
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:EXD3-CPSF7 (FusionGDB2 ID:27868) |
Fusion Gene Summary for EXD3-CPSF7 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: EXD3-CPSF7 | Fusion gene ID: 27868 | Hgene | Tgene | Gene symbol | EXD3 | CPSF7 | Gene ID | 54932 | 79869 |
| Gene name | exonuclease 3'-5' domain containing 3 | cleavage and polyadenylation specific factor 7 | |
| Synonyms | Nbr|mut-7 | CFIm59 | |
| Cytomap | 9q34.3 | 11q12.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | exonuclease mut-7 homologexonuclease 3'-5' domain-containing protein 3nibbler homologprobable exonuclease mut-7 homologtesticular tissue protein Li 125 | cleavage and polyadenylation specificity factor subunit 7CPSF 59 kDa subunitcleavage and polyadenylation specificity factor 59 kDa subunitcleavage factor Im complex 59 kDa subunitpre-mRNA cleavage factor I, 59 kDa subunitpre-mRNA cleavage factor Im 5 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8N9H8 | Q8N684 | |
| Ensembl transtripts involved in fusion gene | ENST00000340951, ENST00000465160, ENST00000479452, ENST00000342129, ENST00000475006, | ENST00000394888, ENST00000439958, ENST00000448745, ENST00000541963, ENST00000494016, ENST00000340437, | |
| Fusion gene scores | * DoF score | 9 X 5 X 7=315 | 9 X 8 X 5=360 |
| # samples | 10 | 10 | |
| ** MAII score | log2(10/315*10)=-1.65535182861255 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(10/360*10)=-1.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: EXD3 [Title/Abstract] AND CPSF7 [Title/Abstract] AND fusion [Title/Abstract] | ||
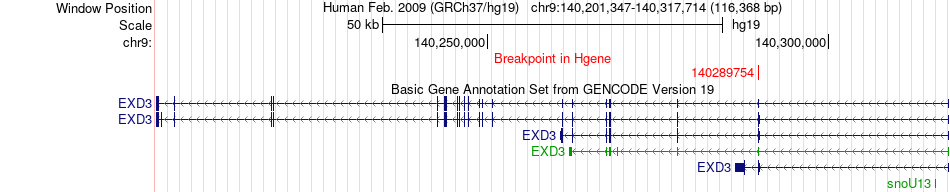
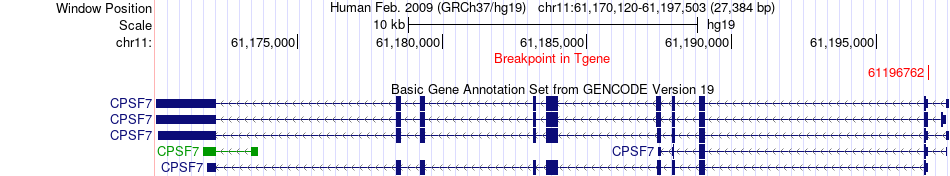
| Most frequent breakpoint | EXD3(140289754)-CPSF7(61196762), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | CPSF7 | GO:0051262 | protein tetramerization | 20695905 |
| Tgene | CPSF7 | GO:0051290 | protein heterotetramerization | 23187700 |
| Tgene | CPSF7 | GO:1990120 | messenger ribonucleoprotein complex assembly | 29276085 |
 Fusion gene breakpoints across EXD3 (5'-gene) Fusion gene breakpoints across EXD3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across CPSF7 (3'-gene) Fusion gene breakpoints across CPSF7 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-L5-A4OH | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
Top |
