
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ETHE1-BMP5 (FusionGDB2 ID:27644) |
Fusion Gene Summary for ETHE1-BMP5 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ETHE1-BMP5 | Fusion gene ID: 27644 | Hgene | Tgene | Gene symbol | ETHE1 | BMP5 | Gene ID | 23474 | 653 |
| Gene name | ETHE1 persulfide dioxygenase | bone morphogenetic protein 5 | |
| Synonyms | HSCO|YF13H12 | - | |
| Cytomap | 19q13.31 | 6p12.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | persulfide dioxygenase ETHE1, mitochondrialethylmalonic encephalopathy 1hepatoma subtracted clone one proteinprotein ETHE1, mitochondrialsulfur dioxygenase ETHE1 | bone morphogenetic protein 5 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | O95571 | P22003 | |
| Ensembl transtripts involved in fusion gene | ENST00000292147, ENST00000600651, | ENST00000370830, ENST00000446683, | |
| Fusion gene scores | * DoF score | 10 X 7 X 8=560 | 3 X 2 X 3=18 |
| # samples | 11 | 3 | |
| ** MAII score | log2(11/560*10)=-2.34792330342031 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: ETHE1 [Title/Abstract] AND BMP5 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ETHE1(44030667)-BMP5(55639041), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ETHE1 | GO:0006749 | glutathione metabolic process | 23144459 |
| Hgene | ETHE1 | GO:0070813 | hydrogen sulfide metabolic process | 23144459 |
| Tgene | BMP5 | GO:0003323 | type B pancreatic cell development | 11865031 |
| Tgene | BMP5 | GO:0008284 | positive regulation of cell proliferation | 15516325 |
| Tgene | BMP5 | GO:0008285 | negative regulation of cell proliferation | 19584291 |
| Tgene | BMP5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation | 11580864 |
| Tgene | BMP5 | GO:0010894 | negative regulation of steroid biosynthetic process | 19584291 |
| Tgene | BMP5 | GO:0032348 | negative regulation of aldosterone biosynthetic process | 19584291 |
| Tgene | BMP5 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway | 19584291 |
| Tgene | BMP5 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 15516325 |
| Tgene | BMP5 | GO:0050679 | positive regulation of epithelial cell proliferation | 11865031 |
| Tgene | BMP5 | GO:1900006 | positive regulation of dendrite development | 11580864 |
