
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ETNK1-ITPR2 (FusionGDB2 ID:27660) |
Fusion Gene Summary for ETNK1-ITPR2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ETNK1-ITPR2 | Fusion gene ID: 27660 | Hgene | Tgene | Gene symbol | ETNK1 | ITPR2 | Gene ID | 55500 | 3709 |
| Gene name | ethanolamine kinase 1 | inositol 1,4,5-trisphosphate receptor type 2 | |
| Synonyms | EKI|EKI 1|EKI1|Nbla10396 | ANHD|CFAP48|INSP3R2|IP3R2 | |
| Cytomap | 12p12.1 | 12p11.23 | |
| Type of gene | protein-coding | protein-coding | |
| Description | ethanolamine kinase 1putative protein product of Nbla10396 | inositol 1,4,5-trisphosphate receptor type 2IP3 receptorIP3R 2cilia and flagella associated protein 48type 2 InsP3 receptor | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9HBU6 | Q14571 | |
| Ensembl transtripts involved in fusion gene | ENST00000266517, ENST00000335148, | ENST00000242737, ENST00000545902, ENST00000381340, | |
| Fusion gene scores | * DoF score | 11 X 7 X 7=539 | 27 X 21 X 12=6804 |
| # samples | 14 | 27 | |
| ** MAII score | log2(14/539*10)=-1.94485844580754 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(27/6804*10)=-4.65535182861255 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ETNK1 [Title/Abstract] AND ITPR2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ETNK1(22826594)-ITPR2(26634174), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ETNK1 | GO:0006646 | phosphatidylethanolamine biosynthetic process | 11044454 |
| Tgene | ITPR2 | GO:0001666 | response to hypoxia | 19120137 |
 Fusion gene breakpoints across ETNK1 (5'-gene) Fusion gene breakpoints across ETNK1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
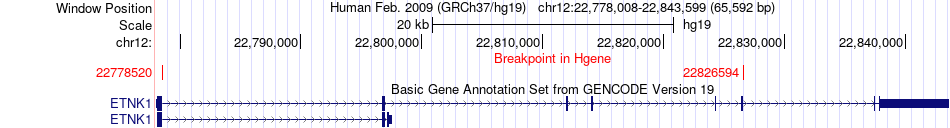 |
 Fusion gene breakpoints across ITPR2 (3'-gene) Fusion gene breakpoints across ITPR2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
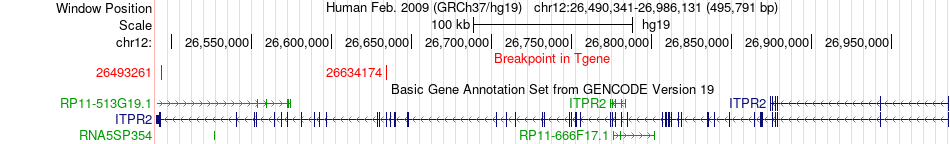 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-25-1329-01A | ETNK1 | chr12 | 22778520 | + | ITPR2 | chr12 | 26493261 | - |
| ChimerDB4 | STAD | TCGA-D7-8575-01A | ETNK1 | chr12 | 22826594 | - | ITPR2 | chr12 | 26634174 | - |
| ChimerDB4 | STAD | TCGA-D7-8575-01A | ETNK1 | chr12 | 22826594 | + | ITPR2 | chr12 | 26634174 | - |
| ChimerDB4 | STAD | TCGA-D7-8575 | ETNK1 | chr12 | 22826594 | + | ITPR2 | chr12 | 26634174 | - |
Top |
Fusion Gene ORF analysis for ETNK1-ITPR2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000266517 | ENST00000242737 | ETNK1 | chr12 | 22778520 | + | ITPR2 | chr12 | 26493261 | - |
| 5CDS-intron | ENST00000266517 | ENST00000242737 | ETNK1 | chr12 | 22826594 | + | ITPR2 | chr12 | 26634174 | - |
| 5CDS-intron | ENST00000266517 | ENST00000545902 | ETNK1 | chr12 | 22778520 | + | ITPR2 | chr12 | 26493261 | - |
| 5CDS-intron | ENST00000266517 | ENST00000545902 | ETNK1 | chr12 | 22826594 | + | ITPR2 | chr12 | 26634174 | - |
| 5CDS-intron | ENST00000335148 | ENST00000242737 | ETNK1 | chr12 | 22778520 | + | ITPR2 | chr12 | 26493261 | - |
| 5CDS-intron | ENST00000335148 | ENST00000545902 | ETNK1 | chr12 | 22778520 | + | ITPR2 | chr12 | 26493261 | - |
| In-frame | ENST00000266517 | ENST00000381340 | ETNK1 | chr12 | 22778520 | + | ITPR2 | chr12 | 26493261 | - |
| In-frame | ENST00000266517 | ENST00000381340 | ETNK1 | chr12 | 22826594 | + | ITPR2 | chr12 | 26634174 | - |
| In-frame | ENST00000335148 | ENST00000381340 | ETNK1 | chr12 | 22778520 | + | ITPR2 | chr12 | 26493261 | - |