Fusion Gene ORF analysis for EXD3-CPSF7 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000340951 | ENST00000394888 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5CDS-5UTR | ENST00000340951 | ENST00000439958 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5CDS-5UTR | ENST00000340951 | ENST00000448745 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5CDS-5UTR | ENST00000340951 | ENST00000541963 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5CDS-5UTR | ENST00000465160 | ENST00000394888 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5CDS-5UTR | ENST00000465160 | ENST00000439958 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5CDS-5UTR | ENST00000465160 | ENST00000448745 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5CDS-5UTR | ENST00000465160 | ENST00000541963 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5CDS-5UTR | ENST00000479452 | ENST00000394888 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5CDS-5UTR | ENST00000479452 | ENST00000439958 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5CDS-5UTR | ENST00000479452 | ENST00000448745 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5CDS-5UTR | ENST00000479452 | ENST00000541963 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5CDS-intron | ENST00000340951 | ENST00000494016 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5CDS-intron | ENST00000465160 | ENST00000494016 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5CDS-intron | ENST00000479452 | ENST00000494016 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5UTR-3CDS | ENST00000342129 | ENST00000340437 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5UTR-3CDS | ENST00000475006 | ENST00000340437 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5UTR-5UTR | ENST00000342129 | ENST00000394888 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5UTR-5UTR | ENST00000342129 | ENST00000439958 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5UTR-5UTR | ENST00000342129 | ENST00000448745 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5UTR-5UTR | ENST00000342129 | ENST00000541963 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5UTR-5UTR | ENST00000475006 | ENST00000394888 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5UTR-5UTR | ENST00000475006 | ENST00000439958 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5UTR-5UTR | ENST00000475006 | ENST00000448745 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5UTR-5UTR | ENST00000475006 | ENST00000541963 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5UTR-intron | ENST00000342129 | ENST00000494016 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| 5UTR-intron | ENST00000475006 | ENST00000494016 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| In-frame | ENST00000340951 | ENST00000340437 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| In-frame | ENST00000465160 | ENST00000340437 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
