
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:EVL-EML1 (FusionGDB2 ID:27802) |
Fusion Gene Summary for EVL-EML1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: EVL-EML1 | Fusion gene ID: 27802 | Hgene | Tgene | Gene symbol | EVL | EML1 | Gene ID | 51466 | 2009 |
| Gene name | Enah/Vasp-like | EMAP like 1 | |
| Synonyms | RNB6 | BH|ELP79|EMAP|EMAP-1|EMAPL | |
| Cytomap | 14q32.2 | 14q32.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | ena/VASP-like proteinena/vasodilator-stimulated phosphoprotein-likeepididymis secretory sperm binding protein | echinoderm microtubule-associated protein-like 1echinoderm microtubule associated protein like 1 | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | Q9UI08 | O00423 | |
| Ensembl transtripts involved in fusion gene | ENST00000555048, ENST00000392920, ENST00000402714, ENST00000544450, | ENST00000262233, ENST00000327921, ENST00000334192, ENST00000556758, | |
| Fusion gene scores | * DoF score | 11 X 11 X 2=242 | 7 X 8 X 5=280 |
| # samples | 11 | 8 | |
| ** MAII score | log2(11/242*10)=-1.13750352374993 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(8/280*10)=-1.8073549220576 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: EVL [Title/Abstract] AND EML1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | EVL(100551192)-EML1(100380903), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across EVL (5'-gene) Fusion gene breakpoints across EVL (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
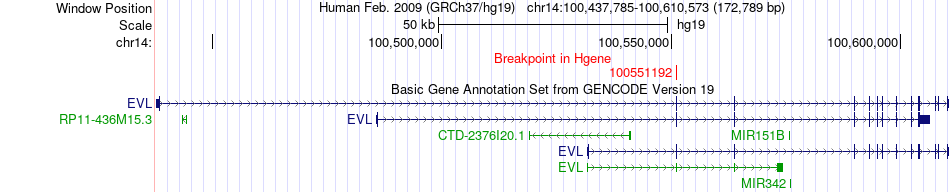 |
 Fusion gene breakpoints across EML1 (3'-gene) Fusion gene breakpoints across EML1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
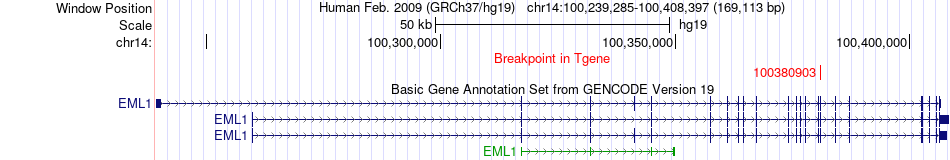 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-A2-A4S3-01A | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + |
Top |
Fusion Gene ORF analysis for EVL-EML1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000555048 | ENST00000262233 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + |
| 3UTR-3CDS | ENST00000555048 | ENST00000327921 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + |
| 3UTR-3CDS | ENST00000555048 | ENST00000334192 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + |
| 3UTR-intron | ENST00000555048 | ENST00000556758 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + |
| 5CDS-intron | ENST00000392920 | ENST00000556758 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + |
| 5CDS-intron | ENST00000402714 | ENST00000556758 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + |
| 5CDS-intron | ENST00000544450 | ENST00000556758 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + |
| In-frame | ENST00000392920 | ENST00000262233 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + |
| In-frame | ENST00000392920 | ENST00000327921 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + |
| In-frame | ENST00000392920 | ENST00000334192 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + |
| In-frame | ENST00000402714 | ENST00000262233 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + |
| In-frame | ENST00000402714 | ENST00000327921 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + |
| In-frame | ENST00000402714 | ENST00000334192 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + |
| In-frame | ENST00000544450 | ENST00000262233 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + |
| In-frame | ENST00000544450 | ENST00000327921 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + |
| In-frame | ENST00000544450 | ENST00000334192 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000402714 | EVL | chr14 | 100551192 | - | ENST00000327921 | EML1 | chr14 | 100380903 | + | 1818 | 778 | 604 | 1605 | 333 |
| ENST00000402714 | EVL | chr14 | 100551192 | - | ENST00000262233 | EML1 | chr14 | 100380903 | + | 3554 | 778 | 604 | 1605 | 333 |
| ENST00000402714 | EVL | chr14 | 100551192 | - | ENST00000334192 | EML1 | chr14 | 100380903 | + | 3031 | 778 | 604 | 1605 | 333 |
| ENST00000544450 | EVL | chr14 | 100551192 | - | ENST00000327921 | EML1 | chr14 | 100380903 | + | 1515 | 475 | 283 | 1302 | 339 |
| ENST00000544450 | EVL | chr14 | 100551192 | - | ENST00000262233 | EML1 | chr14 | 100380903 | + | 3251 | 475 | 283 | 1302 | 339 |
| ENST00000544450 | EVL | chr14 | 100551192 | - | ENST00000334192 | EML1 | chr14 | 100380903 | + | 2728 | 475 | 283 | 1302 | 339 |
| ENST00000392920 | EVL | chr14 | 100551192 | - | ENST00000327921 | EML1 | chr14 | 100380903 | + | 1443 | 403 | 124 | 1230 | 368 |
| ENST00000392920 | EVL | chr14 | 100551192 | - | ENST00000262233 | EML1 | chr14 | 100380903 | + | 3179 | 403 | 124 | 1230 | 368 |
| ENST00000392920 | EVL | chr14 | 100551192 | - | ENST00000334192 | EML1 | chr14 | 100380903 | + | 2656 | 403 | 124 | 1230 | 368 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000402714 | ENST00000327921 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + | 0.004324756 | 0.99567527 |
| ENST00000402714 | ENST00000262233 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + | 0.001176415 | 0.9988236 |
| ENST00000402714 | ENST00000334192 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + | 0.00191529 | 0.9980848 |