| intron-3CDS | ENST00000335148 | ENST00000381340 | ETNK1 | chr12 | 22826594 | + | ITPR2 | chr12 | 26634174 | - |
| intron-intron | ENST00000335148 | ENST00000242737 | ETNK1 | chr12 | 22826594 | + | ITPR2 | chr12 | 26634174 | - |
| intron-intron | ENST00000335148 | ENST00000545902 | ETNK1 | chr12 | 22826594 | + | ITPR2 | chr12 | 26634174 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000266517 | ETNK1 | chr12 | 22826594 | + | ENST00000381340 | ITPR2 | chr12 | 26634174 | - | 5383 | 1301 | 89 | 3394 | 1101 |
| ENST00000266517 | ETNK1 | chr12 | 22778520 | + | ENST00000381340 | ITPR2 | chr12 | 26493261 | - | 2749 | 512 | 89 | 760 | 223 |
| ENST00000335148 | ETNK1 | chr12 | 22778520 | + | ENST00000381340 | ITPR2 | chr12 | 26493261 | - | 2683 | 446 | 23 | 694 | 223 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000266517 | ENST00000381340 | ETNK1 | chr12 | 22826594 | + | ITPR2 | chr12 | 26634174 | - | 0.000238309 | 0.99976164 |
| ENST00000266517 | ENST00000381340 | ETNK1 | chr12 | 22778520 | + | ITPR2 | chr12 | 26493261 | - | 0.23978883 | 0.76021117 |
| ENST00000335148 | ENST00000381340 | ETNK1 | chr12 | 22778520 | + | ITPR2 | chr12 | 26493261 | - | 0.2190329 | 0.78096706 |
Top |
Fusion Genomic Features for ETNK1-ITPR2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
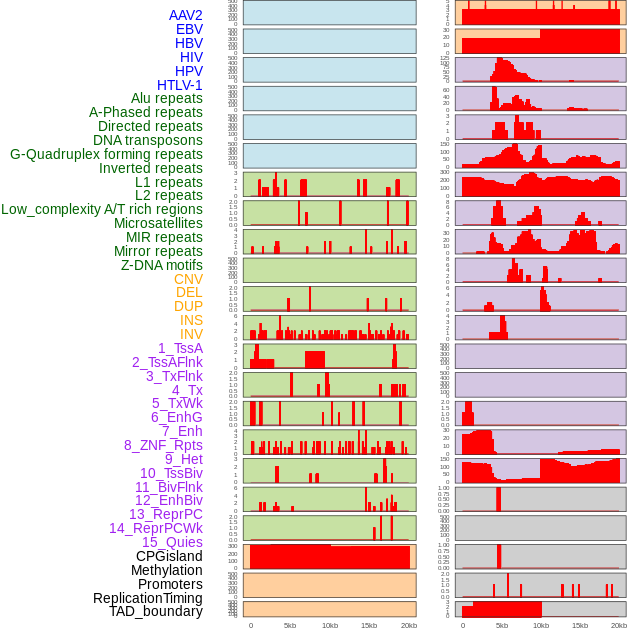 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ETNK1-ITPR2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:22826594/chr12:26634174) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ETNK1 | ITPR2 |
| FUNCTION: Highly specific for ethanolamine phosphorylation. May be a rate-controlling step in phosphatidylethanolamine biosynthesis. {ECO:0000269|PubMed:11044454}. | FUNCTION: Receptor for inositol 1,4,5-trisphosphate, a second messenger that mediates the release of intracellular calcium. This release is regulated by cAMP both dependently and independently of PKA (By similarity). {ECO:0000250|UniProtKB:Q9Z329}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ETNK1 | chr12:22778520 | chr12:26493261 | ENST00000266517 | + | 1 | 8 | 73_83 | 141 | 453.0 | Compositional bias | Note=Poly-Val |
| Hgene | ETNK1 | chr12:22778520 | chr12:26493261 | ENST00000335148 | + | 1 | 3 | 73_83 | 141 | 259.0 | Compositional bias | Note=Poly-Val |
| Hgene | ETNK1 | chr12:22826594 | chr12:26634174 | ENST00000266517 | + | 6 | 8 | 73_83 | 404 | 453.0 | Compositional bias | Note=Poly-Val |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 112_166 | 0 | 182.0 | Domain | MIR 1 | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 173_223 | 0 | 182.0 | Domain | MIR 2 | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 231_287 | 0 | 182.0 | Domain | MIR 3 | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 294_372 | 0 | 182.0 | Domain | MIR 4 | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 378_434 | 0 | 182.0 | Domain | MIR 5 | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 112_166 | 0 | 182.0 | Domain | MIR 1 | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 173_223 | 0 | 182.0 | Domain | MIR 2 | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 231_287 | 0 | 182.0 | Domain | MIR 3 | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 294_372 | 0 | 182.0 | Domain | MIR 4 | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 378_434 | 0 | 182.0 | Domain | MIR 5 | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 265_269 | 0 | 182.