRPFHERMEACFKQLKEKVEKEYGVRIMPSSLDDRRGSRPRSMV RSFTMPSSSRPLSVASVSSLSSDSTPSRPGSDGFALEPLLPKKMHSRSQDKLDKDDLEKEKKDKKKEKRNSKHQEIFEKEFKPTDISLQQ SEAVILSETISPLRPQRPKSQVMNVIGSERRFSVSPSSPSSQQTPPPVTPRAKLSFSMQSSLELNGMTGADVADVPPPLPLKGSVADYGN -------------------------------------------------------------- >27420_27420_2_ERLIN1-DOCK1_ERLIN1_chr10_101933965_ENST00000421367_DOCK1_chr10_129201319_ENST00000280333_length(transcript)=6036nt_BP=3212nt ATTATAATAAAACAGTTGAATATGGCTTAGGAAAATATGAAGGTTCCATGAAGTGGAATTAAGAGCATAGAATAACTGTACTTTCCTTAG GAATAATAGGACTTATGGTAAAGGTAGTATTGGGCAACTTCTTTAAGAGTGTTTTCCTCTGAGATGTCCTATCACCACTATCTACATCTA AAAAACATGCTCGATTCTTGCCCTATAAACTGATGTCACAGCCCCACCATCCCCATTTTTGCTAGTGGTATCATTTTCCTAGTCAACCAA ATTTTTTAGTCATCACATATCACTGTTCGTCATCTATTATAATAGGCAGTCTCTCTCTTGTTTCCCTGGTTTTAGAAGATTTTAGTAATA AACATTTATTGGGTACCTGTTACTTGGAGGGTATTAAGCTAGATGGCAAGATCTGAAACAACATAGGCAGTATAGTAAAAGTGCTTATCT GGGAGTCTGAACATTACAAGCCATCCAAGCATTGCAATTATTGTTAAGGATTATTTTCAATGGTCATGCATTTTCTAATATTTTAATAAT TGGTTAAAGATTTGTTATAGCGTGGGGGCCGCTGCTGGTGTGTGGCTGGGGTTATGTCAGGGCAGCCTGATCTATATAATTTGGGCAGAT GGTGTGAGTCAGAACAGACTATTATAGTGGGATCCCCAAACTTGCTTTTGATGCATGAGTTGGACCACTATTTTTTGGTGGGTAACAACT TTTCAGAGGGGAATGGCAGTTGTGAATTGTATACATTTCATTCTTTAAGCAATATTATGAAAAACTTCAGAGAATGTCTACAGAAAACAG GGTATGGAGCAAGTTATTTTCCATTCTTTTTGCTTTTTGGAAGTTAAAATAGCTAAGCTCTGCAAATATCATTTATTTGGAACAGATAAG GTCCCAGACATTCCTAGATAATAAAAATCAAATGAATGATACAGGTAAGTGTATTTATTGAAGGGTGTGGGTAGAGTGGTCTGGGAAGTC TTGCTTTAATGAAGACGGATTGATTCTATTTACCTCTTCTTACTTCCACTACTAAGTAACCCTGGGCCTGCAGTTTCCTAACCACTTGAC TTCCTTGCCAACAGTCTGTCTCCACTTTCAGCCGTTTTATAGATTCATTGGCCCTAAAGCACTGCTTTCCCATACTTCCCTACCCAAGAG GGCAGGGGTCCCCAAGCCCTGGGCCGCGGACCGGAACCAGACAGTGGCCTGTTAGGAACCAAGCCGGATAGCAGGAGGTGAGCGGCGGGC GAGTGAGCATTATCGCCTTGAGCTCTGCCTGCTGTCAGATTAGCGAGTGGCATCAGATTCTCATAGGCACGCGAACCCTATTGTGAACTG CACATGTGAGGTTTGCCGGCTCCCTGTGAGAATCTAATGCCTGATGATCTGAAGTGGAACAGTTTCATCCCCAACCCACACACACCCCCA CCCCGTCCGCTGGGTTAGAGGACTTTACAACCCTAGTCCACCTTGTCCAGTTATAGTTCCACCTCTAGCCTTTCAAGGCTTAAACCATTA ATGTCCTTAATTTCTCTTGTATTCATCTATCTCCCAACTATACCTTTTTCCTTCCCTTTTTTATTTTTGGCAATATGTGCCCATGGTTTT