| In-frame | ENST00000479452 | ENST00000340437 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000340951 | EXD3 | chr9 | 140289754 | - | ENST00000340437 | CPSF7 | chr11 | 61196762 | - | 3783 | 251 | 267 | 1721 | 484 |
| ENST00000479452 | EXD3 | chr9 | 140289754 | - | ENST00000340437 | CPSF7 | chr11 | 61196762 | - | 3676 | 144 | 160 | 1614 | 484 |
| ENST00000465160 | EXD3 | chr9 | 140289754 | - | ENST00000340437 | CPSF7 | chr11 | 61196762 | - | 3683 | 151 | 167 | 1621 | 484 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000340951 | ENST00000340437 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - | 0.002407378 | 0.9975926 |
| ENST00000479452 | ENST00000340437 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - | 0.002346201 | 0.9976538 |
| ENST00000465160 | ENST00000340437 | EXD3 | chr9 | 140289754 | - | CPSF7 | chr11 | 61196762 | - | 0.002321591 | 0.99767846 |
Top |
Fusion Genomic Features for EXD3-CPSF7 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
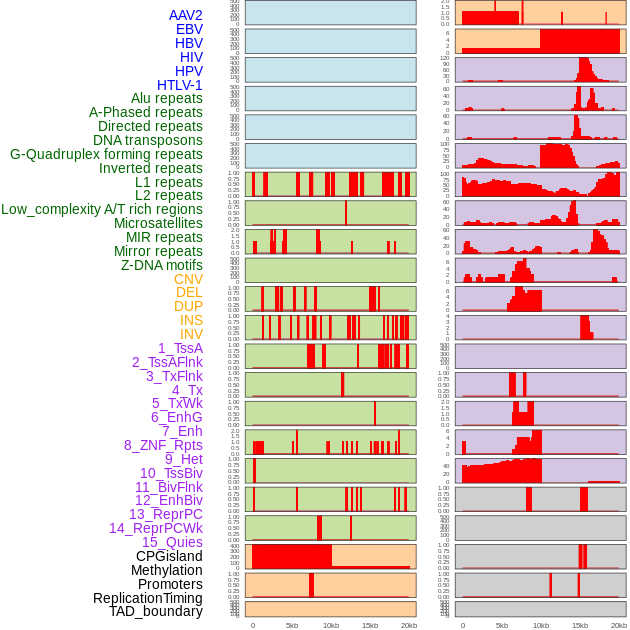
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for EXD3-CPSF7 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:140289754/chr11:61196762) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| EXD3 | CPSF7 |
| FUNCTION: Possesses 3'-5' exoribonuclease activity. Required for 3'-end trimming of AGO1-bound miRNAs (By similarity). {ECO:0000250}. | FUNCTION: Component of the cleavage factor Im (CFIm) complex that functions as an activator of the pre-mRNA 3'-end cleavage and polyadenylation processing required for the maturation of pre-mRNA into functional mRNAs (PubMed:8626397, PubMed:17024186, PubMed:29276085). CFIm contributes to the recruitment of multiprotein complexes on specific sequences on the pre-mRNA 3'-end, so called cleavage and polyadenylation signals (pA signals) (PubMed:8626397, PubMed:17024186). Most pre-mRNAs contain multiple pA signals, resulting in alternative cleavage and polyadenylation (APA) producing mRNAs with variable 3'-end formation (PubMed:23187700, PubMed:29276085). The CFIm complex acts as a key regulator of cleavage and polyadenylation site choice during APA through its binding to 5'-UGUA-3' elements localized in the 3'-untranslated region (UTR) for a huge number of pre-mRNAs (PubMed:20695905, PubMed:29276085). CPSF7 