| Tgene | BMP5 | GO:2000065 | negative regulation of cortisol biosynthetic process | 19584291 |
 Fusion gene breakpoints across ETHE1 (5'-gene) Fusion gene breakpoints across ETHE1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
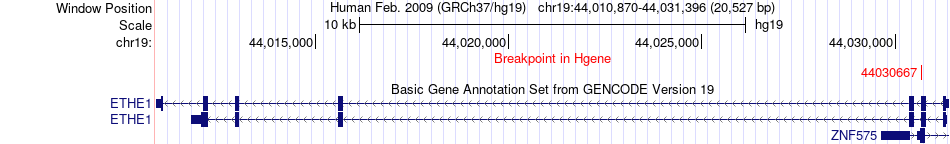 |
 Fusion gene breakpoints across BMP5 (3'-gene) Fusion gene breakpoints across BMP5 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
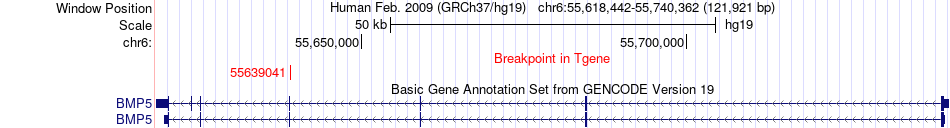 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-55-7815-01A | ETHE1 | chr19 | 44030667 | - | BMP5 | chr6 | 55639041 | - |
Top |
Fusion Gene ORF analysis for ETHE1-BMP5 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000292147 | ENST00000370830 | ETHE1 | chr19 | 44030667 | - | BMP5 | chr6 | 55639041 | - |
| In-frame | ENST00000292147 | ENST00000446683 | ETHE1 | chr19 | 44030667 | - | BMP5 | chr6 | 55639041 | - |
| In-frame | ENST00000600651 | ENST00000370830 | ETHE1 | chr19 | 44030667 | - | BMP5 | chr6 | 55639041 | - |
| In-frame | ENST00000600651 | ENST00000446683 | ETHE1 | chr19 | 44030667 | - | BMP5 | chr6 | 55639041 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000292147 | ETHE1 | chr19 | 44030667 | - | ENST00000370830 | BMP5 | chr6 | 55639041 | - | 2714 | 293 | 67 | 825 | 252 |
| ENST00000292147 | ETHE1 | chr19 | 44030667 | - | ENST00000446683 | BMP5 | chr6 | 55639041 | - | 1321 | 293 | 67 | 714 | 215 |
| ENST00000600651 | ETHE1 | chr19 | 44030667 | - | ENST00000370830 | BMP5 | chr6 | 55639041 | - | 2671 | 250 | 24 | 782 | 252 |
| ENST00000600651 | ETHE1 | chr19 | 44030667 | - | ENST00000446683 | BMP5 | chr6 | 55639041 | - | 1278 | 250 | 24 | 671 | 215 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000292147 | ENST00000370830 | ETHE1 | chr19 | 44030667 | - | BMP5 | chr6 | 55639041 | - | 0.000773109 | 0.9992269 |
| ENST00000292147 | ENST00000446683 | ETHE1 | chr19 | 44030667 | - | BMP5 | chr6 | 55639041 | - | 0.002392899 | 0.9976071 |
| ENST00000600651 | ENST00000370830 | ETHE1 | chr19 | 44030667 | - | BMP5 | chr6 | 55639041 | - | 0.000781122 | 0.9992188 |
| ENST00000600651 | ENST00000446683 | ETHE1 | chr19 | 44030667 | - | BMP5 | chr6 | 55639041 | - | 0.002158546 | 0.9978415 |
Top |
Fusion Genomic Features for ETHE1-BMP5 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