| ENST00000544450 | ENST00000327921 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + | 0.001176833 | 0.9988231 |
| ENST00000544450 | ENST00000262233 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + | 0.000464027 | 0.99953604 |
| ENST00000544450 | ENST00000334192 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + | 0.000616538 | 0.99938345 |
| ENST00000392920 | ENST00000327921 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + | 0.002803707 | 0.9971963 |
| ENST00000392920 | ENST00000262233 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + | 0.001039708 | 0.99896026 |
| ENST00000392920 | ENST00000334192 | EVL | chr14 | 100551192 | - | EML1 | chr14 | 100380903 | + | 0.001536076 | 0.998464 |
Top |
Fusion Genomic Features for EVL-EML1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for EVL-EML1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr14:100551192/chr14:100380903) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| EVL | EML1 |
| FUNCTION: Ena/VASP proteins are actin-associated proteins involved in a range of processes dependent on cytoskeleton remodeling and cell polarity such as axon guidance and lamellipodial and filopodial dynamics in migrating cells. EVL enhances actin nucleation and polymerization. | FUNCTION: Modulates the assembly and organization of the microtubule cytoskeleton, and probably plays a role in regulating the orientation of the mitotic spindle and the orientation of the plane of cell division. Required for normal proliferation of neuronal progenitor cells in the developing brain and for normal brain development. Does not affect neuron migration per se. {ECO:0000250|UniProtKB:Q05BC3}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000262233 | 13 | 22 | 578_613 | 540 | 816.0 | Repeat | Note=WD 8 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000262233 | 13 | 22 | 617_655 | 540 | 816.0 | Repeat | Note=WD 9 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000262233 | 13 | 22 | 664_701 | 540 | 816.0 | Repeat | Note=WD 10 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000262233 | 13 | 22 | 709_768 | 540 | 816.0 | Repeat | Note=WD 11 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000262233 | 13 | 22 | 775_814 | 540 | 816.0 | Repeat | Note=WD 12 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000334192 | 14 | 23 | 578_613 | 559 | 835.0 | Repeat | Note=WD 8 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000334192 | 14 | 23 | 617_655 | 559 | 835.0 | Repeat | Note=WD 9 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000334192 | 14 | 23 | 664_701 | 559 | 835.0 | Repeat | Note=WD 10 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000334192 | 14 | 23 | 709_768 | 559 | 835.0 | Repeat | Note=WD 11 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000334192 | 14 | 23 | 775_814 | 559 | 835.0 | Repeat | Note=WD 12 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EVL | chr14:100551192 | chr14:100380903 | ENST00000392920 | - | 2 | 14 | 162_206 | 60 | 419.0 | Compositional bias | Note=Pro-rich |
| Hgene | EVL | chr14:100551192 | chr14:100380903 | ENST00000402714 | - | 2 | 14 | 162_206 | 58 | 417.0 | Compositional bias | Note=Pro-rich |
| Hgene | EVL | chr14:100551192 | chr14:100380903 | ENST00000392920 | - | 2 | 14 | 1_112 | 60 | 419.0 | Domain | WH1 |
| Hgene | EVL | chr14:100551192 | chr14:100380903 | ENST00000402714 | - | 2 | 14 | 1_112 | 58 | 417.0 | Domain | WH1 |
| Hgene | EVL | chr14:100551192 | chr14:100380903 | ENST00000392920 | - | 2 | 14 | 231_234 | 60 | 419.0 | Motif | Note=KLKR |
| Hgene | EVL | chr14:100551192 | chr14:100380903 | ENST00000402714 | - | 2 | 14 | 231_234 | 58 | 417.0 | Motif | Note=KLKR |
| Hgene | EVL | chr14:100551192 | chr14:100380903 | ENST00000392920 | - | 2 | 14 | 222_242 | 60 | 419.0 | Region | Note=EVH2 block A |
| Hgene | EVL | chr14:100551192 | chr14:100380903 | ENST00000392920 | - | 2 | 14 | 222_413 | 60 | 419.0 | Region | Note=EVH2 |
| Hgene | EVL | chr14:100551192 | chr14:100380903 | ENST00000392920 | - | 2 | 14 | 265_282 | 60 | 419.0 | Region | Note=EVH2 block B |
| Hgene | EVL | chr14:100551192 | chr14:100380903 | ENST00000392920 | - | 2 | 14 | 379_413 | 60 | 419.0 | Region | Note=EVH2 block C |
| Hgene | EVL | chr14:100551192 | chr14:100380903 | ENST00000402714 | - | 2 | 14 | 222_242 | 58 | 417.0 | Region | Note=EVH2 block A |
| Hgene | EVL | chr14:100551192 | chr14:100380903 | ENST00000402714 | - | 2 | 14 | 222_413 | 58 | 417.0 | Region | Note=EVH2 |
| Hgene | EVL | chr14:100551192 | chr14:100380903 | ENST00000402714 | - | 2 | 14 | 265_282 | 58 | 417.0 | Region | Note=EVH2 block B |