activates directly the mRNA 3'-processing machinery (PubMed:29276085). Binds to pA signals in RNA substrates (PubMed:8626397, PubMed:17024186). {ECO:0000269|PubMed:17024186, ECO:0000269|PubMed:20695905, ECO:0000269|PubMed:23187700, ECO:0000269|PubMed:29276085, ECO:0000269|PubMed:8626397}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | CPSF7 | chr9:140289754 | chr11:61196762 | ENST00000340437 | 0 | 10 | 218_329 | 24 | 1168.3333333333333 | Compositional bias | Note=Pro-rich | |
| Tgene | CPSF7 | chr9:140289754 | chr11:61196762 | ENST00000340437 | 0 | 10 | 418_469 | 24 | 1168.3333333333333 | Compositional bias | Note=Arg-rich | |
| Tgene | CPSF7 | chr9:140289754 | chr11:61196762 | ENST00000340437 | 0 | 10 | 51_54 | 24 | 1168.3333333333333 | Compositional bias | Note=Poly-Pro | |
| Tgene | CPSF7 | chr9:140289754 | chr11:61196762 | ENST00000394888 | 0 | 10 | 218_329 | 0 | 1137.6666666666667 | Compositional bias | Note=Pro-rich | |
| Tgene | CPSF7 | chr9:140289754 | chr11:61196762 | ENST00000394888 | 0 | 10 | 418_469 | 0 | 1137.6666666666667 | Compositional bias | Note=Arg-rich | |
| Tgene | CPSF7 | chr9:140289754 | chr11:61196762 | ENST00000394888 | 0 | 10 | 51_54 | 0 | 1137.6666666666667 | Compositional bias | Note=Poly-Pro | |
| Tgene | CPSF7 | chr9:140289754 | chr11:61196762 | ENST00000439958 | 0 | 10 | 218_329 | 0 | 1107.0 | Compositional bias | Note=Pro-rich | |
| Tgene | CPSF7 | chr9:140289754 | chr11:61196762 | ENST00000439958 | 0 | 10 | 418_469 | 0 | 1107.0 | Compositional bias | Note=Arg-rich | |
| Tgene | CPSF7 | chr9:140289754 | chr11:61196762 | ENST00000439958 | 0 | 10 | 51_54 | 0 | 1107.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | CPSF7 | chr9:140289754 | chr11:61196762 | ENST00000448745 | 0 | 9 | 218_329 | 0 | 500.3333333333333 | Compositional bias | Note=Pro-rich | |
| Tgene | CPSF7 | chr9:140289754 | chr11:61196762 | ENST00000448745 | 0 | 9 | 418_469 | 0 | 500.3333333333333 | Compositional bias | Note=Arg-rich | |
| Tgene | CPSF7 | chr9:140289754 | chr11:61196762 | ENST00000448745 | 0 | 9 | 51_54 | 0 | 500.3333333333333 | Compositional bias | Note=Poly-Pro | |
| Tgene | CPSF7 | chr9:140289754 | chr11:61196762 | ENST00000340437 | 0 | 10 | 82_162 | 24 | 1168.3333333333333 | Domain | RRM | |
| Tgene | CPSF7 | chr9:140289754 | chr11:61196762 | ENST00000394888 | 0 | 10 | 82_162 | 0 | 1137.6666666666667 | Domain | RRM | |
| Tgene | CPSF7 | chr9:140289754 | chr11:61196762 | ENST00000439958 | 0 | 10 | 82_162 | 0 | 1107.0 | Domain | RRM | |