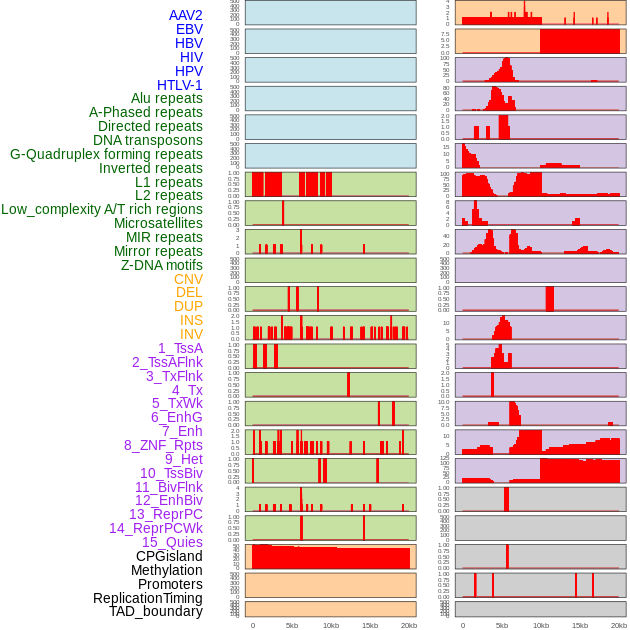 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ETHE1-BMP5 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:44030667/chr6:55639041) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ETHE1 | BMP5 |
| FUNCTION: Sulfur dioxygenase that plays an essential role in hydrogen sulfide catabolism in the mitochondrial matrix. Hydrogen sulfide (H(2)S) is first oxidized by SQRDL, giving rise to cysteine persulfide residues. ETHE1 consumes molecular oxygen to catalyze the oxidation of the persulfide, once it has been transferred to a thiophilic acceptor, such as glutathione (R-SSH). Plays an important role in metabolic homeostasis in mitochondria by metabolizing hydrogen sulfide and preventing the accumulation of supraphysiological H(2)S levels that have toxic effects, due to the inhibition of cytochrome c oxidase. First described as a protein that can shuttle between the nucleus and the cytoplasm and suppress p53-induced apoptosis by sequestering the transcription factor RELA/NFKB3 in the cytoplasm and preventing its accumulation in the nucleus (PubMed:12398897). {ECO:0000269|PubMed:12398897, ECO:0000269|PubMed:14732903, ECO:0000269|PubMed:19136963, ECO:0000269|PubMed:23144459}. | FUNCTION: Induces cartilage and bone formation. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Gene Sequence for ETHE1-BMP5 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >27644_27644_1_ETHE1-BMP5_ETHE1_chr19_44030667_ENST00000292147_BMP5_chr6_55639041_ENST00000370830_length(transcript)=2714nt_BP=293nt AGTGCCGTAGCGCCCGGCTCCTGCAGGCGCTCGGCCTCCGCTCATTCCTGACCCCGCAGTGGGCGCGATGGCGGAGGCTGTACTGAGGGT CGCCCGGCGGCAGCTGAGCCAGCGCGGCGGGTCTGGAGCCCCCATCCTCCTGCGGCAGATGTTCGAGCCTGTGAGCTGCACCTTCACGTA CCTGCTGGGTGACAGAGAGTCCCGGGAGGCCGTTCTGATCGACCCAGTCCTGGAAACAGCGCCTCGGGATGCCCAGCTGATCAAGGAGCT GGGGCTGCGGCTGCTCTATGCTGGACGCAGTATCAACGTAAAATCTGCTGGTCTTGTGGGAAGACAGGGACCTCAGTCAAAACAACCATT CATGGTGGCCTTCTTCAAGGCGAGTGAGGTACTTCTTCGATCCGTGAGAGCAGCCAACAAACGAAAAAATCAAAACCGCAATAAATCCAG CTCTCATCAGGACTCCTCCAGAATGTCCAGTGTTGGAGATTATAACACAAGTGAGCAAAAACAAGCCTGTAAGAAGCACGAACTCTATGT GAGCTTCCGGGATCTGGGATGGCAGGACTGGATTATAGCACCAGAAGGATACGCTGCATTTTATTGTGATGGAGAATGTTCTTTTCCACT TAACGCCCATATGAATGCCACCAACCACGCTATAGTTCAGACTCTGGTTCATCTGATGTTTCCTGACCACGTACCAAAGCCTTGTTGTGC TCCAACCAAATTAAATGCCATCTCTGTTCTGTACTTTGATGACAGCTCCAATGTCATTTTGAAAAAATATAGAAATATGGTAGTACGCTC