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 507_510 | 0 | 182.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 567_569 | 0 | 182.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 265_269 | 0 | 182.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 507_510 | 0 | 182.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 567_569 | 0 | 182.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 1_2227 | 0 | 182.0 | Topological domain | Cytoplasmic | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 2249_2260 | 0 | 182.0 | Topological domain | Extracellular | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 2282_2307 | 0 | 182.0 | Topological domain | Cytoplasmic | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 2329_2351 | 0 | 182.0 | Topological domain | Extracellular | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 2373_2394 | 0 | 182.0 | Topological domain | Cytoplasmic | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 2416_2521 | 0 | 182.0 | Topological domain | Extracellular | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 2543_2701 | 0 | 182.0 | Topological domain | Cytoplasmic | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 1_2227 | 0 | 182.0 | Topological domain | Cytoplasmic | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 2249_2260 | 0 | 182.0 | Topological domain | Extracellular | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 2282_2307 | 0 | 182.0 | Topological domain | Cytoplasmic | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 2329_2351 | 0 | 182.0 | Topological domain | Extracellular | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 2373_2394 | 0 | 182.0 | Topological domain | Cytoplasmic | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 2416_2521 | 0 | 182.0 | Topological domain | Extracellular | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 2543_2701 | 0 | 182.0 | Topological domain | Cytoplasmic | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 2249_2260 | 2004 | 2702.0 | Topological domain | Extracellular | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 2282_2307 | 2004 | 2702.0 | Topological domain | Cytoplasmic | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 2329_2351 | 2004 | 2702.0 | Topological domain | Extracellular | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 2373_2394 | 2004 | 2702.0 | Topological domain | Cytoplasmic | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 2416_2521 | 2004 | 2702.0 | Topological domain | Extracellular | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 2543_2701 | 2004 | 2702.0 | Topological domain | Cytoplasmic | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 2228_2248 | 0 | 182.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 2261_2281 | 0 | 182.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 2308_2328 | 0 | 182.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 2352_2372 | 0 | 182.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 2395_2415 | 0 | 182.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000242737 | 0 | 6 | 2522_2542 | 0 | 182.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 2228_2248 | 0 | 182.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 2261_2281 | 0 | 182.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 2308_2328 | 0 | 182.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 2352_2372 | 0 | 182.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 2395_2415 | 0 | 182.