| Tgene | CPSF7 | chr9:140289754 | chr11:61196762 | ENST00000448745 | 0 | 9 | 82_162 | 0 | 500.3333333333333 | Domain | RRM |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EXD3 | chr9:140289754 | chr11:61196762 | ENST00000340951 | - | 2 | 22 | 517_571 | 18 | 877.0 | Domain | Note=3'-5' exonuclease |
| Hgene | EXD3 | chr9:140289754 | chr11:61196762 | ENST00000342129 | - | 2 | 21 | 517_571 | 0 | 515.0 | Domain | Note=3'-5' exonuclease |
| Hgene | EXD3 | chr9:140289754 | chr11:61196762 | ENST00000479452 | - | 2 | 8 | 517_571 | 18 | 247.0 | Domain | Note=3'-5' exonuclease |
Top |
Fusion Gene Sequence for EXD3-CPSF7 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >27868_27868_1_EXD3-CPSF7_EXD3_chr9_140289754_ENST00000340951_CPSF7_chr11_61196762_ENST00000340437_length(transcript)=3783nt_BP=251nt ACTTCCGCCGGGAGCTTTGTGCGCCCGGCTTCCTCGCCCCGCCCGCGCTGGCCTTGGTGCGCCGGATAAAGCCCTCCGGGCCAGCGGCTG CGGTCGGGCCAAGGTTCCGCGCCGGGCGTGGACGGTGTGGGGCGGTCCCGGCCCCGCAGATCCTCGTTCCTAGCTGCCTGCCAGCGTCCT CGGCCCGGCCCTGAGGATGGACCCAGGAGATCCCGCTGGTGACCCTGCCGCTGGCGAGCGCCACCGCATGGTGGCATCTTCCTTACTTTG TCCATCCTCCGGACTCGCGATCTTCCTTCCGGAGCCATGTCAGAAGGAGTGGACTTGATTGATATATATGCTGACGAGGAGTTCAACCAG GACCCAGAGTTCAACAATACAGATCAGATTGACCTGTATGATGATGTGCTGACAGCCACCTCACAGCCCTCAGATGACAGAAGCAGCAGC ACTGAACCACCTCCTCCTGTTCGCCAGGAGCCATCTCCCAAGCCCAACAACAAGACCCCTGCAATTCTGTATACCTACAGTGGCCTGCGT AATAGACGAGCTGCCGTTTATGTGGGCAGCTTCTCCTGGTGGACCACAGACCAGCAGCTGATCCAGGTTATTCGCTCTATAGGAGTCTAT GATGTGGTGGAGTTGAAATTTGCAGAGAATCGAGCAAATGGCCAGTCCAAAGGGTATGCTGAGGTGGTGGTAGCCTCTGAAAACTCTGTC CACAAATTGTTGGAACTCCTACCAGGGAAAGTTCTTAATGGAGAAAAAGTGGACGTGAGGCCGGCCACCCGGCAGAACCTGTCACAGTTT GAGGCACAGGCTCGGAAACGTGAGTGTGTCCGAGTCCCAAGAGGGGGAATACCTCCACGGGCCCATTCCCGAGATTCTAGTGATTCTGCT GATGGACGGGCCACACCCTCTGAGAACCTTGTACCCTCATCTGCTCGTGTGGATAAGCCCCCCAGTGTGCTGCCCTACTTCAATCGTCCT CCTTCGGCCCTTCCCCTGATGGGTCTGCCCCCACCACCAATTCCACCCCCACCACCTCTCTCCTCAAGCTTTGGGGTCCCTCCTCCTCCT CCTGGTATCCACTACCAGCATCTCATGCCCCCACCTCCTCGATTACCTCCTCATCTTGCTGTACCTCCCCCTGGGGCCATCCCACCTGCC CTTCACCTCAATCCAGCCTTCTTCCCCCCACCAAACGCTACAGTGGGGCCTCCACCAGATACTTACATGAAGGCCTCTGCCCCCTATAAC CACCATGGCAGCCGAGATTCGGGCCCTCCACCCTCTACAGTGAGTGAAGCCGAATTTGAAGATATCATGAAGCGAAACAGAGCAATTTCC AGCAGTGCCATTTCCAAAGCAGTATCTGGAGCCAGTGCAGGGGATTACAGTGACGCAATTGAGACGCTGCTCACAGCCATTGCGGTTATC AAACAGTCCCGGGTTGCCAATGATGAGCGTTGCCGTGTCCTCATCTCCTCTCTTAAGGACTGTCTTCATGGCATCGAAGCCAAGTCCTAC AGTGTGGGTGCCAGTGGGAGCTCTTCCAGGAAAAGACATCGCTCCCGGGAAAGGTCACCTAGCCGGTCCCGGGAGAGCAGCAGGAGGCAC CGGGATCTGCTTCATAATGAAGATCGGCATGATGATTATTTCCAAGAAAGGAACCGGGAGCATGAGAGACACCGGGATAGAGAACGGGAC