| Hgene | EVL | chr14:100551192 | chr14:100380903 | ENST00000402714 | - | 2 | 14 | 379_413 | 58 | 417.0 | Region | Note=EVH2 block C |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000262233 | 13 | 22 | 160_166 | 540 | 816.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000334192 | 14 | 23 | 160_166 | 559 | 835.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000262233 | 13 | 22 | 176_815 | 540 | 816.0 | Region | Note=Tandem atypical propeller in EMLs | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000334192 | 14 | 23 | 176_815 | 559 | 835.0 | Region | Note=Tandem atypical propeller in EMLs | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000262233 | 13 | 22 | 261_310 | 540 | 816.0 | Repeat | Note=WD 1 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000262233 | 13 | 22 | 315_358 | 540 | 816.0 | Repeat | Note=WD 2 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000262233 | 13 | 22 | 363_400 | 540 | 816.0 | Repeat | Note=WD 3 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000262233 | 13 | 22 | 409_446 | 540 | 816.0 | Repeat | Note=WD 4 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000262233 | 13 | 22 | 450_489 | 540 | 816.0 | Repeat | Note=WD 5 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000262233 | 13 | 22 | 493_530 | 540 | 816.0 | Repeat | Note=WD 6 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000262233 | 13 | 22 | 535_572 | 540 | 816.0 | Repeat | Note=WD 7 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000334192 | 14 | 23 | 261_310 | 559 | 835.0 | Repeat | Note=WD 1 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000334192 | 14 | 23 | 315_358 | 559 | 835.0 | Repeat | Note=WD 2 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000334192 | 14 | 23 | 363_400 | 559 | 835.0 | Repeat | Note=WD 3 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000334192 | 14 | 23 | 409_446 | 559 | 835.0 | Repeat | Note=WD 4 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000334192 | 14 | 23 | 450_489 | 559 | 835.0 | Repeat | Note=WD 5 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000334192 | 14 | 23 | 493_530 | 559 | 835.0 | Repeat | Note=WD 6 | |
| Tgene | EML1 | chr14:100551192 | chr14:100380903 | ENST00000334192 | 14 | 23 | 535_572 | 559 | 835.0 | Repeat | Note=WD 7 |
Top |
Fusion Gene Sequence for EVL-EML1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >27802_27802_1_EVL-EML1_EVL_chr14_100551192_ENST00000392920_EML1_chr14_100380903_ENST00000262233_length(transcript)=3179nt_BP=403nt GTGTTAGCTTCTTGTTTCTCTATTTTCTACCTAGGTTTCAGTTTTTCACTTGGGTTTGTTTCCCCTTTGTCTCTGGTTCAGCTTCCTTTT CCTGTTTGGTTTTAAGTAGGCTATAAAAATCAAGTTGCTGTCTTCAGAGGGTCTGTGGTCCTCTGATCAACATAGGCTGGTGGGAGTACA GGACTCGCCTCCTCAGGGTTCCCTGTGCTGCCACTTTTCAGCCATGGCCACAAGTGAACAGAGTATCTGCCAAGCCCGGGCTTCCGTGAT GGTCTACGATGACACCAGTAAGAAATGGGTACCAATCAAACCTGGCCAGCAGGGATTCAGCCGGATCAACATCTACCACAACACTGCCAG CAACACCTTCAGAGTCGTTGGAGTCAAGTTGCAGGATCAGCAGGGTCACACTGATGAGCTCTGGGGACTGGCCATCCATGCCTCAAAATC TCAGTTCTTGACCTGTGGGCATGACAAGCATGCCACTCTCTGGGACGCTGTGGGTCACCGTCCCGTCTGGGACAAAATAATAGAGGATCC AGCTCAGTCTTCTGGTTTTCATCCTTCAGGGTCTGTGGTTGCAGTCGGAACACTCACTGGGAGGTGGTTTGTGTTTGACACAGAAACAAA AGACTTGGTCACCGTTCACACAGATGGAAACGAACAGCTCTCTGTAATGCGATACTCACCAGATGGGAATTTCTTAGCCATAGGCTCACA TGACAACTGCATCTATATATATGGCGTTAGTGACAACGGGAGGAAGTACACGCGAGTGGGCAAGTGCTCGGGTCATTCCAGCTTCATTAC TCACCTGGACTGGTCTGTAAACTCACAGTTCCTCGTGTCAAATTCCGGAGACTACGAAATCCTCTACTGGGTTCCCTCTGCCTGTAAGCA AGTCGTAAGTGTGGAAACTACAAGAGACATTGAATGGGCTACCTATACCTGCACTTTGGGATTCCATGTTTTTGGAGTGTGGCCAGAAGG CTCGGACGGAACCGACATCAATGCCGTCTGTCGGGCCCATGAGAAGAAACTCCTGTCAACAGGCGACGACTTTGGCAAAGTGCACCTCTT CTCATACCCCTGCTCGCAGTTCAGGGCTCCAAGCCACATCTACGGCGGGCACAGCAGCCATGTCACCAATGTCGATTTCCTCTGTGAAGA CAGCCACCTCATCTCCACGGGCGGGAAAGACACAAGCATCATGCAGTGGCGCGTCATTTAGTACCCACCGAGAGCTGTGGGGAGCAGCAT GGGCAAGGAAGACACAGACTCGCATTACCCTTGGTCACTGTGATTTCTGTTTTGTTTAAAAAATTCTTACAAACCTCAGGAAAACTGTGC CCTCCGCCGGCTACCTTAGCTTAGCGTGTCAGCGGGCGCCACAGCGGATCAGCGGTTCCGTGTTCACTTTTGTTGTACAATATATGACAC AGTGCACATTGAATACCAACAAGGTTGCAACGTTTACATTATAGCCACATCAACAGAAGTAACTGGTATATTCTTAGTAACTTTTCTATG AACTCTTCAAAAATGGTCACAGAATGCCTTTTAAAACATTGTATATAATCTTCACTGTTTCACCATCTAGCTTGCTAAGTCAAATATTTA TGATGATAATGAGGTACTGAACCACGATGGCTGTTGAGGAATTGGTCCTAAAAGGACAGATCACTTCAGAAGAGTGAATAACTGATTTGC ACAGCTGAATCAGGAGACACAAAGATGAGACTGTGTTTGGTTACATTTTCCAAAGTTTCATTGCATTCTCCCTTGGGGAGGCTGTGAGAG AGGGCTTGTATCCCTCTTGTGCTAAGCAGACTCTACTCCTAACTGACTTCAATATTTCAGCAGGGTACACAGGCGTTTCCAAGTTTCAGT GACACCGTCCTGCCTAACCAGATGCGGTCAGCCTCTTCACACCCACCTGGCTTGCATCCCCCATCCCTTGTTCACACGCCCTGATTCACG GTGAGACATTTTGCCACCTTCTTGTGTATATTACTTGGCATGAGATGATATTGTACTTGTATAGGATTCTAGCAATTCATAATAAATATG TAAGACTAGGCTTTACTGTCTTATGCTTATGGACATTGTATATTTGTATTTTATGACCAAGTAGACCAAGTCAGAAAGATCTCTCTCGAG CGTACCATAAACCTGCAGAGAGAAGTCTCGAAAGGCTCCACCAGGTACCAAGGGCAGCTGCTTTTCCTGTCTTTTGTGCATGGGCGACCC ATTACAGTATGAGATAAGATTGAGTTCTGATGCGTTAAACGGAGGTGGCAGAAATTTGTCAAGAAGGCCTTATCCATTTCGATTGTGTGA CAGATTGAAATTTATTGTTTACATTGGGGAATGTATCTCAAATTTTTAAATAGAAGAGTAATAAACAGACTTTAAAGCAAATATTAAGAT TTTTACTCATTCAAGGCAAGTAAATGAATGGAATTATCTGAGCTCTATGGCACTGGTTGTTTAGAGTGACTGATGAAGTGCAACTTTCAA AAACATTTTTGATGACATCACCAGCCTACTGCAGAAGTGCAGGGCACCAGTAAACACCATGTATTATTGAAGATGAATCTGTTTGTATGT ATCCTTGTCAAATATATTCTATAATGAAATAAAATCTGAAAAGTGGATTTCTTATTGACCTATATTCATGAAAGCATATAAATTAAAATA TTTAAAATTAGATATGATTCACACTATATTCTGTTTCATATGCAGATTTTATTCTCACCTGGGCCATTTGCAGATGAGACTGTAGTTTGC AGATGAGACTGTAGTTTGCAGATGGCGTGGAAGCATTCATCAGGGGAGATAACCATAAAGGATTTGGCCTAATTACCATACTCAATTGTC AGTTTACGTGGTTTTGTGAATACTGGCAAAAGCAATTGTTTTTAAATTAACAATGGAGAGAATGATAAGATGAGGGAAGGAAAAGGCATT CATTATTGACTTACATGTCAGTAAGGTCTGCTTTTATTTCTATGTACTCCTGTTTGCCAAGCTCAATAATGGACAAAGGATACAAACACA