TTAATTTAAAACGAACAGAATATGTAGTGACGCCTACCATAGCACTCCTTCCCATCACACAAGCCTTCAAGGAAACTGAAGTACTTACTT TGGTCTCCCTGGAGGATTCCCTCCGCCTCCCGCCCCATGTGCTTAGCAATTCTGTTCCTGTAGTCTGGATGGCCTTCATAAAGCCCTCAC TGGACCAGCATTCTGGCACATAATAGGAACACTTAAAAAACGAGAGAATGATGCCGTTATATCTAATATCTTCCTCTTCTGAACTTTCAC AGCACTTTATTTGCAACAAGTTTAGTTGCTCCTGAAGGGCAGGATCTCTGCCGGAAGCAAGTGCGTGCCACACAGTGGGGCTCCGCATAC ACTGCAAAAGGACAAATAAACCGAACAGCTACCGTTTGAGAGTGAGCGAGTGGGTTCTCTGCACAAGAACAAACCAACCAGTCCCTTGTC CGAAAGGGCGTCTCCTTTTCTCTGCTTCGCTGCTCACTCCAGACTGCGGGCTGTCCTCTTCCGAAGCAGTTAACCAGCAGTGTACAGAAA GCGACTTGCCTCCAAAGGAGCCTGCGCGGCCCGCGGCTAGGAGAATTTTGTCCCATGCGCTCCCCGTCTCACTAGCCGCGGGCCGGGGCT ACGCCGTGTGCGTCCCCGCGCAGCCGCAGTGCTGGGCGAGTGGGCGGGGCCGGCTGTTGGCGGCGGTTGGCTCGGCGCGGGAGTCGGCTG CACGTGCGGGCGGGGGCGATGCGTCACTGATCGGTGAGGCGCGGCCGAGGGGTCGGCTTTCCTCGCGAGCCTGCGGCTGGGCTTCTTCTC AGTTAGTGCCTTCCACCCGGGAGCGACCCTTGGGAGAGGGAGTTTCAGGAAGCTCACCGAGCAGGGGCGGCCCACTGGCCTCCGGGGGCG GAGGAGTTGGCAAGGGGTCAGCGGGCTCAGCCAGAAGGGAAGAATGAGGGGACAGGGGTACTGGACTCCCCGGCTCAGCCTGCGAGAGAG CGCCAAGTTTCCGGAGGGAGAGGGTAGAAACTGGAGGGGGTGGACCTGTCACTCACGGGACTGAGGGTCCTTTTCTCCCGCTCCCAGGAG GAACGAGAATGAATATGACTCAAGCCCGGGTTCTGGTGGCTGCAGTGGTGGGGTTGGTGGCTGTCCTGCTCTACGCCTCCATCCACAAGA TTGAGGAGGGCCATCTGGCTGTGTACTACAGGGGAGGAGCTTTACTAACTAGCCCCAGTGGACCAGGCTATCATATCATGTTGCCTTTCA TTACTACGTTCAGATCTGTGCAGACAACACTACAAACTGATGAAGTTAAAAATGTGCCTTGTGGAACAAGTGGTGGGGTCATGATCTATA TTGACCGAATAGAAGTGGTTAATATGTTGGCTCCTTATGCAGTGTTTGATATCGTGAGGAACTATACTGCAGATTATGACAAGACCTTAA TCTTCAATAAAATCCACCATGAGCTGAACCAGTTCTGCAGTGCCCACACACTTCAGGAAGTTTACATTGAATTGTTTGATCAAATAGATG AAAACCTGAAGCAAGCTCTGCAGAAAGACTTAAACCTCATGGCCCCAGGTCTCACTATACAGATGTGGGAGGAGGCCATTGCCTTGGGCA AGGAGCTAGCCGAGCAGTATGAGAACGAAATGTTTGATTATGAGCAACTCAGCGAATTGCTGAAAAAACAGGCTCAGTTTTATGAAAACA TCGTCAAAGTGATCAGGCCCAAGCCTGACTATTTTGCTGTTGGCTACTACGGACAAGGGTTCCCCACATTCCTGCGGGGAAAAGTTTTCA TTTACCGAGGGAAAGAGTATGAGCGCCGGGAAGATTTTGAGGCTCGGCTCTTAACTCAGTTTCCAAACGCCGAGAAAATGAAGACAACAT CTCCACCAGGCGACGATATTAAAAACTCTCCTGGCCAGTATATTCAGTGCTTCACAGTGAAGCCCAAACTCGATCTGCCTCCTAAGTTTC