CGGCACCACTGAGAAAGGAGTCTGGTTGGAAGCAAATGTTTTTTTAATGGACTTGCATCTCCTCACCTTGATCAGGACTAAAGGACGGAG GCCGCCCCACCCCCTTCCCTTTCCTCCAAACCCCTAACTCCCTCCAGACACCCAGGGAATACCCTCTGCCCCACAGGATTGAAGACTGCT TGGCAGTCCTCCCAATCCCACACCTCCTGTTTGCCAGGGGAAAGAACCTAAAGACTTCGTGTGATTGGGAGGGGTGGCAGACAGGAAGAA AACATGTCCAGGCCCCTGGTCTCCATAGAGAATGGTGCTTTGTCCAAGAAAACGTATGAGTTTCTGATTCTCCGGGAGCCGTTCAATGGT GAGGTTGATGGGAAGACTTCCTTCCCAAAGAAAATAGATCCTCCATGCAGGATCTAGGAGAGTGACTGGGTGTGCCAAAATATGCCCAGG GTCCTGCCCTCAGCACTAGATTTAATGGGGCCAAGAGGGTCCAAACCCCTTGCTAACATACCACTTCTTTGTTTAACTCCTTTACCTTTC CAGCCCTTTGAGGAGGGACCATGAGAACAGAAATTACCTTATGAAAAGCTACTTCTGTTCCTGCTTTCCCTCTCACGTATTGACGGTTTA TTTCTTTGACCTCCCAGAGGGCTGAACTCTTTCAACTCTGCGCTGCCCAGCCTTCTCAGTGGACTTGCCCCTCCTAAGCAGAGAAGGCCT ATGAGGTTGCTTGCTGCTGGGAAGCCTGGCAGAGCCAATTACCACCCTCTGCTGCTTAGTGCTTGGGTACCTCTTGCAATAACCAGCTCT TAGTTGTTCCCTTTCCCTGGGGCTTTTCCATTTAACACATGGAGCCCTTCCCCCAGAAGGCTACTTCCTTGTTTTAGAGGAAGGTACTGC CCATTGGGAGATGGGGACATTGGGACCTCAGCAATGAAGAACCCTTGTGAAGTAACCAGGAGGAATGGGGAAAGAAGCAAGTTGGGCAGG ATATGGCCTACTTCCATAGGCTTTTCTTTTTTCAGGTTTGATGTAAGCATGGGCTTACATCCCCCAGGTACATACTTTTACTTATTGTGG GATAACCTGGCACTAGTAGGCAGGTAAAGTCACAAATTTGGTGTCTTTTCACCTTTTGACTGTTGACTTAATAGCTCCTCTCACTCTGCC TGGAGATACTTCCTGCCTCAGATGAGGAGCCAGAAGAAACAGAGCCCGACTTGAATGAACTCAGCTCAGAGTTCTAAGGACCAGCATTCT GGGGGCCATTTTCTCTACAGGCAAATGGAATTGCTTTTCCATAACATCCAAATTGTAATGTGGTTGCTGCTGAAGGAGGAGGCAGCAGCG AGGTCCTGCGGTACCCATGGGGTGATGCTACTTCTGCATGCATCTACAGGGCATCTGACACCTAACATGAGACGTGGCATGTGAGATGAG ACTTGGCATGTGAGACATAGGGTCACTAGAGACCCTTCTGGGTCAGAGGAGAGAGACTGAATTGGACTAAACCCGTCCTCTGTTCCCAGC ACGTTTCTCATATAGCCCTCAGTCACTGAGGGAGTCCCCCGCAGGATTGGAGAGGCACATTCCCTTGGGACAGAGGCTACAGGTTGGAGC TTTTTTTCCCCTGTCCCCCAACCCCATCCCCACCTCCACTTCAGAACATGGCACCCCACCCAACTGGCCAAGTGTTAAGTGATGTGCTTA TTGAGAGCAACTCCGGGTGTCTTTTAAAATGTAGAGAAAAGGTGACAGTTTAAGGAAAAATATATATAGAATACCAGAAATGCCGTTTAC CCGGAGAATTTTTTTCTCCCCATTTGTTTTGTTTTTACTCAATGACACCATTTTTAGTTTTATTTCCTGATAGCAAAAGGAAAAAAAACA CCCATCCCTCAAAAAGGCCAAGGTCCCGTCCCCCTGTTGTCGGTGATTTGTTTGTCTTTCTGATAGGTTGAAAATTGTGTAATAAACTTG ATGACGCTGTCAATCTTTTATACTGCATTGTATTTTTTTCCTTTTGTAACAAAATATTTTTAAATAATAAATGGGGTGTGAGCTGTTTTA >27868_27868_1_EXD3-CPSF7_EXD3_chr9_140289754_ENST00000340951_CPSF7_chr11_61196762_ENST00000340437_length(amino acids)=484AA_BP= MSILRTRDLPSGAMSEGVDLIDIYADEEFNQDPEFNNTDQIDLYDDVLTATSQPSDDRSSSTEPPPPVRQEPSPKPNNKTPAILYTYSGL RNRRAAVYVGSFSWWTTDQQLIQVIRSIGVYDVVELKFAENRANGQSKGYAEVVVASENSVHKLLELLPGKVLNGEKVDVRPATRQNLSQ FEAQARKRECVRVPRGGIPPRAHSRDSSDSADGRATPSENLVPSSARVDKPPSVLPYFNRPPSALPLMGLPPPPIPPPPPLSSSFGVPPP