ATGTGGCTGCCACTAATATTAAATAATATTGATAATAACAAAAAGATCTGTATTAAGGTTTATGGCTGCAATAAAAAGCATACTTTCAGA CAAACGGGGAATTTCCTAAAATTAGTCTGGCTCATTTTGACTCTTTTCCTATGTACAATATCATGTATATAGTCATTTTTATTTATTTAA ACGTATATATTCTCTTCTGTGGAGGCTTTATCAATAAAATCTATTTCATAGGCCACTACATTATACAATAGATCATCTAGTATAATTTTC AATGGGTTATGCTGAGTCCATTCAATTCCATTTCAACATTTTTTTCTAATACAATTTTACATAATTTTCTGAGATTAGACAGAACTCCTT TGACTGTTTAATATTGTGGAAGGAAATTGAATAATTTAGTTATATTGTGCCAATGAAAACCATGATAGAATATTTATTTAGAAATAAGAC TTAAAAAAATAACAACCCAAACAAAAATTCTTGGATAAACACAATTTCTCTTTTAGAGTTCCTGCTTCATTAGATAAAGTGAAATGTTGA ACTGGATTTGAATAAATAAACGGTAGGCAGAACATAGAAGCTGAAAGATTGCACAGAATTGTTTTAACTGTTTGCTTTAACAGAAGTGGT GTAACTGTAGGAGCATGTTCCTTTTTGGCTTGTGGATTGTAGATTTTGCAAAGAGCTCAAAGCTTCAGGAGACAGGCTTGAGCCTGTGTC CATAGCTTTCCAAGTTAACACTAATTGTAGTACAACCTTCTTACCTCAAATAAATCATGTCTGCCTATTATCTAGAACACTACTGCTGCA GAATTTTTTTTTCAGATTTAAAAACCTAGTCCAAAATAAAGAAACATTTACATGTGCTCCACTTTCCTCCCCACAGGCAAGAGTCACTGC ATGCGTAAAATGTACAAGAGCAGAAGATGCCGAAAGAACCATTTTCTTTAGCTCTCCTTTATAACTGAGTTTGCTGATTGATGTGATGCA AGGGATTTTATTTAGAAGTAAAAGCACTGAAACTAAACTGGAGGAAATTACCAAAAGTCCCAGAATGGCTTTGACAGAATTGCCATTTCT AAAGAACAGCTGAAATTCATTTTTCATATCTTTGAGAGTGAGCTCAATGAAAACCTTATTTTGTAAAGGCATTTTGTAAACCTTCTTCCA AGCACAAATAATTCCAAACAGGAAGGTTCTGTTGTGTGTGTGTGTGTGTGTGTGTCTGTGTGTGCATGTGTGTTCTTCCCTCTAAAATTA ATTTATTTTCTCTATCTATTAGAATGCCCTCTAAGGCCTTTCTTTAACACACACTGACTTTTTGAAGAATGATCTTTAGATTGATAACTT GCTATTTATGGTGACTCTGTGGTTAGGCTTATTTACCGTTAATCTAAAGTATTCATAAAGCACCATTTATTAATCATCCTTTTGGATCTT TCAAACCAAACGTATACCAAAAATGTCTTCAAAAATGTTATGGCTTTGTTTGTAATTCGGGTATATTAAAATGTTGTTTTGAAATACAGT TTATCTTAAAGAAAATTTGACAGTAACTATATTAAGGGGTACATTAAGTAATATATTTTTAATTAATGGTGTACCATTGAGAGAATAAGA AATGGAAGTCTATTGTTTTATTCTTTAAATGTGCTGTTTAAATATATTTCCATATTTTAAAATATATTAAACAGATGCTAATTTCTGCTG TTCTTGATTTGGTTCTTTATTTAAAAGGAATACAAAATGTAAGTATATCATAAATTGAGACAATAAGAATATTTATCAAATGTAAACTGT TTGTGGATCACATGATTCTTTATGTTGTTCTTAATTGCCAAAATTGTTTTTTGTGTTGTTAATAATTATCTCACTATTAAATGTGCAAAG >27644_27644_1_ETHE1-BMP5_ETHE1_chr19_44030667_ENST00000292147_BMP5_chr6_55639041_ENST00000370830_length(amino acids)=252AA_BP=75 MAEAVLRVARRQLSQRGGSGAPILLRQMFEPVSCTFTYLLGDRESREAVLIDPVLETAPRDAQLIKELGLRLLYAGRSINVKSAGLVGRQ GPQSKQPFMVAFFKASEVLLRSVRAANKRKNQNRNKSSSHQDSSRMSSVGDYNTSEQKQACKKHELYVSFRDLGWQDWIIAPEGYAAFYC -------------------------------------------------------------- >27644_27644_2_ETHE1-BMP5_ETHE1_chr19_44030667_ENST00000292147_BMP5_chr6_55639041_ENST00000446683_length(transcript)=1321nt_BP=293nt AGTGCCGTAGCGCCCGGCTCCTGCAGGCGCTCGGCCTCCGCTCATTCCTGACCCCGCAGTGGGCGCGATGGCGGAGGCTGTACTGAGGGT CGCCCGGCGGCAGCTGAGCCAGCGCGGCGGGTCTGGAGCCCCCATCCTCCTGCGGCAGATGTTCGAGCCTGTGAGCTGCACCTTCACGTA CCTGCTGGGTGACAGAGAGTCCCGGGAGGCCGTTCTGATCGACCCAGTCCTGGAAACAGCGCCTCGGGATGCCCAGCTGATCAAGGAGCT GGGGCTGCGGCTGCTCTATGCTGGACGCAGTATCAACGTAAAATCTGCTGGTCTTGTGGGAAGACAGGGACCTCAGTCAAAACAACCATT CATGGTGGCCTTCTTCAAGGCGAGTGAGGTACTTCTTCGATCCGTGAGAGCAGCCAACAAACGAAAAAATCAAAACCGCAATAAATCCAG CTCTCATCAGGACTCCTCCAGAATGTCCAGTGTTGGAGATTATAACACAAGTGAGCAAAAACAAGCCTGTAAGAAGCACGAACTCTATGT GAGCTTCCGGGATCTGGGATGGCAGGTTCATCTGATGTTTCCTGACCACGTACCAAAGCCTTGTTGTGCTCCAACCAAATTAAATGCCAT CTCTGTTCTGTACTTTGATGACAGCTCCAATGTCATTTTGAAAAAATATAGAAATATGGTAGTACGCTCATGTGGCTGCCACTAATATTA AATAATATTGATAATAACAAAAAGATCTGTATTAAGGTTTATGGCTGCAATAAAAAGCATACTTTCAGACAAACGGGGAATTTCCTAAAA TTAGTCTGGCTCATTTTGACTCTTTTCCTATGTACAATATCATGTATATAGTCATTTTTATTTATTTAAACGTATATATTCTCTTCTGTG GAGGCTTTATCAATAAAATCTATTTCATAGGCCACTACATTATACAATAGATCATCTAGTATAATTTTCAATGGGTTATGCTGAGTCCAT TCAATTCCATTTCAACATTTTTTTCTAATACAATTTTACATAATTTTCTGAGATTAGACAGAACTCCTTTGACTGTTTAATATTGTGGAA GGAAATTGAATAATTTAGTTATATTGTGCCAATGAAAACCATGATAGAATATTTATTTAGAAATAAGACTTAAAAAAATAACAACCCAAA CAAAAATTCTTGGATAAACACAATTTCTCTTTTAGAGTTCCTGCTTCATTAGATAAAGTGAAATGTTGAACTGGATTTGAATAAATAAAC >27644_27644_2_ETHE1-BMP5_ETHE1_chr19_44030667_ENST00000292147_BMP5_chr6_55639041_ENST00000446683_length(amino acids)=215AA_BP=75 MAEAVLRVARRQLSQRGGSGAPILLRQMFEPVSCTFTYLLGDRESREAVLIDPVLETAPRDAQLIKELGLRLLYAGRSINVKSAGLVGRQ GPQSKQPFMVAFFKASEVLLRSVRAANKRKNQNRNKSSSHQDSSRMSSVGDYNTSEQKQACKKHELYVSFRDLGWQVHLMFPDHVPKPCC -------------------------------------------------------------- >27644_27644_3_ETHE1-BMP5_ETHE1_chr19_44030667_ENST00000600651_BMP5_chr6_55639041_ENST00000370830_length(transcript)=2671nt_BP=250nt ATTCCTGACCCCGCAGTGGGCGCGATGGCGGAGGCTGTACTGAGGGTCGCCCGGCGGCAGCTGAGCCAGCGCGGCGGGTCTGGAGCCCCC ATCCTCCTGCGGCAGATGTTCGAGCCTGTGAGCTGCACCTTCACGTACCTGCTGGGTGACAGAGAGTCCCGGGAGGCCGTTCTGATCGAC CCAGTCCTGGAAACAGCGCCTCGGGATGCCCAGCTGATCAAGGAGCTGGGGCTGCGGCTGCTCTATGCTGGACGCAGTATCAACGTAAAA TCTGCTGGTCTTGTGGGAAGACAGGGACCTCAGTCAAAACAACCATTCATGGTGGCCTTCTTCAAGGCGAGTGAGGTACTTCTTCGATCC