CACACATCTACTATTTTAGATAAATGTACTGTTATATATATATGTAAACTACTATTGCTCTCTTTATAATGATTAATCACTTTATTATGA >27802_27802_1_EVL-EML1_EVL_chr14_100551192_ENST00000392920_EML1_chr14_100380903_ENST00000262233_length(amino acids)=368AA_BP=93 MLSSEGLWSSDQHRLVGVQDSPPQGSLCCHFSAMATSEQSICQARASVMVYDDTSKKWVPIKPGQQGFSRINIYHNTASNTFRVVGVKLQ DQQGHTDELWGLAIHASKSQFLTCGHDKHATLWDAVGHRPVWDKIIEDPAQSSGFHPSGSVVAVGTLTGRWFVFDTETKDLVTVHTDGNE QLSVMRYSPDGNFLAIGSHDNCIYIYGVSDNGRKYTRVGKCSGHSSFITHLDWSVNSQFLVSNSGDYEILYWVPSACKQVVSVETTRDIE WATYTCTLGFHVFGVWPEGSDGTDINAVCRAHEKKLLSTGDDFGKVHLFSYPCSQFRAPSHIYGGHSSHVTNVDFLCEDSHLISTGGKDT -------------------------------------------------------------- >27802_27802_2_EVL-EML1_EVL_chr14_100551192_ENST00000392920_EML1_chr14_100380903_ENST00000327921_length(transcript)=1443nt_BP=403nt GTGTTAGCTTCTTGTTTCTCTATTTTCTACCTAGGTTTCAGTTTTTCACTTGGGTTTGTTTCCCCTTTGTCTCTGGTTCAGCTTCCTTTT CCTGTTTGGTTTTAAGTAGGCTATAAAAATCAAGTTGCTGTCTTCAGAGGGTCTGTGGTCCTCTGATCAACATAGGCTGGTGGGAGTACA GGACTCGCCTCCTCAGGGTTCCCTGTGCTGCCACTTTTCAGCCATGGCCACAAGTGAACAGAGTATCTGCCAAGCCCGGGCTTCCGTGAT GGTCTACGATGACACCAGTAAGAAATGGGTACCAATCAAACCTGGCCAGCAGGGATTCAGCCGGATCAACATCTACCACAACACTGCCAG CAACACCTTCAGAGTCGTTGGAGTCAAGTTGCAGGATCAGCAGGGTCACACTGATGAGCTCTGGGGACTGGCCATCCATGCCTCAAAATC TCAGTTCTTGACCTGTGGGCATGACAAGCATGCCACTCTCTGGGACGCTGTGGGTCACCGTCCCGTCTGGGACAAAATAATAGAGGATCC AGCTCAGTCTTCTGGTTTTCATCCTTCAGGGTCTGTGGTTGCAGTCGGAACACTCACTGGGAGGTGGTTTGTGTTTGACACAGAAACAAA AGACTTGGTCACCGTTCACACAGATGGAAACGAACAGCTCTCTGTAATGCGATACTCACCAGATGGGAATTTCTTAGCCATAGGCTCACA TGACAACTGCATCTATATATATGGCGTTAGTGACAACGGGAGGAAGTACACGCGAGTGGGCAAGTGCTCGGGTCATTCCAGCTTCATTAC TCACCTGGACTGGTCTGTAAACTCACAGTTCCTCGTGTCAAATTCCGGAGACTACGAAATCCTCTACTGGGTTCCCTCTGCCTGTAAGCA AGTCGTAAGTGTGGAAACTACAAGAGACATTGAATGGGCTACCTATACCTGCACTTTGGGATTCCATGTTTTTGGAGTGTGGCCAGAAGG CTCGGACGGAACCGACATCAATGCCGTCTGTCGGGCCCATGAGAAGAAACTCCTGTCAACAGGCGACGACTTTGGCAAAGTGCACCTCTT CTCATACCCCTGCTCGCAGTTCAGGGCTCCAAGCCACATCTACGGCGGGCACAGCAGCCATGTCACCAATGTCGATTTCCTCTGTGAAGA CAGCCACCTCATCTCCACGGGCGGGAAAGACACAAGCATCATGCAGTGGCGCGTCATTTAGTACCCACCGAGAGCTGTGGGGAGCAGCAT GGGCAAGGAAGACACAGACTCGCATTACCCTTGGTCACTGTGATTTCTGTTTTGTTTAAAAAATTCTTACAAACCTCAGGAAAACTGTGC CCTCCGCCGGCTACCTTAGCTTAGCGTGTCAGCGGGCGCCACAGCGGATCAGCGGTTCCGTGTTCACTTTTGTTGTACAATATATGACAC >27802_27802_2_EVL-EML1_EVL_chr14_100551192_ENST00000392920_EML1_chr14_100380903_ENST00000327921_length(amino acids)=368AA_BP=93 MLSSEGLWSSDQHRLVGVQDSPPQGSLCCHFSAMATSEQSICQARASVMVYDDTSKKWVPIKPGQQGFSRINIYHNTASNTFRVVGVKLQ DQQGHTDELWGLAIHASKSQFLTCGHDKHATLWDAVGHRPVWDKIIEDPAQSSGFHPSGSVVAVGTLTGRWFVFDTETKDLVTVHTDGNE QLSVMRYSPDGNFLAIGSHDNCIYIYGVSDNGRKYTRVGKCSGHSSFITHLDWSVNSQFLVSNSGDYEILYWVPSACKQVVSVETTRDIE WATYTCTLGFHVFGVWPEGSDGTDINAVCRAHEKKLLSTGDDFGKVHLFSYPCSQFRAPSHIYGGHSSHVTNVDFLCEDSHLISTGGKDT -------------------------------------------------------------- >27802_27802_3_EVL-EML1_EVL_chr14_100551192_ENST00000392920_EML1_chr14_100380903_ENST00000334192_length(transcript)=2656nt_BP=403nt GTGTTAGCTTCTTGTTTCTCTATTTTCTACCTAGGTTTCAGTTTTTCACTTGGGTTTGTTTCCCCTTTGTCTCTGGTTCAGCTTCCTTTT CCTGTTTGGTTTTAAGTAGGCTATAAAAATCAAGTTGCTGTCTTCAGAGGGTCTGTGGTCCTCTGATCAACATAGGCTGGTGGGAGTACA GGACTCGCCTCCTCAGGGTTCCCTGTGCTGCCACTTTTCAGCCATGGCCACAAGTGAACAGAGTATCTGCCAAGCCCGGGCTTCCGTGAT GGTCTACGATGACACCAGTAAGAAATGGGTACCAATCAAACCTGGCCAGCAGGGATTCAGCCGGATCAACATCTACCACAACACTGCCAG CAACACCTTCAGAGTCGTTGGAGTCAAGTTGCAGGATCAGCAGGGTCACACTGATGAGCTCTGGGGACTGGCCATCCATGCCTCAAAATC TCAGTTCTTGACCTGTGGGCATGACAAGCATGCCACTCTCTGGGACGCTGTGGGTCACCGTCCCGTCTGGGACAAAATAATAGAGGATCC AGCTCAGTCTTCTGGTTTTCATCCTTCAGGGTCTGTGGTTGCAGTCGGAACACTCACTGGGAGGTGGTTTGTGTTTGACACAGAAACAAA AGACTTGGTCACCGTTCACACAGATGGAAACGAACAGCTCTCTGTAATGCGATACTCACCAGATGGGAATTTCTTAGCCATAGGCTCACA TGACAACTGCATCTATATATATGGCGTTAGTGACAACGGGAGGAAGTACACGCGAGTGGGCAAGTGCTCGGGTCATTCCAGCTTCATTAC TCACCTGGACTGGTCTGTAAACTCACAGTTCCTCGTGTCAAATTCCGGAGACTACGAAATCCTCTACTGGGTTCCCTCTGCCTGTAAGCA AGTCGTAAGTGTGGAAACTACAAGAGACATTGAATGGGCTACCTATACCTGCACTTTGGGATTCCATGTTTTTGGAGTGTGGCCAGAAGG CTCGGACGGAACCGACATCAATGCCGTCTGTCGGGCCCATGAGAAGAAACTCCTGTCAACAGGCGACGACTTTGGCAAAGTGCACCTCTT CTCATACCCCTGCTCGCAGTTCAGGGCTCCAAGCCACATCTACGGCGGGCACAGCAGCCATGTCACCAATGTCGATTTCCTCTGTGAAGA CAGCCACCTCATCTCCACGGGCGGGAAAGACACAAGCATCATGCAGTGGCGCGTCATTTAGTACCCACCGAGAGCTGTGGGGAGCAGCAT GGGCAAGGAAGACACAGACTCGCATTACCCTTGGTCACTGTGATTTCTGTTTTGTTTAAAAAATTCTTACAAACCTCAGGAAAACTGTGC CCTCCGCCGGCTACCTTAGCTTAGCGTGTCAGCGGGCGCCACAGCGGATCAGCGGTTCCGTGTTCACTTTTGTTGTACAATATATGACAC AGTGCACATTGAATACCAACAAGGTTGCAACGTTTACATTATAGCCACATCAACAGAAGTAACTGGTATATTCTTAGTAACTTTTCTATG AACTCTTCAAAAATGGTCACAGAATGCCTTTTAAAACATTGTATATAATCTTCACTGTTTCACCATCTAGCTTGCTAAGTCAAATATTTA TGATGATAATGAGGTACTGAACCACGATGGCTGTTGAGGAATTGGTCCTAAAAGGACAGATCACTTCAGAAGAGTGAATAACTGATTTGC ACAGCTGAATCAGGAGACACAAAGATGAGACTGTGTTTGGTTACATTTTCCAAAGTTTCATTGCATTCTCCCTTGGGGAGGCTGTGAGAG AGGGCTTGTATCCCTCTTGTGCTAAGCAGACTCTACTCCTAACTGACTTCAATATTTCAGCAGGGTACACAGGCGTTTCCAAGTTTCAGT GACACCGTCCTGCCTAACCAGATGCGGTCAGCCTCTTCACACCCACCTGGCTTGCATCCCCCATCCCTTGTTCACACGCCCTGATTCACG GTGAGACATTTTGCCACCTTCTTGTGTATATTACTTGGCATGAGATGATATTGTACTTGTATAGGATTCTAGCAATTCATAATAAATATG TAAGACTAGGCTTTACTGTCTTATGCTTATGGACATTGTATATTTGTATTTTATGACCAAGTAGACCAAGTCAGAAAGATCTCTCTCGAG CGTACCATAAACCTGCAGAGAGAAGTCTCGAAAGGCTCCACCAGGTACCAAGGGCAGCTGCTTTTCCTGTCTTTTGTGCATGGGCGACCC