PPGIHYQHLMPPPPRLPPHLAVPPPGAIPPALHLNPAFFPPPNATVGPPPDTYMKASAPYNHHGSRDSGPPPSTVSEAEFEDIMKRNRAI SSSAISKAVSGASAGDYSDAIETLLTAIAVIKQSRVANDERCRVLISSLKDCLHGIEAKSYSVGASGSSSRKRHRSRERSPSRSRESSRR -------------------------------------------------------------- >27868_27868_2_EXD3-CPSF7_EXD3_chr9_140289754_ENST00000465160_CPSF7_chr11_61196762_ENST00000340437_length(transcript)=3683nt_BP=151nt AAGGTTCCGCGCCGGGCGTGGACGGTGTGGGGCGGTCCCGGCCCCGCAGATCCTCGTTCCTAGCTGCCTGCCAGCGTCCTCGGCCCGGCC CTGAGGATGGACCCAGGAGATCCCGCTGGTGACCCTGCCGCTGGCGAGCGCCACCGCATGGTGGCATCTTCCTTACTTTGTCCATCCTCC GGACTCGCGATCTTCCTTCCGGAGCCATGTCAGAAGGAGTGGACTTGATTGATATATATGCTGACGAGGAGTTCAACCAGGACCCAGAGT TCAACAATACAGATCAGATTGACCTGTATGATGATGTGCTGACAGCCACCTCACAGCCCTCAGATGACAGAAGCAGCAGCACTGAACCAC CTCCTCCTGTTCGCCAGGAGCCATCTCCCAAGCCCAACAACAAGACCCCTGCAATTCTGTATACCTACAGTGGCCTGCGTAATAGACGAG CTGCCGTTTATGTGGGCAGCTTCTCCTGGTGGACCACAGACCAGCAGCTGATCCAGGTTATTCGCTCTATAGGAGTCTATGATGTGGTGG AGTTGAAATTTGCAGAGAATCGAGCAAATGGCCAGTCCAAAGGGTATGCTGAGGTGGTGGTAGCCTCTGAAAACTCTGTCCACAAATTGT TGGAACTCCTACCAGGGAAAGTTCTTAATGGAGAAAAAGTGGACGTGAGGCCGGCCACCCGGCAGAACCTGTCACAGTTTGAGGCACAGG CTCGGAAACGTGAGTGTGTCCGAGTCCCAAGAGGGGGAATACCTCCACGGGCCCATTCCCGAGATTCTAGTGATTCTGCTGATGGACGGG CCACACCCTCTGAGAACCTTGTACCCTCATCTGCTCGTGTGGATAAGCCCCCCAGTGTGCTGCCCTACTTCAATCGTCCTCCTTCGGCCC TTCCCCTGATGGGTCTGCCCCCACCACCAATTCCACCCCCACCACCTCTCTCCTCAAGCTTTGGGGTCCCTCCTCCTCCTCCTGGTATCC ACTACCAGCATCTCATGCCCCCACCTCCTCGATTACCTCCTCATCTTGCTGTACCTCCCCCTGGGGCCATCCCACCTGCCCTTCACCTCA ATCCAGCCTTCTTCCCCCCACCAAACGCTACAGTGGGGCCTCCACCAGATACTTACATGAAGGCCTCTGCCCCCTATAACCACCATGGCA GCCGAGATTCGGGCCCTCCACCCTCTACAGTGAGTGAAGCCGAATTTGAAGATATCATGAAGCGAAACAGAGCAATTTCCAGCAGTGCCA TTTCCAAAGCAGTATCTGGAGCCAGTGCAGGGGATTACAGTGACGCAATTGAGACGCTGCTCACAGCCATTGCGGTTATCAAACAGTCCC GGGTTGCCAATGATGAGCGTTGCCGTGTCCTCATCTCCTCTCTTAAGGACTGTCTTCATGGCATCGAAGCCAAGTCCTACAGTGTGGGTG CCAGTGGGAGCTCTTCCAGGAAAAGACATCGCTCCCGGGAAAGGTCACCTAGCCGGTCCCGGGAGAGCAGCAGGAGGCACCGGGATCTGC TTCATAATGAAGATCGGCATGATGATTATTTCCAAGAAAGGAACCGGGAGCATGAGAGACACCGGGATAGAGAACGGGACCGGCACCACT GAGAAAGGAGTCTGGTTGGAAGCAAATGTTTTTTTAATGGACTTGCATCTCCTCACCTTGATCAGGACTAAAGGACGGAGGCCGCCCCAC CCCCTTCCCTTTCCTCCAAACCCCTAACTCCCTCCAGACACCCAGGGAATACCCTCTGCCCCACAGGATTGAAGACTGCTTGGCAGTCCT CCCAATCCCACACCTCCTGTTTGCCAGGGGAAAGAACCTAAAGACTTCGTGTGATTGGGAGGGGTGGCAGACAGGAAGAAAACATGTCCA GGCCCCTGGTCTCCATAGAGAATGGTGCTTTGTCCAAGAAAACGTATGAGTTTCTGATTCTCCGGGAGCCGTTCAATGGTGAGGTTGATG GGAAGACTTCCTTCCCAAAGAAAATAGATCCTCCATGCAGGATCTAGGAGAGTGACTGGGTGTGCCAAAATATGCCCAGGGTCCTGCCCT CAGCACTAGATTTAATGGGGCCAAGAGGGTCCAAACCCCTTGCTAACATACCACTTCTTTGTTTAACTCCTTTACCTTTCCAGCCCTTTG