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000242737 | 0 | 6 | 2522_2542 | 0 | 182.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 2228_2248 | 2004 | 2702.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 2261_2281 | 2004 | 2702.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 2308_2328 | 2004 | 2702.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 2352_2372 | 2004 | 2702.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 2395_2415 | 2004 | 2702.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 2522_2542 | 2004 | 2702.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ETNK1 | chr12:22826594 | chr12:26634174 | ENST00000335148 | + | 1 | 3 | 73_83 | 0 | 259.0 | Compositional bias | Note=Poly-Val |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 112_166 | 2619 | 2702.0 | Domain | MIR 1 | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 173_223 | 2619 | 2702.0 | Domain | MIR 2 | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 231_287 | 2619 | 2702.0 | Domain | MIR 3 | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 294_372 | 2619 | 2702.0 | Domain | MIR 4 | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 378_434 | 2619 | 2702.0 | Domain | MIR 5 | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 112_166 | 2004 | 2702.0 | Domain | MIR 1 | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 173_223 | 2004 | 2702.0 | Domain | MIR 2 | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 231_287 | 2004 | 2702.0 | Domain | MIR 3 | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 294_372 | 2004 | 2702.0 | Domain | MIR 4 | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 378_434 | 2004 | 2702.0 | Domain | MIR 5 | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 265_269 | 2619 | 2702.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 507_510 | 2619 | 2702.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 567_569 | 2619 | 2702.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 265_269 | 2004 | 2702.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 507_510 | 2004 | 2702.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 567_569 | 2004 | 2702.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 1_2227 | 2619 | 2702.0 | Topological domain | Cytoplasmic | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 2249_2260 | 2619 | 2702.0 | Topological domain | Extracellular | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 2282_2307 | 2619 | 2702.0 | Topological domain | Cytoplasmic | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 2329_2351 | 2619 | 2702.0 | Topological domain | Extracellular | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 2373_2394 | 2619 | 2702.0 | Topological domain | Cytoplasmic | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 2416_2521 | 2619 | 2702.0 | Topological domain | Extracellular | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 2543_2701 | 2619 | 2702.0 | Topological domain | Cytoplasmic | |
| Tgene | ITPR2 | chr12:22826594 | chr12:26634174 | ENST00000381340 | 41 | 57 | 1_2227 | 2004 | 2702.0 | Topological domain | Cytoplasmic | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 2228_2248 | 2619 | 2702.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 2261_2281 | 2619 | 2702.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 2308_2328 | 2619 | 2702.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 2352_2372 | 2619 | 2702.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 2395_2415 | 2619 | 2702.0 | Transmembrane | Helical | |