ATTACAGTATGAGATAAGATTGAGTTCTGATGCGTTAAACGGAGGTGGCAGAAATTTGTCAAGAAGGCCTTATCCATTTCGATTGTGTGA CAGATTGAAATTTATTGTTTACATTGGGGAATGTATCTCAAATTTTTAAATAGAAGAGTAATAAACAGACTTTAAAGCAAATATTAAGAT TTTTACTCATTCAAGGCAAGTAAATGAATGGAATTATCTGAGCTCTATGGCACTGGTTGTTTAGAGTGACTGATGAAGTGCAACTTTCAA AAACATTTTTGATGACATCACCAGCCTACTGCAGAAGTGCAGGGCACCAGTAAACACCATGTATTATTGAAGATGAATCTGTTTGTATGT >27802_27802_3_EVL-EML1_EVL_chr14_100551192_ENST00000392920_EML1_chr14_100380903_ENST00000334192_length(amino acids)=368AA_BP=93 MLSSEGLWSSDQHRLVGVQDSPPQGSLCCHFSAMATSEQSICQARASVMVYDDTSKKWVPIKPGQQGFSRINIYHNTASNTFRVVGVKLQ DQQGHTDELWGLAIHASKSQFLTCGHDKHATLWDAVGHRPVWDKIIEDPAQSSGFHPSGSVVAVGTLTGRWFVFDTETKDLVTVHTDGNE QLSVMRYSPDGNFLAIGSHDNCIYIYGVSDNGRKYTRVGKCSGHSSFITHLDWSVNSQFLVSNSGDYEILYWVPSACKQVVSVETTRDIE WATYTCTLGFHVFGVWPEGSDGTDINAVCRAHEKKLLSTGDDFGKVHLFSYPCSQFRAPSHIYGGHSSHVTNVDFLCEDSHLISTGGKDT -------------------------------------------------------------- >27802_27802_4_EVL-EML1_EVL_chr14_100551192_ENST00000402714_EML1_chr14_100380903_ENST00000262233_length(transcript)=3554nt_BP=778nt GGTGGGGCCCCGCGCCTTTTGTGTCGGGCTTTGACCGCCGCGCGCGCTCATTCACCTCAGCGGCCGCGGGCGGAGGGGCGCACGCGCGCG CAGTCCCCTCGGAGGGCGGTGGGGCGAACAGCAACGCGCGTACCCTAGCTCTTGGCTGCTGTGCTGAGACTAGCCCGCGCGCCCGCGCGC CTCTCCCTCGAGGACTCGGAGCCGCCCGCGCACTCGCCAGGGGCCGCCGCCCCGAGTCTCAGGCGGACGCGCACGCGTTCCCGCCGCCCG GACCCCAGCCGGCTCGCCCCCCGCCCCCCCATCCCCCGGCGGCGGCCCCCAGGCACGCGCCCGCGCACACGCCGGCCCGGGCACGCGCCC GCCAAGATGGCAGGGGCCGGGGCCCAGCTGTCAGTCGCCGCCGCCGCGGCCCTTCCCCGCAGCTAGCGGCCCCGACGCCCACCCGGAGAA GATGAGCCCCCGCTGCCCCCCGCAGCCCAGTCAGACGCGGAGCCGCCGCGCCCGGGGCCGGCTCCCCCGCTAGCGCCCGCGTCCCGCGCC CCGCCGCCGCCGCCGCCGCCGCCGGGTCGCCCCTGGCCCGGCGCGCCCGTCCCCGGCAGCGACAATGAGTGAACAGAGTATCTGCCAAGC CCGGGCTTCCGTGATGGTCTACGATGACACCAGTAAGAAATGGGTACCAATCAAACCTGGCCAGCAGGGATTCAGCCGGATCAACATCTA CCACAACACTGCCAGCAACACCTTCAGAGTCGTTGGAGTCAAGTTGCAGGATCAGCAGGGTCACACTGATGAGCTCTGGGGACTGGCCAT CCATGCCTCAAAATCTCAGTTCTTGACCTGTGGGCATGACAAGCATGCCACTCTCTGGGACGCTGTGGGTCACCGTCCCGTCTGGGACAA AATAATAGAGGATCCAGCTCAGTCTTCTGGTTTTCATCCTTCAGGGTCTGTGGTTGCAGTCGGAACACTCACTGGGAGGTGGTTTGTGTT TGACACAGAAACAAAAGACTTGGTCACCGTTCACACAGATGGAAACGAACAGCTCTCTGTAATGCGATACTCACCAGATGGGAATTTCTT AGCCATAGGCTCACATGACAACTGCATCTATATATATGGCGTTAGTGACAACGGGAGGAAGTACACGCGAGTGGGCAAGTGCTCGGGTCA TTCCAGCTTCATTACTCACCTGGACTGGTCTGTAAACTCACAGTTCCTCGTGTCAAATTCCGGAGACTACGAAATCCTCTACTGGGTTCC CTCTGCCTGTAAGCAAGTCGTAAGTGTGGAAACTACAAGAGACATTGAATGGGCTACCTATACCTGCACTTTGGGATTCCATGTTTTTGG AGTGTGGCCAGAAGGCTCGGACGGAACCGACATCAATGCCGTCTGTCGGGCCCATGAGAAGAAACTCCTGTCAACAGGCGACGACTTTGG CAAAGTGCACCTCTTCTCATACCCCTGCTCGCAGTTCAGGGCTCCAAGCCACATCTACGGCGGGCACAGCAGCCATGTCACCAATGTCGA TTTCCTCTGTGAAGACAGCCACCTCATCTCCACGGGCGGGAAAGACACAAGCATCATGCAGTGGCGCGTCATTTAGTACCCACCGAGAGC TGTGGGGAGCAGCATGGGCAAGGAAGACACAGACTCGCATTACCCTTGGTCACTGTGATTTCTGTTTTGTTTAAAAAATTCTTACAAACC TCAGGAAAACTGTGCCCTCCGCCGGCTACCTTAGCTTAGCGTGTCAGCGGGCGCCACAGCGGATCAGCGGTTCCGTGTTCACTTTTGTTG TACAATATATGACACAGTGCACATTGAATACCAACAAGGTTGCAACGTTTACATTATAGCCACATCAACAGAAGTAACTGGTATATTCTT AGTAACTTTTCTATGAACTCTTCAAAAATGGTCACAGAATGCCTTTTAAAACATTGTATATAATCTTCACTGTTTCACCATCTAGCTTGC TAAGTCAAATATTTATGATGATAATGAGGTACTGAACCACGATGGCTGTTGAGGAATTGGTCCTAAAAGGACAGATCACTTCAGAAGAGT GAATAACTGATTTGCACAGCTGAATCAGGAGACACAAAGATGAGACTGTGTTTGGTTACATTTTCCAAAGTTTCATTGCATTCTCCCTTG GGGAGGCTGTGAGAGAGGGCTTGTATCCCTCTTGTGCTAAGCAGACTCTACTCCTAACTGACTTCAATATTTCAGCAGGGTACACAGGCG TTTCCAAGTTTCAGTGACACCGTCCTGCCTAACCAGATGCGGTCAGCCTCTTCACACCCACCTGGCTTGCATCCCCCATCCCTTGTTCAC ACGCCCTGATTCACGGTGAGACATTTTGCCACCTTCTTGTGTATATTACTTGGCATGAGATGATATTGTACTTGTATAGGATTCTAGCAA TTCATAATAAATATGTAAGACTAGGCTTTACTGTCTTATGCTTATGGACATTGTATATTTGTATTTTATGACCAAGTAGACCAAGTCAGA AAGATCTCTCTCGAGCGTACCATAAACCTGCAGAGAGAAGTCTCGAAAGGCTCCACCAGGTACCAAGGGCAGCTGCTTTTCCTGTCTTTT GTGCATGGGCGACCCATTACAGTATGAGATAAGATTGAGTTCTGATGCGTTAAACGGAGGTGGCAGAAATTTGTCAAGAAGGCCTTATCC ATTTCGATTGTGTGACAGATTGAAATTTATTGTTTACATTGGGGAATGTATCTCAAATTTTTAAATAGAAGAGTAATAAACAGACTTTAA AGCAAATATTAAGATTTTTACTCATTCAAGGCAAGTAAATGAATGGAATTATCTGAGCTCTATGGCACTGGTTGTTTAGAGTGACTGATG AAGTGCAACTTTCAAAAACATTTTTGATGACATCACCAGCCTACTGCAGAAGTGCAGGGCACCAGTAAACACCATGTATTATTGAAGATG AATCTGTTTGTATGTATCCTTGTCAAATATATTCTATAATGAAATAAAATCTGAAAAGTGGATTTCTTATTGACCTATATTCATGAAAGC ATATAAATTAAAATATTTAAAATTAGATATGATTCACACTATATTCTGTTTCATATGCAGATTTTATTCTCACCTGGGCCATTTGCAGAT GAGACTGTAGTTTGCAGATGAGACTGTAGTTTGCAGATGGCGTGGAAGCATTCATCAGGGGAGATAACCATAAAGGATTTGGCCTAATTA CCATACTCAATTGTCAGTTTACGTGGTTTTGTGAATACTGGCAAAAGCAATTGTTTTTAAATTAACAATGGAGAGAATGATAAGATGAGG GAAGGAAAAGGCATTCATTATTGACTTACATGTCAGTAAGGTCTGCTTTTATTTCTATGTACTCCTGTTTGCCAAGCTCAATAATGGACA AAGGATACAAACACACACACATCTACTATTTTAGATAAATGTACTGTTATATATATATGTAAACTACTATTGCTCTCTTTATAATGATTA >27802_27802_4_EVL-EML1_EVL_chr14_100551192_ENST00000402714_EML1_chr14_100380903_ENST00000262233_length(amino acids)=333AA_BP=58 MSEQSICQARASVMVYDDTSKKWVPIKPGQQGFSRINIYHNTASNTFRVVGVKLQDQQGHTDELWGLAIHASKSQFLTCGHDKHATLWDA VGHRPVWDKIIEDPAQSSGFHPSGSVVAVGTLTGRWFVFDTETKDLVTVHTDGNEQLSVMRYSPDGNFLAIGSHDNCIYIYGVSDNGRKY TRVGKCSGHSSFITHLDWSVNSQFLVSNSGDYEILYWVPSACKQVVSVETTRDIEWATYTCTLGFHVFGVWPEGSDGTDINAVCRAHEKK -------------------------------------------------------------- >27802_27802_5_EVL-EML1_EVL_chr14_100551192_ENST00000402714_EML1_chr14_100380903_ENST00000327921_length(transcript)=1818nt_BP=778nt