GTGAGAGCAGCCAACAAACGAAAAAATCAAAACCGCAATAAATCCAGCTCTCATCAGGACTCCTCCAGAATGTCCAGTGTTGGAGATTAT AACACAAGTGAGCAAAAACAAGCCTGTAAGAAGCACGAACTCTATGTGAGCTTCCGGGATCTGGGATGGCAGGACTGGATTATAGCACCA GAAGGATACGCTGCATTTTATTGTGATGGAGAATGTTCTTTTCCACTTAACGCCCATATGAATGCCACCAACCACGCTATAGTTCAGACT CTGGTTCATCTGATGTTTCCTGACCACGTACCAAAGCCTTGTTGTGCTCCAACCAAATTAAATGCCATCTCTGTTCTGTACTTTGATGAC AGCTCCAATGTCATTTTGAAAAAATATAGAAATATGGTAGTACGCTCATGTGGCTGCCACTAATATTAAATAATATTGATAATAACAAAA AGATCTGTATTAAGGTTTATGGCTGCAATAAAAAGCATACTTTCAGACAAACGGGGAATTTCCTAAAATTAGTCTGGCTCATTTTGACTC TTTTCCTATGTACAATATCATGTATATAGTCATTTTTATTTATTTAAACGTATATATTCTCTTCTGTGGAGGCTTTATCAATAAAATCTA TTTCATAGGCCACTACATTATACAATAGATCATCTAGTATAATTTTCAATGGGTTATGCTGAGTCCATTCAATTCCATTTCAACATTTTT TTCTAATACAATTTTACATAATTTTCTGAGATTAGACAGAACTCCTTTGACTGTTTAATATTGTGGAAGGAAATTGAATAATTTAGTTAT ATTGTGCCAATGAAAACCATGATAGAATATTTATTTAGAAATAAGACTTAAAAAAATAACAACCCAAACAAAAATTCTTGGATAAACACA ATTTCTCTTTTAGAGTTCCTGCTTCATTAGATAAAGTGAAATGTTGAACTGGATTTGAATAAATAAACGGTAGGCAGAACATAGAAGCTG AAAGATTGCACAGAATTGTTTTAACTGTTTGCTTTAACAGAAGTGGTGTAACTGTAGGAGCATGTTCCTTTTTGGCTTGTGGATTGTAGA TTTTGCAAAGAGCTCAAAGCTTCAGGAGACAGGCTTGAGCCTGTGTCCATAGCTTTCCAAGTTAACACTAATTGTAGTACAACCTTCTTA CCTCAAATAAATCATGTCTGCCTATTATCTAGAACACTACTGCTGCAGAATTTTTTTTTCAGATTTAAAAACCTAGTCCAAAATAAAGAA ACATTTACATGTGCTCCACTTTCCTCCCCACAGGCAAGAGTCACTGCATGCGTAAAATGTACAAGAGCAGAAGATGCCGAAAGAACCATT TTCTTTAGCTCTCCTTTATAACTGAGTTTGCTGATTGATGTGATGCAAGGGATTTTATTTAGAAGTAAAAGCACTGAAACTAAACTGGAG GAAATTACCAAAAGTCCCAGAATGGCTTTGACAGAATTGCCATTTCTAAAGAACAGCTGAAATTCATTTTTCATATCTTTGAGAGTGAGC TCAATGAAAACCTTATTTTGTAAAGGCATTTTGTAAACCTTCTTCCAAGCACAAATAATTCCAAACAGGAAGGTTCTGTTGTGTGTGTGT GTGTGTGTGTGTCTGTGTGTGCATGTGTGTTCTTCCCTCTAAAATTAATTTATTTTCTCTATCTATTAGAATGCCCTCTAAGGCCTTTCT TTAACACACACTGACTTTTTGAAGAATGATCTTTAGATTGATAACTTGCTATTTATGGTGACTCTGTGGTTAGGCTTATTTACCGTTAAT CTAAAGTATTCATAAAGCACCATTTATTAATCATCCTTTTGGATCTTTCAAACCAAACGTATACCAAAAATGTCTTCAAAAATGTTATGG CTTTGTTTGTAATTCGGGTATATTAAAATGTTGTTTTGAAATACAGTTTATCTTAAAGAAAATTTGACAGTAACTATATTAAGGGGTACA TTAAGTAATATATTTTTAATTAATGGTGTACCATTGAGAGAATAAGAAATGGAAGTCTATTGTTTTATTCTTTAAATGTGCTGTTTAAAT ATATTTCCATATTTTAAAATATATTAAACAGATGCTAATTTCTGCTGTTCTTGATTTGGTTCTTTATTTAAAAGGAATACAAAATGTAAG TATATCATAAATTGAGACAATAAGAATATTTATCAAATGTAAACTGTTTGTGGATCACATGATTCTTTATGTTGTTCTTAATTGCCAAAA >27644_27644_3_ETHE1-BMP5_ETHE1_chr19_44030667_ENST00000600651_BMP5_chr6_55639041_ENST00000370830_length(amino acids)=252AA_BP=75 MAEAVLRVARRQLSQRGGSGAPILLRQMFEPVSCTFTYLLGDRESREAVLIDPVLETAPRDAQLIKELGLRLLYAGRSINVKSAGLVGRQ GPQSKQPFMVAFFKASEVLLRSVRAANKRKNQNRNKSSSHQDSSRMSSVGDYNTSEQKQACKKHELYVSFRDLGWQDWIIAPEGYAAFYC -------------------------------------------------------------- >27644_27644_4_ETHE1-BMP5_ETHE1_chr19_44030667_ENST00000600651_BMP5_chr6_55639041_ENST00000446683_length(transcript)=1278nt_BP=250nt ATTCCTGACCCCGCAGTGGGCGCGATGGCGGAGGCTGTACTGAGGGTCGCCCGGCGGCAGCTGAGCCAGCGCGGCGGGTCTGGAGCCCCC ATCCTCCTGCGGCAGATGTTCGAGCCTGTGAGCTGCACCTTCACGTACCTGCTGGGTGACAGAGAGTCCCGGGAGGCCGTTCTGATCGAC CCAGTCCTGGAAACAGCGCCTCGGGATGCCCAGCTGATCAAGGAGCTGGGGCTGCGGCTGCTCTATGCTGGACGCAGTATCAACGTAAAA TCTGCTGGTCTTGTGGGAAGACAGGGACCTCAGTCAAAACAACCATTCATGGTGGCCTTCTTCAAGGCGAGTGAGGTACTTCTTCGATCC GTGAGAGCAGCCAACAAACGAAAAAATCAAAACCGCAATAAATCCAGCTCTCATCAGGACTCCTCCAGAATGTCCAGTGTTGGAGATTAT AACACAAGTGAGCAAAAACAAGCCTGTAAGAAGCACGAACTCTATGTGAGCTTCCGGGATCTGGGATGGCAGGTTCATCTGATGTTTCCT GACCACGTACCAAAGCCTTGTTGTGCTCCAACCAAATTAAATGCCATCTCTGTTCTGTACTTTGATGACAGCTCCAATGTCATTTTGAAA AAATATAGAAATATGGTAGTACGCTCATGTGGCTGCCACTAATATTAAATAATATTGATAATAACAAAAAGATCTGTATTAAGGTTTATG GCTGCAATAAAAAGCATACTTTCAGACAAACGGGGAATTTCCTAAAATTAGTCTGGCTCATTTTGACTCTTTTCCTATGTACAATATCAT GTATATAGTCATTTTTATTTATTTAAACGTATATATTCTCTTCTGTGGAGGCTTTATCAATAAAATCTATTTCATAGGCCACTACATTAT ACAATAGATCATCTAGTATAATTTTCAATGGGTTATGCTGAGTCCATTCAATTCCATTTCAACATTTTTTTCTAATACAATTTTACATAA TTTTCTGAGATTAGACAGAACTCCTTTGACTGTTTAATATTGTGGAAGGAAATTGAATAATTTAGTTATATTGTGCCAATGAAAACCATG ATAGAATATTTATTTAGAAATAAGACTTAAAAAAATAACAACCCAAACAAAAATTCTTGGATAAACACAATTTCTCTTTTAGAGTTCCTG CTTCATTAGATAAAGTGAAATGTTGAACTGGATTTGAATAAATAAACGGTAGGCAGAACATAGAAGCTGAAAGATTGCACAGAATTGTTT >27644_27644_4_ETHE1-BMP5_ETHE1_chr19_44030667_ENST00000600651_BMP5_chr6_55639041_ENST00000446683_length(amino acids)=215AA_BP=75 MAEAVLRVARRQLSQRGGSGAPILLRQMFEPVSCTFTYLLGDRESREAVLIDPVLETAPRDAQLIKELGLRLLYAGRSINVKSAGLVGRQ GPQSKQPFMVAFFKASEVLLRSVRAANKRKNQNRNKSSSHQDSSRMSSVGDYNTSEQKQACKKHELYVSFRDLGWQVHLMFPDHVPKPCC -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ETHE1-BMP5 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ETHE1-BMP5 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ETHE1-BMP5 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