| Tgene | ITPR2 | chr12:22778520 | chr12:26493261 | ENST00000381340 | 54 | 57 | 2522_2542 | 2619 | 2702.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for ETNK1-ITPR2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >27660_27660_1_ETNK1-ITPR2_ETNK1_chr12_22778520_ENST00000266517_ITPR2_chr12_26493261_ENST00000381340_length(transcript)=2749nt_BP=512nt GTGACCGGAGGCGAGAAACCCCGCCTCGGCACCCTGACGCAGCGCAGGACCCGCCCCGCGCGTGACGCCAGCGTCAGGCCAGCCCCGGCA TGCTCTGCGGCCGCCCGCGGTCCAGCTCCGACAACAGGAATTTTCTCCGAGAGCGGGCCGGGCTCAGTTCAGCTGCTGTCCAGACCCGGA TCGGCAACAGTGCCGCCTCCAGACGTTCTCCTGCCGCTCGCCCGCCCGTCCCAGCGCCCCCAGCCCTCCCGCGAGGGCGCCCCGGGACGG AAGGATCCACCAGTCTGTCGGCGCCCGCCGTTCTCGTGGTCGCCGTCGCCGTCGTCGTGGTGGTAGTCTCCGCCGTCGCCTGGGCCATGG CCAATTACATCCACGTCCCTCCCGGCTCCCCGGAGGTGCCCAAGCTGAACGTCACCGTTCAGGATCAGGAGGAGCATCGCTGCCGGGAGG GGGCCCTGAGCCTCCTGCAACACCTGCGGCCTCACTGGGACCCCCAGGAGGTGACCCTGCAGGAGAAGAATTTGGATTGGTTTCCTCGGA TGCGAGCCATGTCCCTCGTTAGCAATGAAGGCGACAGTGAGCAAAATGAAATTCGGAGCCTTCAGGAGAAGTTGGAATCGACCATGAGTC TGGTCAAACAGCTGTCGGGTCAGCTGGCGGAGCTCAAGGAGCAGATGACAGAACAAAGGAAGAATAAGCAGAGACTGGGCTTCCTCGGAT CAAACACACCCCATGTGAATCATCACATGCCACCACACTGATACCATGGGGGGAAGCCGTGACTAGCCTTTCATCAGTGTCCTGCCTGAT CACTGAATAAAGAACTGAGATGGAGGGGAGTGAACAGTGCCTATTGTTGAAAAGTTAAAAACAACCAAGTGCCAAGATGTTGAGTGGGTT AGCTCCGAGAACAATTTATAACTGTGTTTTCATGGTTGCGAAGACCTAACCTCAAATGCATCTGCTAGAAAGCGTACATCACACATTCGC AATGCATCAGGAAGAAAAGGCTTGCCCAAAAGGCTGGAGAGGGCAGGGAGCGGCAGGATGGAAGGAGACACGGGGCAGGGAGAACTCTCT TCTGCTAAATCGATAGGAGTCAGTTTTGTCTTAAATGCTGACTACAGCCACTGACATGGTTGGCTGGAATTTCTTTCTTTTAATTGTGGC ATATAGGTTTGTGACACAAGAAGTCATACTTTGGTGGCTAAGTTTTACTAAGGAAAATAACTGAAAAGATTAAAAGTGAGAGCTGAAAAG AGAAATGATAATGCTTCCAAACTGTAGCTGTCACAGGGCAATTTCTTTATTTATAACATGAAGCACAATGGATTTACAGCTCTAGGAACT TAGTACTTTGGAGCTTTTGCCTCTCACACTGACAACATAACAGGATGTGATTGCCTTCTCTGGGATTCAGACAGGCTCTGTCAATGTGGA GCACAAAAGGAGATTTTCATATAACTTGTTAAAAACATGTTCTAAGTCATGTATAGGCTAAGATTTTAAGAAGATCTGGGGGAATAAAAA GCCAACAGTAAGCCCCAGGAAAGGGTTTTTTGAGACCATATGTATGCTATTAAAATATATTGAGATTAATACATGAAAATGCTTAAAAGT GATAGGAATTATTGAGAAACTTATGATGGTGGCATTGCCTTTTATAAACATGGAGTGAAAGCCATTTGACTCAACGTTTGCTGTGCTTAA AGAATTGCTTCAGGACCCGAGCGTTTTTATGTATGCTGTTCCTGCAGTTAGGAAAAAAAAATCCAAGAAATGTATTGAATACTCAAGAAA ATGCCACATTTCAATTACATATTTAAAACTGTATCTGTAAGGGCTTTTCAAAATGTAGCAAGATAAAAATCTCTTATTCAAATTGTTTTT GTATTAAATCATGCACTCAGAATTTGTCGAGGGAGAGAATAACCTGGTGTGTTCAGGTTATTCTTAGAGACTACACATTTGGAATAGCAG AGCAAAATATGGATAATGAAAGTGTTGGGAAAAAAGTGATGTGACAGGGAGTGAAGACACTTGATACCAGACTCTGGGAGATACTCTGCA AGTTGACCTGGCCTCTCCCCCACAGGAACAAAACACTGCCTCCAGAGTCTTTAAATTCTCAGTTATCAACGCCAAGGTTTAAGGTCTAGA TAGGGTTTGCTATAGGGTTTGCTCTGACAATTTTTAAAGCTTTTGGATTGTTCTAACATAGCTTAAGCTATTGGTTCCTAAAAATCCCAA ATCAAGATCTATGTAGAATATAAAGGAAGCCTGAACCAATCCTTCCACATACTGTTAAGATGTAGACTTGGAACAAAGCTGTTGGGACCC AGAGCAATGAATTTTTGAACTGAAGCTACTGGTACTCCCCAGCACCACCTCATACTAAGAATTCCTCACTCTGACATGACAGTATTTTTT CTGACCAGGAGCTGAAAGACCCTGACATTCATGATCCAAAGATATAAGAAATTTTGATATGTCTGCATGCAGCACAGAACTTTGAACCTA GGAGGCAGTCAATAAATACATGAGAAATTCAGCTGTCATTCAGTTACTCTTAATTCTTTATAAAACTTTAAATGATACCACATATTTGTT TGTTTAAAATGGCTTTCCCAAAATCAAAGTAGACTAAAGCAGCCATCTTTAAAATCCAGGATCATCAATGCTATTAACAGTATCAGGTAA >27660_27660_1_ETNK1-ITPR2_ETNK1_chr12_22778520_ENST00000266517_ITPR2_chr12_26493261_ENST00000381340_length(amino acids)=223AA_BP=140 MLCGRPRSSSDNRNFLRERAGLSSAAVQTRIGNSAASRRSPAARPPVPAPPALPRGRPGTEGSTSLSAPAVLVVAVAVVVVVVSAVAWAM