GGTGGGGCCCCGCGCCTTTTGTGTCGGGCTTTGACCGCCGCGCGCGCTCATTCACCTCAGCGGCCGCGGGCGGAGGGGCGCACGCGCGCG CAGTCCCCTCGGAGGGCGGTGGGGCGAACAGCAACGCGCGTACCCTAGCTCTTGGCTGCTGTGCTGAGACTAGCCCGCGCGCCCGCGCGC CTCTCCCTCGAGGACTCGGAGCCGCCCGCGCACTCGCCAGGGGCCGCCGCCCCGAGTCTCAGGCGGACGCGCACGCGTTCCCGCCGCCCG GACCCCAGCCGGCTCGCCCCCCGCCCCCCCATCCCCCGGCGGCGGCCCCCAGGCACGCGCCCGCGCACACGCCGGCCCGGGCACGCGCCC GCCAAGATGGCAGGGGCCGGGGCCCAGCTGTCAGTCGCCGCCGCCGCGGCCCTTCCCCGCAGCTAGCGGCCCCGACGCCCACCCGGAGAA GATGAGCCCCCGCTGCCCCCCGCAGCCCAGTCAGACGCGGAGCCGCCGCGCCCGGGGCCGGCTCCCCCGCTAGCGCCCGCGTCCCGCGCC CCGCCGCCGCCGCCGCCGCCGCCGGGTCGCCCCTGGCCCGGCGCGCCCGTCCCCGGCAGCGACAATGAGTGAACAGAGTATCTGCCAAGC CCGGGCTTCCGTGATGGTCTACGATGACACCAGTAAGAAATGGGTACCAATCAAACCTGGCCAGCAGGGATTCAGCCGGATCAACATCTA CCACAACACTGCCAGCAACACCTTCAGAGTCGTTGGAGTCAAGTTGCAGGATCAGCAGGGTCACACTGATGAGCTCTGGGGACTGGCCAT CCATGCCTCAAAATCTCAGTTCTTGACCTGTGGGCATGACAAGCATGCCACTCTCTGGGACGCTGTGGGTCACCGTCCCGTCTGGGACAA AATAATAGAGGATCCAGCTCAGTCTTCTGGTTTTCATCCTTCAGGGTCTGTGGTTGCAGTCGGAACACTCACTGGGAGGTGGTTTGTGTT TGACACAGAAACAAAAGACTTGGTCACCGTTCACACAGATGGAAACGAACAGCTCTCTGTAATGCGATACTCACCAGATGGGAATTTCTT AGCCATAGGCTCACATGACAACTGCATCTATATATATGGCGTTAGTGACAACGGGAGGAAGTACACGCGAGTGGGCAAGTGCTCGGGTCA TTCCAGCTTCATTACTCACCTGGACTGGTCTGTAAACTCACAGTTCCTCGTGTCAAATTCCGGAGACTACGAAATCCTCTACTGGGTTCC CTCTGCCTGTAAGCAAGTCGTAAGTGTGGAAACTACAAGAGACATTGAATGGGCTACCTATACCTGCACTTTGGGATTCCATGTTTTTGG AGTGTGGCCAGAAGGCTCGGACGGAACCGACATCAATGCCGTCTGTCGGGCCCATGAGAAGAAACTCCTGTCAACAGGCGACGACTTTGG CAAAGTGCACCTCTTCTCATACCCCTGCTCGCAGTTCAGGGCTCCAAGCCACATCTACGGCGGGCACAGCAGCCATGTCACCAATGTCGA TTTCCTCTGTGAAGACAGCCACCTCATCTCCACGGGCGGGAAAGACACAAGCATCATGCAGTGGCGCGTCATTTAGTACCCACCGAGAGC TGTGGGGAGCAGCATGGGCAAGGAAGACACAGACTCGCATTACCCTTGGTCACTGTGATTTCTGTTTTGTTTAAAAAATTCTTACAAACC TCAGGAAAACTGTGCCCTCCGCCGGCTACCTTAGCTTAGCGTGTCAGCGGGCGCCACAGCGGATCAGCGGTTCCGTGTTCACTTTTGTTG >27802_27802_5_EVL-EML1_EVL_chr14_100551192_ENST00000402714_EML1_chr14_100380903_ENST00000327921_length(amino acids)=333AA_BP=58 MSEQSICQARASVMVYDDTSKKWVPIKPGQQGFSRINIYHNTASNTFRVVGVKLQDQQGHTDELWGLAIHASKSQFLTCGHDKHATLWDA VGHRPVWDKIIEDPAQSSGFHPSGSVVAVGTLTGRWFVFDTETKDLVTVHTDGNEQLSVMRYSPDGNFLAIGSHDNCIYIYGVSDNGRKY TRVGKCSGHSSFITHLDWSVNSQFLVSNSGDYEILYWVPSACKQVVSVETTRDIEWATYTCTLGFHVFGVWPEGSDGTDINAVCRAHEKK -------------------------------------------------------------- >27802_27802_6_EVL-EML1_EVL_chr14_100551192_ENST00000402714_EML1_chr14_100380903_ENST00000334192_length(transcript)=3031nt_BP=778nt GGTGGGGCCCCGCGCCTTTTGTGTCGGGCTTTGACCGCCGCGCGCGCTCATTCACCTCAGCGGCCGCGGGCGGAGGGGCGCACGCGCGCG CAGTCCCCTCGGAGGGCGGTGGGGCGAACAGCAACGCGCGTACCCTAGCTCTTGGCTGCTGTGCTGAGACTAGCCCGCGCGCCCGCGCGC CTCTCCCTCGAGGACTCGGAGCCGCCCGCGCACTCGCCAGGGGCCGCCGCCCCGAGTCTCAGGCGGACGCGCACGCGTTCCCGCCGCCCG GACCCCAGCCGGCTCGCCCCCCGCCCCCCCATCCCCCGGCGGCGGCCCCCAGGCACGCGCCCGCGCACACGCCGGCCCGGGCACGCGCCC GCCAAGATGGCAGGGGCCGGGGCCCAGCTGTCAGTCGCCGCCGCCGCGGCCCTTCCCCGCAGCTAGCGGCCCCGACGCCCACCCGGAGAA GATGAGCCCCCGCTGCCCCCCGCAGCCCAGTCAGACGCGGAGCCGCCGCGCCCGGGGCCGGCTCCCCCGCTAGCGCCCGCGTCCCGCGCC CCGCCGCCGCCGCCGCCGCCGCCGGGTCGCCCCTGGCCCGGCGCGCCCGTCCCCGGCAGCGACAATGAGTGAACAGAGTATCTGCCAAGC CCGGGCTTCCGTGATGGTCTACGATGACACCAGTAAGAAATGGGTACCAATCAAACCTGGCCAGCAGGGATTCAGCCGGATCAACATCTA CCACAACACTGCCAGCAACACCTTCAGAGTCGTTGGAGTCAAGTTGCAGGATCAGCAGGGTCACACTGATGAGCTCTGGGGACTGGCCAT CCATGCCTCAAAATCTCAGTTCTTGACCTGTGGGCATGACAAGCATGCCACTCTCTGGGACGCTGTGGGTCACCGTCCCGTCTGGGACAA AATAATAGAGGATCCAGCTCAGTCTTCTGGTTTTCATCCTTCAGGGTCTGTGGTTGCAGTCGGAACACTCACTGGGAGGTGGTTTGTGTT TGACACAGAAACAAAAGACTTGGTCACCGTTCACACAGATGGAAACGAACAGCTCTCTGTAATGCGATACTCACCAGATGGGAATTTCTT AGCCATAGGCTCACATGACAACTGCATCTATATATATGGCGTTAGTGACAACGGGAGGAAGTACACGCGAGTGGGCAAGTGCTCGGGTCA TTCCAGCTTCATTACTCACCTGGACTGGTCTGTAAACTCACAGTTCCTCGTGTCAAATTCCGGAGACTACGAAATCCTCTACTGGGTTCC CTCTGCCTGTAAGCAAGTCGTAAGTGTGGAAACTACAAGAGACATTGAATGGGCTACCTATACCTGCACTTTGGGATTCCATGTTTTTGG AGTGTGGCCAGAAGGCTCGGACGGAACCGACATCAATGCCGTCTGTCGGGCCCATGAGAAGAAACTCCTGTCAACAGGCGACGACTTTGG CAAAGTGCACCTCTTCTCATACCCCTGCTCGCAGTTCAGGGCTCCAAGCCACATCTACGGCGGGCACAGCAGCCATGTCACCAATGTCGA TTTCCTCTGTGAAGACAGCCACCTCATCTCCACGGGCGGGAAAGACACAAGCATCATGCAGTGGCGCGTCATTTAGTACCCACCGAGAGC TGTGGGGAGCAGCATGGGCAAGGAAGACACAGACTCGCATTACCCTTGGTCACTGTGATTTCTGTTTTGTTTAAAAAATTCTTACAAACC TCAGGAAAACTGTGCCCTCCGCCGGCTACCTTAGCTTAGCGTGTCAGCGGGCGCCACAGCGGATCAGCGGTTCCGTGTTCACTTTTGTTG TACAATATATGACACAGTGCACATTGAATACCAACAAGGTTGCAACGTTTACATTATAGCCACATCAACAGAAGTAACTGGTATATTCTT AGTAACTTTTCTATGAACTCTTCAAAAATGGTCACAGAATGCCTTTTAAAACATTGTATATAATCTTCACTGTTTCACCATCTAGCTTGC TAAGTCAAATATTTATGATGATAATGAGGTACTGAACCACGATGGCTGTTGAGGAATTGGTCCTAAAAGGACAGATCACTTCAGAAGAGT GAATAACTGATTTGCACAGCTGAATCAGGAGACACAAAGATGAGACTGTGTTTGGTTACATTTTCCAAAGTTTCATTGCATTCTCCCTTG GGGAGGCTGTGAGAGAGGGCTTGTATCCCTCTTGTGCTAAGCAGACTCTACTCCTAACTGACTTCAATATTTCAGCAGGGTACACAGGCG TTTCCAAGTTTCAGTGACACCGTCCTGCCTAACCAGATGCGGTCAGCCTCTTCACACCCACCTGGCTTGCATCCCCCATCCCTTGTTCAC ACGCCCTGATTCACGGTGAGACATTTTGCCACCTTCTTGTGTATATTACTTGGCATGAGATGATATTGTACTTGTATAGGATTCTAGCAA TTCATAATAAATATGTAAGACTAGGCTTTACTGTCTTATGCTTATGGACATTGTATATTTGTATTTTATGACCAAGTAGACCAAGTCAGA AAGATCTCTCTCGAGCGTACCATAAACCTGCAGAGAGAAGTCTCGAAAGGCTCCACCAGGTACCAAGGGCAGCTGCTTTTCCTGTCTTTT