ACAGGCCAGTGTCAGAGCAGATTGTAAGTTTTTACAGGGTGAACGAGGTCCAGCGATTTGAATATTCTCGGCCAATCCGGAAGGGAGAGA AAAACCCAGACAATGAATTTGCGAATATGTGGATCGAGAGAACCATATATACAACTGCATATAAATTACCTGGAATTTTAAGGTGGTTTG AGGTCAAGTCTGTTTTCATGGTGGAAATCAGCCCCCTGGAGAATGCCATTGAGACCATGCAGCTGACGAACGACAAGATCAACAGCATGG TGCAGCAGCACCTGGATGACCCCAGCCTGCCCATCAACCCGCTCTCCATGCTCCTGAACGGCATCGTGGACCCAGCTGTCATGGGGGGCT TCGCAAACTACGAAAAGGCCTTCTTTACAGACCGGTACCTGCAGGAGCACCCTGAGGCCCATGAAAAGATCGAGAAGCTCAAGGACCTGA TTGCTTGGCAGATTCCTTTTCTGGCCGAAGGGATCAGAATCCATGGAGACAAAGTCACGGAGGCACTGAGGCCGTTCCACGAGAGGATGG AGGCCTGTTTCAAACAGCTGAAGGAAAAGGTGGAGAAAGAGTACGGCGTCCGAATCATGCCCTCAAGTCTGGATGATAGAAGAGGCAGCC GCCCCCGGTCCATGGTGCGGTCCTTCACGATGCCTTCCTCATCCCGCCCTCTGTCTGTGGCCTCTGTCTCTTCCCTCTCATCGGACAGCA CCCCTTCCAGACCAGGCTCCGACGGGTTTGCCCTGGAGCCTCTCCTGCCAAAGAAAATGCACTCCAGGTCCCAGGACAAGCTGGACAAGG ATGACCTGGAGAAGGAGAAGAAGGACAAGAAGAAGGAAAAAAGGAACAGCAAACATCAAGAGATATTTGAGAAAGAATTTAAACCCACCG ACATTTCCCTGCAGCAGTCTGAGGCTGTGATCCTTTCGGAAACGATAAGTCCCCTGCGGCCCCAGAGACCGAAGAGCCAGGTGATGAACG TCATTGGAAGCGAAAGGCGCTTCTCGGTGTCCCCCTCGTCACCGTCCTCCCAGCAAACACCCCCTCCAGTTACACCAAGGGCCAAGCTCA GCTTCAGCATGCAGTCGAGCTTGGAGCTGAACGGCATGACGGGGGCGGACGTGGCCGATGTCCCACCCCCTCTGCCTCTCAAAGGCAGCG TGGCAGATTACGGGAATTTGATGGAAAACCAGGACTTGCTGGGCTCGCCAACACCTCCACCTCCCCCTCCACACCAGAGGCATCTGCCAC CTCCACTGCCCAGCAAAACTCCGCCTCCTCCCCCTCCAAAGACAACTCGCAAGCAGGCATCGGTGGACTCCGGGATCGTGCAGTGACGTC GCAAGCCTCTCTGGAAAGAGTGTGCTGCCCCTCCCCATCTCCATGCCCTCTCCTTCTGTGTCCCCTGAGTCTGCTGTTTACCTCATTGGG CCTGTGATGTTAACATTTCGTGCGACTGCTTTTTCTTCAAAGGAGTTCAGTTCTCACCATGGAGTGAGTGGCCTTTAGCGTCATGGAGCA AGGTGGGTCTGGGAGGTAGATATGGGTCCGGGATGTGCTATCGTAGTTATCAGAGTTGGGGGCCTCTGAGTGTGTCTGGCTCTGAGAGAG TCTGAGTCTTGCCCAAACATTCTTTCTTTTTGTGCCAAATGACTTGCATTTGCAAAGAGCTCAATTGCTCTGAGCTCAGCCAAGTAGGAG AGGCTAGGCCATCACTCTTAGGAAGCTGTGTAGTGATGATGTATAAGAATCCTCCTCACTGTCATGGGATGTTGTATCCAGCCCCTCCTT GTTCCAGCCGGTGGTGTGACTTCGTTGGTTGAGGTGTGTCTCCAACCTACATCAGACCATGAAGTTCAACCCCTCCAGGGAAGCTCCTGA TTTCCCCTGCATAATTGAAAATAGGATATTTCTCAGCTATTGAACAGTTACTAATTTATGGGGTGGAAACAGCATTAAGAATACTGAATC