ANYIHVPPGSPEVPKLNVTVQDQEEHRCREGALSLLQHLRPHWDPQEVTLQEKNLDWFPRMRAMSLVSNEGDSEQNEIRSLQEKLESTMS -------------------------------------------------------------- >27660_27660_2_ETNK1-ITPR2_ETNK1_chr12_22778520_ENST00000335148_ITPR2_chr12_26493261_ENST00000381340_length(transcript)=2683nt_BP=446nt GCCAGCGTCAGGCCAGCCCCGGCATGCTCTGCGGCCGCCCGCGGTCCAGCTCCGACAACAGGAATTTTCTCCGAGAGCGGGCCGGGCTCA GTTCAGCTGCTGTCCAGACCCGGATCGGCAACAGTGCCGCCTCCAGACGTTCTCCTGCCGCTCGCCCGCCCGTCCCAGCGCCCCCAGCCC TCCCGCGAGGGCGCCCCGGGACGGAAGGATCCACCAGTCTGTCGGCGCCCGCCGTTCTCGTGGTCGCCGTCGCCGTCGTCGTGGTGGTAG TCTCCGCCGTCGCCTGGGCCATGGCCAATTACATCCACGTCCCTCCCGGCTCCCCGGAGGTGCCCAAGCTGAACGTCACCGTTCAGGATC AGGAGGAGCATCGCTGCCGGGAGGGGGCCCTGAGCCTCCTGCAACACCTGCGGCCTCACTGGGACCCCCAGGAGGTGACCCTGCAGGAGA AGAATTTGGATTGGTTTCCTCGGATGCGAGCCATGTCCCTCGTTAGCAATGAAGGCGACAGTGAGCAAAATGAAATTCGGAGCCTTCAGG AGAAGTTGGAATCGACCATGAGTCTGGTCAAACAGCTGTCGGGTCAGCTGGCGGAGCTCAAGGAGCAGATGACAGAACAAAGGAAGAATA AGCAGAGACTGGGCTTCCTCGGATCAAACACACCCCATGTGAATCATCACATGCCACCACACTGATACCATGGGGGGAAGCCGTGACTAG CCTTTCATCAGTGTCCTGCCTGATCACTGAATAAAGAACTGAGATGGAGGGGAGTGAACAGTGCCTATTGTTGAAAAGTTAAAAACAACC AAGTGCCAAGATGTTGAGTGGGTTAGCTCCGAGAACAATTTATAACTGTGTTTTCATGGTTGCGAAGACCTAACCTCAAATGCATCTGCT AGAAAGCGTACATCACACATTCGCAATGCATCAGGAAGAAAAGGCTTGCCCAAAAGGCTGGAGAGGGCAGGGAGCGGCAGGATGGAAGGA GACACGGGGCAGGGAGAACTCTCTTCTGCTAAATCGATAGGAGTCAGTTTTGTCTTAAATGCTGACTACAGCCACTGACATGGTTGGCTG GAATTTCTTTCTTTTAATTGTGGCATATAGGTTTGTGACACAAGAAGTCATACTTTGGTGGCTAAGTTTTACTAAGGAAAATAACTGAAA AGATTAAAAGTGAGAGCTGAAAAGAGAAATGATAATGCTTCCAAACTGTAGCTGTCACAGGGCAATTTCTTTATTTATAACATGAAGCAC AATGGATTTACAGCTCTAGGAACTTAGTACTTTGGAGCTTTTGCCTCTCACACTGACAACATAACAGGATGTGATTGCCTTCTCTGGGAT TCAGACAGGCTCTGTCAATGTGGAGCACAAAAGGAGATTTTCATATAACTTGTTAAAAACATGTTCTAAGTCATGTATAGGCTAAGATTT TAAGAAGATCTGGGGGAATAAAAAGCCAACAGTAAGCCCCAGGAAAGGGTTTTTTGAGACCATATGTATGCTATTAAAATATATTGAGAT TAATACATGAAAATGCTTAAAAGTGATAGGAATTATTGAGAAACTTATGATGGTGGCATTGCCTTTTATAAACATGGAGTGAAAGCCATT TGACTCAACGTTTGCTGTGCTTAAAGAATTGCTTCAGGACCCGAGCGTTTTTATGTATGCTGTTCCTGCAGTTAGGAAAAAAAAATCCAA GAAATGTATTGAATACTCAAGAAAATGCCACATTTCAATTACATATTTAAAACTGTATCTGTAAGGGCTTTTCAAAATGTAGCAAGATAA AAATCTCTTATTCAAATTGTTTTTGTATTAAATCATGCACTCAGAATTTGTCGAGGGAGAGAATAACCTGGTGTGTTCAGGTTATTCTTA GAGACTACACATTTGGAATAGCAGAGCAAAATATGGATAATGAAAGTGTTGGGAAAAAAGTGATGTGACAGGGAGTGAAGACACTTGATA CCAGACTCTGGGAGATACTCTGCAAGTTGACCTGGCCTCTCCCCCACAGGAACAAAACACTGCCTCCAGAGTCTTTAAATTCTCAGTTAT CAACGCCAAGGTTTAAGGTCTAGATAGGGTTTGCTATAGGGTTTGCTCTGACAATTTTTAAAGCTTTTGGATTGTTCTAACATAGCTTAA GCTATTGGTTCCTAAAAATCCCAAATCAAGATCTATGTAGAATATAAAGGAAGCCTGAACCAATCCTTCCACATACTGTTAAGATGTAGA CTTGGAACAAAGCTGTTGGGACCCAGAGCAATGAATTTTTGAACTGAAGCTACTGGTACTCCCCAGCACCACCTCATACTAAGAATTCCT CACTCTGACATGACAGTATTTTTTCTGACCAGGAGCTGAAAGACCCTGACATTCATGATCCAAAGATATAAGAAATTTTGATATGTCTGC ATGCAGCACAGAACTTTGAACCTAGGAGGCAGTCAATAAATACATGAGAAATTCAGCTGTCATTCAGTTACTCTTAATTCTTTATAAAAC TTTAAATGATACCACATATTTGTTTGTTTAAAATGGCTTTCCCAAAATCAAAGTAGACTAAAGCAGCCATCTTTAAAATCCAGGATCATC >27660_27660_2_ETNK1-ITPR2_ETNK1_chr12_22778520_ENST00000335148_ITPR2_chr12_26493261_ENST00000381340_length(amino acids)=223AA_BP=140 MLCGRPRSSSDNRNFLRERAGLSSAAVQTRIGNSAASRRSPAARPPVPAPPALPRGRPGTEGSTSLSAPAVLVVAVAVVVVVVSAVAWAM ANYIHVPPGSPEVPKLNVTVQDQEEHRCREGALSLLQHLRPHWDPQEVTLQEKNLDWFPRMRAMSLVSNEGDSEQNEIRSLQEKLESTMS -------------------------------------------------------------- >27660_27660_3_ETNK1-ITPR2_ETNK1_chr12_22826594_ENST00000266517_ITPR2_chr12_26634174_ENST00000381340_length(transcript)=5383nt_BP=1301nt