GTGCATGGGCGACCCATTACAGTATGAGATAAGATTGAGTTCTGATGCGTTAAACGGAGGTGGCAGAAATTTGTCAAGAAGGCCTTATCC ATTTCGATTGTGTGACAGATTGAAATTTATTGTTTACATTGGGGAATGTATCTCAAATTTTTAAATAGAAGAGTAATAAACAGACTTTAA AGCAAATATTAAGATTTTTACTCATTCAAGGCAAGTAAATGAATGGAATTATCTGAGCTCTATGGCACTGGTTGTTTAGAGTGACTGATG AAGTGCAACTTTCAAAAACATTTTTGATGACATCACCAGCCTACTGCAGAAGTGCAGGGCACCAGTAAACACCATGTATTATTGAAGATG >27802_27802_6_EVL-EML1_EVL_chr14_100551192_ENST00000402714_EML1_chr14_100380903_ENST00000334192_length(amino acids)=333AA_BP=58 MSEQSICQARASVMVYDDTSKKWVPIKPGQQGFSRINIYHNTASNTFRVVGVKLQDQQGHTDELWGLAIHASKSQFLTCGHDKHATLWDA VGHRPVWDKIIEDPAQSSGFHPSGSVVAVGTLTGRWFVFDTETKDLVTVHTDGNEQLSVMRYSPDGNFLAIGSHDNCIYIYGVSDNGRKY TRVGKCSGHSSFITHLDWSVNSQFLVSNSGDYEILYWVPSACKQVVSVETTRDIEWATYTCTLGFHVFGVWPEGSDGTDINAVCRAHEKK -------------------------------------------------------------- >27802_27802_7_EVL-EML1_EVL_chr14_100551192_ENST00000544450_EML1_chr14_100380903_ENST00000262233_length(transcript)=3251nt_BP=475nt AGTTCCCAGCCTGGAGCCTAAGCATACACTGCATTAATCAGACTTGCTGGAATGATCACATGGTTTTCTAGCTGCCTCCCCTGTTAGCTG CCGCTTGTCAGCAGCTGGCGGAGCTGGAGGCTGCAGGCCCCTGTCTGCAGAAAGCCTTTTACTTCAGAATATTGGGGGGTGTGGAAACCC CGCCTCATTTTTCTGCACTTTGCACCATCTGACTAACTAAATGGTTGTCCTCCATACCAAAAAGACTACCACACAATCTAAGGAAATTGT CTCTGACTAGGTCATGTTTGCATTTGAAGAATTCAGTGAACAGAGTATCTGCCAAGCCCGGGCTTCCGTGATGGTCTACGATGACACCAG TAAGAAATGGGTACCAATCAAACCTGGCCAGCAGGGATTCAGCCGGATCAACATCTACCACAACACTGCCAGCAACACCTTCAGAGTCGT TGGAGTCAAGTTGCAGGATCAGCAGGGTCACACTGATGAGCTCTGGGGACTGGCCATCCATGCCTCAAAATCTCAGTTCTTGACCTGTGG GCATGACAAGCATGCCACTCTCTGGGACGCTGTGGGTCACCGTCCCGTCTGGGACAAAATAATAGAGGATCCAGCTCAGTCTTCTGGTTT TCATCCTTCAGGGTCTGTGGTTGCAGTCGGAACACTCACTGGGAGGTGGTTTGTGTTTGACACAGAAACAAAAGACTTGGTCACCGTTCA CACAGATGGAAACGAACAGCTCTCTGTAATGCGATACTCACCAGATGGGAATTTCTTAGCCATAGGCTCACATGACAACTGCATCTATAT ATATGGCGTTAGTGACAACGGGAGGAAGTACACGCGAGTGGGCAAGTGCTCGGGTCATTCCAGCTTCATTACTCACCTGGACTGGTCTGT AAACTCACAGTTCCTCGTGTCAAATTCCGGAGACTACGAAATCCTCTACTGGGTTCCCTCTGCCTGTAAGCAAGTCGTAAGTGTGGAAAC TACAAGAGACATTGAATGGGCTACCTATACCTGCACTTTGGGATTCCATGTTTTTGGAGTGTGGCCAGAAGGCTCGGACGGAACCGACAT CAATGCCGTCTGTCGGGCCCATGAGAAGAAACTCCTGTCAACAGGCGACGACTTTGGCAAAGTGCACCTCTTCTCATACCCCTGCTCGCA GTTCAGGGCTCCAAGCCACATCTACGGCGGGCACAGCAGCCATGTCACCAATGTCGATTTCCTCTGTGAAGACAGCCACCTCATCTCCAC GGGCGGGAAAGACACAAGCATCATGCAGTGGCGCGTCATTTAGTACCCACCGAGAGCTGTGGGGAGCAGCATGGGCAAGGAAGACACAGA CTCGCATTACCCTTGGTCACTGTGATTTCTGTTTTGTTTAAAAAATTCTTACAAACCTCAGGAAAACTGTGCCCTCCGCCGGCTACCTTA GCTTAGCGTGTCAGCGGGCGCCACAGCGGATCAGCGGTTCCGTGTTCACTTTTGTTGTACAATATATGACACAGTGCACATTGAATACCA ACAAGGTTGCAACGTTTACATTATAGCCACATCAACAGAAGTAACTGGTATATTCTTAGTAACTTTTCTATGAACTCTTCAAAAATGGTC ACAGAATGCCTTTTAAAACATTGTATATAATCTTCACTGTTTCACCATCTAGCTTGCTAAGTCAAATATTTATGATGATAATGAGGTACT GAACCACGATGGCTGTTGAGGAATTGGTCCTAAAAGGACAGATCACTTCAGAAGAGTGAATAACTGATTTGCACAGCTGAATCAGGAGAC ACAAAGATGAGACTGTGTTTGGTTACATTTTCCAAAGTTTCATTGCATTCTCCCTTGGGGAGGCTGTGAGAGAGGGCTTGTATCCCTCTT GTGCTAAGCAGACTCTACTCCTAACTGACTTCAATATTTCAGCAGGGTACACAGGCGTTTCCAAGTTTCAGTGACACCGTCCTGCCTAAC CAGATGCGGTCAGCCTCTTCACACCCACCTGGCTTGCATCCCCCATCCCTTGTTCACACGCCCTGATTCACGGTGAGACATTTTGCCACC TTCTTGTGTATATTACTTGGCATGAGATGATATTGTACTTGTATAGGATTCTAGCAATTCATAATAAATATGTAAGACTAGGCTTTACTG TCTTATGCTTATGGACATTGTATATTTGTATTTTATGACCAAGTAGACCAAGTCAGAAAGATCTCTCTCGAGCGTACCATAAACCTGCAG AGAGAAGTCTCGAAAGGCTCCACCAGGTACCAAGGGCAGCTGCTTTTCCTGTCTTTTGTGCATGGGCGACCCATTACAGTATGAGATAAG ATTGAGTTCTGATGCGTTAAACGGAGGTGGCAGAAATTTGTCAAGAAGGCCTTATCCATTTCGATTGTGTGACAGATTGAAATTTATTGT TTACATTGGGGAATGTATCTCAAATTTTTAAATAGAAGAGTAATAAACAGACTTTAAAGCAAATATTAAGATTTTTACTCATTCAAGGCA AGTAAATGAATGGAATTATCTGAGCTCTATGGCACTGGTTGTTTAGAGTGACTGATGAAGTGCAACTTTCAAAAACATTTTTGATGACAT CACCAGCCTACTGCAGAAGTGCAGGGCACCAGTAAACACCATGTATTATTGAAGATGAATCTGTTTGTATGTATCCTTGTCAAATATATT CTATAATGAAATAAAATCTGAAAAGTGGATTTCTTATTGACCTATATTCATGAAAGCATATAAATTAAAATATTTAAAATTAGATATGAT TCACACTATATTCTGTTTCATATGCAGATTTTATTCTCACCTGGGCCATTTGCAGATGAGACTGTAGTTTGCAGATGAGACTGTAGTTTG CAGATGGCGTGGAAGCATTCATCAGGGGAGATAACCATAAAGGATTTGGCCTAATTACCATACTCAATTGTCAGTTTACGTGGTTTTGTG AATACTGGCAAAAGCAATTGTTTTTAAATTAACAATGGAGAGAATGATAAGATGAGGGAAGGAAAAGGCATTCATTATTGACTTACATGT CAGTAAGGTCTGCTTTTATTTCTATGTACTCCTGTTTGCCAAGCTCAATAATGGACAAAGGATACAAACACACACACATCTACTATTTTA GATAAATGTACTGTTATATATATATGTAAACTACTATTGCTCTCTTTATAATGATTAATCACTTTATTATGAATGAATGAATGAATTTGA >27802_27802_7_EVL-EML1_EVL_chr14_100551192_ENST00000544450_EML1_chr14_100380903_ENST00000262233_length(amino acids)=339AA_BP=64 MFAFEEFSEQSICQARASVMVYDDTSKKWVPIKPGQQGFSRINIYHNTASNTFRVVGVKLQDQQGHTDELWGLAIHASKSQFLTCGHDKH ATLWDAVGHRPVWDKIIEDPAQSSGFHPSGSVVAVGTLTGRWFVFDTETKDLVTVHTDGNEQLSVMRYSPDGNFLAIGSHDNCIYIYGVS DNGRKYTRVGKCSGHSSFITHLDWSVNSQFLVSNSGDYEILYWVPSACKQVVSVETTRDIEWATYTCTLGFHVFGVWPEGSDGTDINAVC -------------------------------------------------------------- >27802_27802_8_EVL-EML1_EVL_chr14_100551192_ENST00000544450_EML1_chr14_100380903_ENST00000327921_length(transcript)=1515nt_BP=475nt AGTTCCCAGCCTGGAGCCTAAGCATACACTGCATTAATCAGACTTGCTGGAATGATCACATGGTTTTCTAGCTGCCTCCCCTGTTAGCTG CCGCTTGTCAGCAGCTGGCGGAGCTGGAGGCTGCAGGCCCCTGTCTGCAGAAAGCCTTTTACTTCAGAATATTGGGGGGTGTGGAAACCC CGCCTCATTTTTCTGCACTTTGCACCATCTGACTAACTAAATGGTTGTCCTCCATACCAAAAAGACTACCACACAATCTAAGGAAATTGT