AAATGGAAAAACAAATGAATACAGGAAGATAAGTGTTCGTTCTTTTCTGAAAAAGAGTATGTGTACCACAAGAGCTGGTTTTAATTGGGT GAATTGTTTTTGTCCTCATTCTGTACAGAAATTTGTATATATGATGGTTCTTAGAACTTGTTTTAATTTTTGTGGTCCTTCCGTTTATTA TAATAGGCGTCCACCAATGATTATCCATATGTGTTCTTAATTTTTAACTGCTGGAAGTGTTAAAACACACACACACACACACACACATTT TTTTTTTGAGAACTCCAAAGTCCTGAAAATTTTGGTGGACAATGATTTTTAAAAAACTAACTTTGTATAACCTAATATTTGTATTCTCTC ATCTATATTTTTTTATTCACTATAATCATGATACTCTTCTAATAGACTTAAAAGTTAATCATTGACAACGAAAAATAAAATGTTATTTTA >27420_27420_2_ERLIN1-DOCK1_ERLIN1_chr10_101933965_ENST00000421367_DOCK1_chr10_129201319_ENST00000280333_length(amino acids)=745AA_BP=167 MNMTQARVLVAAVVGLVAVLLYASIHKIEEGHLAVYYRGGALLTSPSGPGYHIMLPFITTFRSVQTTLQTDEVKNVPCGTSGGVMIYIDR IEVVNMLAPYAVFDIVRNYTADYDKTLIFNKIHHELNQFCSAHTLQEVYIELFDQIDENLKQALQKDLNLMAPGLTIQMWEEAIALGKEL AEQYENEMFDYEQLSELLKKQAQFYENIVKVIRPKPDYFAVGYYGQGFPTFLRGKVFIYRGKEYERREDFEARLLTQFPNAEKMKTTSPP GDDIKNSPGQYIQCFTVKPKLDLPPKFHRPVSEQIVSFYRVNEVQRFEYSRPIRKGEKNPDNEFANMWIERTIYTTAYKLPGILRWFEVK SVFMVEISPLENAIETMQLTNDKINSMVQQHLDDPSLPINPLSMLLNGIVDPAVMGGFANYEKAFFTDRYLQEHPEAHEKIEKLKDLIAW QIPFLAEGIRIHGDKVTEALRPFHERMEACFKQLKEKVEKEYGVRIMPSSLDDRRGSRPRSMVRSFTMPSSSRPLSVASVSSLSSDSTPS RPGSDGFALEPLLPKKMHSRSQDKLDKDDLEKEKKDKKKEKRNSKHQEIFEKEFKPTDISLQQSEAVILSETISPLRPQRPKSQVMNVIG SERRFSVSPSSPSSQQTPPPVTPRAKLSFSMQSSLELNGMTGADVADVPPPLPLKGSVADYGNLMENQDLLGSPTPPPPPPHQRHLPPPL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ERLIN1-DOCK1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | DOCK1 | chr10:101933965 | chr10:129201319 | ENST00000280333 | 37 | 52 | 1837_1852 | 1288.0 | 1866.0 | NCK2 (minor) | |
| Tgene | DOCK1 | chr10:101933965 | chr10:129201319 | ENST00000280333 | 37 | 52 | 1793_1819 | 1288.0 | 1866.0 | NCK2 second and third SH3 domain (minor) | |
| Tgene | DOCK1 | chr10:101933965 | chr10:129201319 | ENST00000280333 | 37 | 52 | 1820_1836 | 1288.0 | 1866.0 | NCK2 third SH3 domain (major) |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ERLIN1-DOCK1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ERLIN1-DOCK1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