GTGACCGGAGGCGAGAAACCCCGCCTCGGCACCCTGACGCAGCGCAGGACCCGCCCCGCGCGTGACGCCAGCGTCAGGCCAGCCCCGGCA TGCTCTGCGGCCGCCCGCGGTCCAGCTCCGACAACAGGAATTTTCTCCGAGAGCGGGCCGGGCTCAGTTCAGCTGCTGTCCAGACCCGGA TCGGCAACAGTGCCGCCTCCAGACGTTCTCCTGCCGCTCGCCCGCCCGTCCCAGCGCCCCCAGCCCTCCCGCGAGGGCGCCCCGGGACGG AAGGATCCACCAGTCTGTCGGCGCCCGCCGTTCTCGTGGTCGCCGTCGCCGTCGTCGTGGTGGTAGTCTCCGCCGTCGCCTGGGCCATGG CCAATTACATCCACGTCCCTCCCGGCTCCCCGGAGGTGCCCAAGCTGAACGTCACCGTTCAGGATCAGGAGGAGCATCGCTGCCGGGAGG GGGCCCTGAGCCTCCTGCAACACCTGCGGCCTCACTGGGACCCCCAGGAGGTGACCCTGCAGCTCTTCACAGATGGAATCACAAATAAAC TTATTGGCTGTTACGTGGGAAACACCATGGAGGATGTAGTCCTGGTGAGAATTTATGGCAATAAGACTGAGTTATTAGTCGATCGAGATG AGGAAGTAAAGAGTTTTCGAGTGTTGCAGGCTCATGGGTGTGCACCACAACTCTACTGTACCTTCAATAATGGACTATGCTATGAATTTA TACAAGGAGAAGCACTGGATCCAAAGCATGTCTGCAACCCAGCCATTTTCAGGCTAATAGCTCGTCAGCTTGCTAAAATCCATGCTATTC ATGCACACAATGGCTGGATCCCCAAATCTAATCTTTGGCTAAAGATGGGAAAGTATTTCTCTCTCATTCCCACAGGATTTGCAGATGAAG ACATTAATAAAAGGTTCCTAAGTGATATCCCAAGCTCTCAGATTCTCCAGGAAGAGATGACTTGGATGAAGGAGATTCTTTCCAACCTGG GCTCACCTGTTGTGCTTTGCCATAATGACCTATTGTGTAAGAATATAATCTACAATGAGAAACAAGGTGATGTACAGTTCATTGATTATG AATATTCTGGATACAACTACCTGGCATATGATATTGGAAATCATTTCAATGAATTTGCAGGTGTGAGTGATGTAGACTATAGTCTGTATC CAGATAGAGAACTACAGAGTCAGTGGCTGCGTGCTTACCTTGAAGCCTACAAAGAATTTAAGGGCTTTGGGACTGAAGTTACTGAAAAGG AGGTAGAAATACTCTTCATTCAAGTCAATCAGTTTGCATTGACCTGTATCGCTACACATGAGTCTAATGGGATTGATATCATCATTGCTT TGATTCTGAATGACATAAACCCTCTTGGTAAATACCGAATGGACCTGGTGCTCCAGCTAAAGAACAATGCATCTAAACTTTTGCTGGCCA TTATGGAAAGCAGACATGACAGTGAGAATGCAGAAAGAATTCTTTTTAACATGAGACCCAGAGAACTGGTGGATGTGATGAAGAATGCCT ATAACCAAGGATTGGAATGTGACCATGGGGATGATGAGGGTGGAGATGATGGTGTTTCTCCAAAAGATGTTGGACACAATATCTATATTC TGGCCCATCAGTTGGCCCGCCACAATAAACTGTTGCAGCAGATGCTCAAACCAGGATCGGATCCAGATGAAGGAGATGAAGCCTTAAAGT ATTATGCCAACCACACTGCACAGATTGAGATTGTCCGGCATGATAGGACCATGGAACAAATAGTTTTTCCTGTCCCCAATATATGTGAAT ACCTCACTCGAGAATCCAAGTGCCGTGTGTTCAATACAACTGAAAGGGATGAACAAGGAAGTAAAGTGAATGACTTTTTCCAGCAAACAG AAGATCTCTACAATGAAATGAAGTGGCAGAAGAAAATCAGGAATAACCCTGCACTGTTCTGGTTCTCGAGGCACATCTCTCTCTGGGGGA GCATTTCCTTCAACCTGGCTGTGTTCATCAATTTAGCTGTTGCTCTCTTCTACCCATTTGGGGATGATGGAGATGAAGGTACACTTTCTC CATTGTTCTCGGTTCTTCTTTGGATAGCAGTTGCGATCTGCACATCTATGCTGTTTTTCTTCTCCAAGCCTGTGGGTATTCGGCCGTTTC TTGTATCAATAATGCTCAGATCAATATATACAATAGGTCTTGGGCCTACATTAATACTTCTTGGTGCAGCTAATCTTTGTAATAAAATTG TTTTTCTGGTGAGTTTTGTTGGAAATCGTGGCACGTTCACCCGTGGGTACCGAGCAGTCATCCTGGATATGGCCTTTCTCTATCACGTGG CGTATGTCCTGGTTTGCATGCTGGGCCTTTTTGTCCATGAATTCTTCTATAGCTTCCTGCTTTTTGATTTGGTGTACAGGGAAGAGACTT TGCTGAATGTCATAAAAAGTGTCACACGAAATGGCCGCTCTATTATTCTAACTGCAGTCCTGGCTCTCATCCTCGTCTACCTGTTTTCCA TTATTGGGTTCCTTTTTTTGAAGGATGACTTCACTATGGAAGTTGATAGGCTGAAAAACCGAACTCCTGTTACAGGCAGTCATCAAGTGC CTACTATGACTTTAACTACCATGATGGAAGCATGTGCCAAGGAGAACTGTTCACCCACAATTCCAGCTTCAAATACAGCTGATGAAGAGT ATGAAGATGGAATTGAAAGGACGTGTGACACTCTCCTTATGTGCATTGTCACCGTGCTGAACCAGGGCCTCAGGAATGGCGGTGGTGTGG GGGATGTGCTAAGAAGGCCATCGAAAGATGAGCCCTTGTTTGCTGCCCGAGTGGTTTATGACCTTCTTTTTTATTTCATTGTTATCATTA TTGTTCTGAACTTGATTTTTGGTGTTATCATCGATACTTTTGCTGATCTCAGAAGCGAAAAACAGAAAAAAGAAGAAATTCTAAAGACAA CTTGTTTCATCTGTGGACTTGAGAGAGACAAGTTTGATAATAAAACGGTTTCATTTGAGGAGCACATTAAGTCAGAACACAATATGTGGC ATTATTTGTACTTCATAGTCCTGGTGAAAGTTAAAGACCCAACAGAATACACTGGACCTGAAAGTTATGTGGCTCAAATGATTGTGGAGA AGAATTTGGATTGGTTTCCTCGGATGCGAGCCATGTCCCTCGTTAGCAATGAAGGCGACAGTGAGCAAAATGAAATTCGGAGCCTTCAGG AGAAGTTGGAATCGACCATGAGTCTGGTCAAACAGCTGTCGGGTCAGCTGGCGGAGCTCAAGGAGCAGATGACAGAACAAAGGAAGAATA