CTCTGACTAGGTCATGTTTGCATTTGAAGAATTCAGTGAACAGAGTATCTGCCAAGCCCGGGCTTCCGTGATGGTCTACGATGACACCAG TAAGAAATGGGTACCAATCAAACCTGGCCAGCAGGGATTCAGCCGGATCAACATCTACCACAACACTGCCAGCAACACCTTCAGAGTCGT TGGAGTCAAGTTGCAGGATCAGCAGGGTCACACTGATGAGCTCTGGGGACTGGCCATCCATGCCTCAAAATCTCAGTTCTTGACCTGTGG GCATGACAAGCATGCCACTCTCTGGGACGCTGTGGGTCACCGTCCCGTCTGGGACAAAATAATAGAGGATCCAGCTCAGTCTTCTGGTTT TCATCCTTCAGGGTCTGTGGTTGCAGTCGGAACACTCACTGGGAGGTGGTTTGTGTTTGACACAGAAACAAAAGACTTGGTCACCGTTCA CACAGATGGAAACGAACAGCTCTCTGTAATGCGATACTCACCAGATGGGAATTTCTTAGCCATAGGCTCACATGACAACTGCATCTATAT ATATGGCGTTAGTGACAACGGGAGGAAGTACACGCGAGTGGGCAAGTGCTCGGGTCATTCCAGCTTCATTACTCACCTGGACTGGTCTGT AAACTCACAGTTCCTCGTGTCAAATTCCGGAGACTACGAAATCCTCTACTGGGTTCCCTCTGCCTGTAAGCAAGTCGTAAGTGTGGAAAC TACAAGAGACATTGAATGGGCTACCTATACCTGCACTTTGGGATTCCATGTTTTTGGAGTGTGGCCAGAAGGCTCGGACGGAACCGACAT CAATGCCGTCTGTCGGGCCCATGAGAAGAAACTCCTGTCAACAGGCGACGACTTTGGCAAAGTGCACCTCTTCTCATACCCCTGCTCGCA GTTCAGGGCTCCAAGCCACATCTACGGCGGGCACAGCAGCCATGTCACCAATGTCGATTTCCTCTGTGAAGACAGCCACCTCATCTCCAC GGGCGGGAAAGACACAAGCATCATGCAGTGGCGCGTCATTTAGTACCCACCGAGAGCTGTGGGGAGCAGCATGGGCAAGGAAGACACAGA CTCGCATTACCCTTGGTCACTGTGATTTCTGTTTTGTTTAAAAAATTCTTACAAACCTCAGGAAAACTGTGCCCTCCGCCGGCTACCTTA >27802_27802_8_EVL-EML1_EVL_chr14_100551192_ENST00000544450_EML1_chr14_100380903_ENST00000327921_length(amino acids)=339AA_BP=64 MFAFEEFSEQSICQARASVMVYDDTSKKWVPIKPGQQGFSRINIYHNTASNTFRVVGVKLQDQQGHTDELWGLAIHASKSQFLTCGHDKH ATLWDAVGHRPVWDKIIEDPAQSSGFHPSGSVVAVGTLTGRWFVFDTETKDLVTVHTDGNEQLSVMRYSPDGNFLAIGSHDNCIYIYGVS DNGRKYTRVGKCSGHSSFITHLDWSVNSQFLVSNSGDYEILYWVPSACKQVVSVETTRDIEWATYTCTLGFHVFGVWPEGSDGTDINAVC -------------------------------------------------------------- >27802_27802_9_EVL-EML1_EVL_chr14_100551192_ENST00000544450_EML1_chr14_100380903_ENST00000334192_length(transcript)=2728nt_BP=475nt AGTTCCCAGCCTGGAGCCTAAGCATACACTGCATTAATCAGACTTGCTGGAATGATCACATGGTTTTCTAGCTGCCTCCCCTGTTAGCTG CCGCTTGTCAGCAGCTGGCGGAGCTGGAGGCTGCAGGCCCCTGTCTGCAGAAAGCCTTTTACTTCAGAATATTGGGGGGTGTGGAAACCC CGCCTCATTTTTCTGCACTTTGCACCATCTGACTAACTAAATGGTTGTCCTCCATACCAAAAAGACTACCACACAATCTAAGGAAATTGT CTCTGACTAGGTCATGTTTGCATTTGAAGAATTCAGTGAACAGAGTATCTGCCAAGCCCGGGCTTCCGTGATGGTCTACGATGACACCAG TAAGAAATGGGTACCAATCAAACCTGGCCAGCAGGGATTCAGCCGGATCAACATCTACCACAACACTGCCAGCAACACCTTCAGAGTCGT TGGAGTCAAGTTGCAGGATCAGCAGGGTCACACTGATGAGCTCTGGGGACTGGCCATCCATGCCTCAAAATCTCAGTTCTTGACCTGTGG GCATGACAAGCATGCCACTCTCTGGGACGCTGTGGGTCACCGTCCCGTCTGGGACAAAATAATAGAGGATCCAGCTCAGTCTTCTGGTTT TCATCCTTCAGGGTCTGTGGTTGCAGTCGGAACACTCACTGGGAGGTGGTTTGTGTTTGACACAGAAACAAAAGACTTGGTCACCGTTCA CACAGATGGAAACGAACAGCTCTCTGTAATGCGATACTCACCAGATGGGAATTTCTTAGCCATAGGCTCACATGACAACTGCATCTATAT ATATGGCGTTAGTGACAACGGGAGGAAGTACACGCGAGTGGGCAAGTGCTCGGGTCATTCCAGCTTCATTACTCACCTGGACTGGTCTGT AAACTCACAGTTCCTCGTGTCAAATTCCGGAGACTACGAAATCCTCTACTGGGTTCCCTCTGCCTGTAAGCAAGTCGTAAGTGTGGAAAC TACAAGAGACATTGAATGGGCTACCTATACCTGCACTTTGGGATTCCATGTTTTTGGAGTGTGGCCAGAAGGCTCGGACGGAACCGACAT CAATGCCGTCTGTCGGGCCCATGAGAAGAAACTCCTGTCAACAGGCGACGACTTTGGCAAAGTGCACCTCTTCTCATACCCCTGCTCGCA GTTCAGGGCTCCAAGCCACATCTACGGCGGGCACAGCAGCCATGTCACCAATGTCGATTTCCTCTGTGAAGACAGCCACCTCATCTCCAC GGGCGGGAAAGACACAAGCATCATGCAGTGGCGCGTCATTTAGTACCCACCGAGAGCTGTGGGGAGCAGCATGGGCAAGGAAGACACAGA CTCGCATTACCCTTGGTCACTGTGATTTCTGTTTTGTTTAAAAAATTCTTACAAACCTCAGGAAAACTGTGCCCTCCGCCGGCTACCTTA GCTTAGCGTGTCAGCGGGCGCCACAGCGGATCAGCGGTTCCGTGTTCACTTTTGTTGTACAATATATGACACAGTGCACATTGAATACCA ACAAGGTTGCAACGTTTACATTATAGCCACATCAACAGAAGTAACTGGTATATTCTTAGTAACTTTTCTATGAACTCTTCAAAAATGGTC ACAGAATGCCTTTTAAAACATTGTATATAATCTTCACTGTTTCACCATCTAGCTTGCTAAGTCAAATATTTATGATGATAATGAGGTACT GAACCACGATGGCTGTTGAGGAATTGGTCCTAAAAGGACAGATCACTTCAGAAGAGTGAATAACTGATTTGCACAGCTGAATCAGGAGAC ACAAAGATGAGACTGTGTTTGGTTACATTTTCCAAAGTTTCATTGCATTCTCCCTTGGGGAGGCTGTGAGAGAGGGCTTGTATCCCTCTT GTGCTAAGCAGACTCTACTCCTAACTGACTTCAATATTTCAGCAGGGTACACAGGCGTTTCCAAGTTTCAGTGACACCGTCCTGCCTAAC CAGATGCGGTCAGCCTCTTCACACCCACCTGGCTTGCATCCCCCATCCCTTGTTCACACGCCCTGATTCACGGTGAGACATTTTGCCACC TTCTTGTGTATATTACTTGGCATGAGATGATATTGTACTTGTATAGGATTCTAGCAATTCATAATAAATATGTAAGACTAGGCTTTACTG TCTTATGCTTATGGACATTGTATATTTGTATTTTATGACCAAGTAGACCAAGTCAGAAAGATCTCTCTCGAGCGTACCATAAACCTGCAG AGAGAAGTCTCGAAAGGCTCCACCAGGTACCAAGGGCAGCTGCTTTTCCTGTCTTTTGTGCATGGGCGACCCATTACAGTATGAGATAAG ATTGAGTTCTGATGCGTTAAACGGAGGTGGCAGAAATTTGTCAAGAAGGCCTTATCCATTTCGATTGTGTGACAGATTGAAATTTATTGT TTACATTGGGGAATGTATCTCAAATTTTTAAATAGAAGAGTAATAAACAGACTTTAAAGCAAATATTAAGATTTTTACTCATTCAAGGCA AGTAAATGAATGGAATTATCTGAGCTCTATGGCACTGGTTGTTTAGAGTGACTGATGAAGTGCAACTTTCAAAAACATTTTTGATGACAT CACCAGCCTACTGCAGAAGTGCAGGGCACCAGTAAACACCATGTATTATTGAAGATGAATCTGTTTGTATGTATCCTTGTCAAATATATT >27802_27802_9_EVL-EML1_EVL_chr14_100551192_ENST00000544450_EML1_chr14_100380903_ENST00000334192_length(amino acids)=339AA_BP=64 MFAFEEFSEQSICQARASVMVYDDTSKKWVPIKPGQQGFSRINIYHNTASNTFRVVGVKLQDQQGHTDELWGLAIHASKSQFLTCGHDKH ATLWDAVGHRPVWDKIIEDPAQSSGFHPSGSVVAVGTLTGRWFVFDTETKDLVTVHTDGNEQLSVMRYSPDGNFLAIGSHDNCIYIYGVS DNGRKYTRVGKCSGHSSFITHLDWSVNSQFLVSNSGDYEILYWVPSACKQVVSVETTRDIEWATYTCTLGFHVFGVWPEGSDGTDINAVC -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for EVL-EML1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for EVL-EML1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for EVL-EML1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