AGCAGAGACTGGGCTTCCTCGGATCAAACACACCCCATGTGAATCATCACATGCCACCACACTGATACCATGGGGGGAAGCCGTGACTAG CCTTTCATCAGTGTCCTGCCTGATCACTGAATAAAGAACTGAGATGGAGGGGAGTGAACAGTGCCTATTGTTGAAAAGTTAAAAACAACC AAGTGCCAAGATGTTGAGTGGGTTAGCTCCGAGAACAATTTATAACTGTGTTTTCATGGTTGCGAAGACCTAACCTCAAATGCATCTGCT AGAAAGCGTACATCACACATTCGCAATGCATCAGGAAGAAAAGGCTTGCCCAAAAGGCTGGAGAGGGCAGGGAGCGGCAGGATGGAAGGA GACACGGGGCAGGGAGAACTCTCTTCTGCTAAATCGATAGGAGTCAGTTTTGTCTTAAATGCTGACTACAGCCACTGACATGGTTGGCTG GAATTTCTTTCTTTTAATTGTGGCATATAGGTTTGTGACACAAGAAGTCATACTTTGGTGGCTAAGTTTTACTAAGGAAAATAACTGAAA AGATTAAAAGTGAGAGCTGAAAAGAGAAATGATAATGCTTCCAAACTGTAGCTGTCACAGGGCAATTTCTTTATTTATAACATGAAGCAC AATGGATTTACAGCTCTAGGAACTTAGTACTTTGGAGCTTTTGCCTCTCACACTGACAACATAACAGGATGTGATTGCCTTCTCTGGGAT TCAGACAGGCTCTGTCAATGTGGAGCACAAAAGGAGATTTTCATATAACTTGTTAAAAACATGTTCTAAGTCATGTATAGGCTAAGATTT TAAGAAGATCTGGGGGAATAAAAAGCCAACAGTAAGCCCCAGGAAAGGGTTTTTTGAGACCATATGTATGCTATTAAAATATATTGAGAT TAATACATGAAAATGCTTAAAAGTGATAGGAATTATTGAGAAACTTATGATGGTGGCATTGCCTTTTATAAACATGGAGTGAAAGCCATT TGACTCAACGTTTGCTGTGCTTAAAGAATTGCTTCAGGACCCGAGCGTTTTTATGTATGCTGTTCCTGCAGTTAGGAAAAAAAAATCCAA GAAATGTATTGAATACTCAAGAAAATGCCACATTTCAATTACATATTTAAAACTGTATCTGTAAGGGCTTTTCAAAATGTAGCAAGATAA AAATCTCTTATTCAAATTGTTTTTGTATTAAATCATGCACTCAGAATTTGTCGAGGGAGAGAATAACCTGGTGTGTTCAGGTTATTCTTA GAGACTACACATTTGGAATAGCAGAGCAAAATATGGATAATGAAAGTGTTGGGAAAAAAGTGATGTGACAGGGAGTGAAGACACTTGATA CCAGACTCTGGGAGATACTCTGCAAGTTGACCTGGCCTCTCCCCCACAGGAACAAAACACTGCCTCCAGAGTCTTTAAATTCTCAGTTAT CAACGCCAAGGTTTAAGGTCTAGATAGGGTTTGCTATAGGGTTTGCTCTGACAATTTTTAAAGCTTTTGGATTGTTCTAACATAGCTTAA GCTATTGGTTCCTAAAAATCCCAAATCAAGATCTATGTAGAATATAAAGGAAGCCTGAACCAATCCTTCCACATACTGTTAAGATGTAGA CTTGGAACAAAGCTGTTGGGACCCAGAGCAATGAATTTTTGAACTGAAGCTACTGGTACTCCCCAGCACCACCTCATACTAAGAATTCCT CACTCTGACATGACAGTATTTTTTCTGACCAGGAGCTGAAAGACCCTGACATTCATGATCCAAAGATATAAGAAATTTTGATATGTCTGC ATGCAGCACAGAACTTTGAACCTAGGAGGCAGTCAATAAATACATGAGAAATTCAGCTGTCATTCAGTTACTCTTAATTCTTTATAAAAC TTTAAATGATACCACATATTTGTTTGTTTAAAATGGCTTTCCCAAAATCAAAGTAGACTAAAGCAGCCATCTTTAAAATCCAGGATCATC >27660_27660_3_ETNK1-ITPR2_ETNK1_chr12_22826594_ENST00000266517_ITPR2_chr12_26634174_ENST00000381340_length(amino acids)=1101AA_BP=404 MLCGRPRSSSDNRNFLRERAGLSSAAVQTRIGNSAASRRSPAARPPVPAPPALPRGRPGTEGSTSLSAPAVLVVAVAVVVVVVSAVAWAM ANYIHVPPGSPEVPKLNVTVQDQEEHRCREGALSLLQHLRPHWDPQEVTLQLFTDGITNKLIGCYVGNTMEDVVLVRIYGNKTELLVDRD EEVKSFRVLQAHGCAPQLYCTFNNGLCYEFIQGEALDPKHVCNPAIFRLIARQLAKIHAIHAHNGWIPKSNLWLKMGKYFSLIPTGFADE DINKRFLSDIPSSQILQEEMTWMKEILSNLGSPVVLCHNDLLCKNIIYNEKQGDVQFIDYEYSGYNYLAYDIGNHFNEFAGVSDVDYSLY PDRELQSQWLRAYLEAYKEFKGFGTEVTEKEVEILFIQVNQFALTCIATHESNGIDIIIALILNDINPLGKYRMDLVLQLKNNASKLLLA IMESRHDSENAERILFNMRPRELVDVMKNAYNQGLECDHGDDEGGDDGVSPKDVGHNIYILAHQLARHNKLLQQMLKPGSDPDEGDEALK YYANHTAQIEIVRHDRTMEQIVFPVPNICEYLTRESKCRVFNTTERDEQGSKVNDFFQQTEDLYNEMKWQKKIRNNPALFWFSRHISLWG SISFNLAVFINLAVALFYPFGDDGDEGTLSPLFSVLLWIAVAICTSMLFFFSKPVGIRPFLVSIMLRSIYTIGLGPTLILLGAANLCNKI VFLVSFVGNRGTFTRGYRAVILDMAFLYHVAYVLVCMLGLFVHEFFYSFLLFDLVYREETLLNVIKSVTRNGRSIILTAVLALILVYLFS IIGFLFLKDDFTMEVDRLKNRTPVTGSHQVPTMTLTTMMEACAKENCSPTIPASNTADEEYEDGIERTCDTLLMCIVTVLNQGLRNGGGV GDVLRRPSKDEPLFAARVVYDLLFYFIVIIIVLNLIFGVIIDTFADLRSEKQKKEEILKTTCFICGLERDKFDNKTVSFEEHIKSEHNMW HYLYFIVLVKVKDPTEYTGPESYVAQMIVEKNLDWFPRMRAMSLVSNEGDSEQNEIRSLQEKLESTMSLVKQLSGQLAELKEQMTEQRKN -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ETNK1-ITPR2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ETNK1-ITPR2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ETNK1-ITPR2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