AGGAGGGACCATGAGAACAGAAATTACCTTATGAAAAGCTACTTCTGTTCCTGCTTTCCCTCTCACGTATTGACGGTTTATTTCTTTGAC CTCCCAGAGGGCTGAACTCTTTCAACTCTGCGCTGCCCAGCCTTCTCAGTGGACTTGCCCCTCCTAAGCAGAGAAGGCCTATGAGGTTGC TTGCTGCTGGGAAGCCTGGCAGAGCCAATTACCACCCTCTGCTGCTTAGTGCTTGGGTACCTCTTGCAATAACCAGCTCTTAGTTGTTCC CTTTCCCTGGGGCTTTTCCATTTAACACATGGAGCCCTTCCCCCAGAAGGCTACTTCCTTGTTTTAGAGGAAGGTACTGCCCATTGGGAG ATGGGGACATTGGGACCTCAGCAATGAAGAACCCTTGTGAAGTAACCAGGAGGAATGGGGAAAGAAGCAAGTTGGGCAGGATATGGCCTA CTTCCATAGGCTTTTCTTTTTTCAGGTTTGATGTAAGCATGGGCTTACATCCCCCAGGTACATACTTTTACTTATTGTGGGATAACCTGG CACTAGTAGGCAGGTAAAGTCACAAATTTGGTGTCTTTTCACCTTTTGACTGTTGACTTAATAGCTCCTCTCACTCTGCCTGGAGATACT TCCTGCCTCAGATGAGGAGCCAGAAGAAACAGAGCCCGACTTGAATGAACTCAGCTCAGAGTTCTAAGGACCAGCATTCTGGGGGCCATT TTCTCTACAGGCAAATGGAATTGCTTTTCCATAACATCCAAATTGTAATGTGGTTGCTGCTGAAGGAGGAGGCAGCAGCGAGGTCCTGCG GTACCCATGGGGTGATGCTACTTCTGCATGCATCTACAGGGCATCTGACACCTAACATGAGACGTGGCATGTGAGATGAGACTTGGCATG TGAGACATAGGGTCACTAGAGACCCTTCTGGGTCAGAGGAGAGAGACTGAATTGGACTAAACCCGTCCTCTGTTCCCAGCACGTTTCTCA TATAGCCCTCAGTCACTGAGGGAGTCCCCCGCAGGATTGGAGAGGCACATTCCCTTGGGACAGAGGCTACAGGTTGGAGCTTTTTTTCCC CTGTCCCCCAACCCCATCCCCACCTCCACTTCAGAACATGGCACCCCACCCAACTGGCCAAGTGTTAAGTGATGTGCTTATTGAGAGCAA CTCCGGGTGTCTTTTAAAATGTAGAGAAAAGGTGACAGTTTAAGGAAAAATATATATAGAATACCAGAAATGCCGTTTACCCGGAGAATT TTTTTCTCCCCATTTGTTTTGTTTTTACTCAATGACACCATTTTTAGTTTTATTTCCTGATAGCAAAAGGAAAAAAAACACCCATCCCTC AAAAAGGCCAAGGTCCCGTCCCCCTGTTGTCGGTGATTTGTTTGTCTTTCTGATAGGTTGAAAATTGTGTAATAAACTTGATGACGCTGT >27868_27868_2_EXD3-CPSF7_EXD3_chr9_140289754_ENST00000465160_CPSF7_chr11_61196762_ENST00000340437_length(amino acids)=484AA_BP= MSILRTRDLPSGAMSEGVDLIDIYADEEFNQDPEFNNTDQIDLYDDVLTATSQPSDDRSSSTEPPPPVRQEPSPKPNNKTPAILYTYSGL RNRRAAVYVGSFSWWTTDQQLIQVIRSIGVYDVVELKFAENRANGQSKGYAEVVVASENSVHKLLELLPGKVLNGEKVDVRPATRQNLSQ FEAQARKRECVRVPRGGIPPRAHSRDSSDSADGRATPSENLVPSSARVDKPPSVLPYFNRPPSALPLMGLPPPPIPPPPPLSSSFGVPPP PPGIHYQHLMPPPPRLPPHLAVPPPGAIPPALHLNPAFFPPPNATVGPPPDTYMKASAPYNHHGSRDSGPPPSTVSEAEFEDIMKRNRAI SSSAISKAVSGASAGDYSDAIETLLTAIAVIKQSRVANDERCRVLISSLKDCLHGIEAKSYSVGASGSSSRKRHRSRERSPSRSRESSRR -------------------------------------------------------------- >27868_27868_3_EXD3-CPSF7_EXD3_chr9_140289754_ENST00000479452_CPSF7_chr11_61196762_ENST00000340437_length(transcript)=3676nt_BP=144nt CGCGCCGGGCGTGGACGGTGTGGGGCGGTCCCGGCCCCGCAGATCCTCGTTCCTAGCTGCCTGCCAGCGTCCTCGGCCCGGCCCTGAGGA TGGACCCAGGAGATCCCGCTGGTGACCCTGCCGCTGGCGAGCGCCACCGCATGGTGGCATCTTCCTTACTTTGTCCATCCTCCGGACTCG CGATCTTCCTTCCGGAGCCATGTCAGAAGGAGTGGACTTGATTGATATATATGCTGACGAGGAGTTCAACCAGGACCCAGAGTTCAACAA TACAGATCAGATTGACCTGTATGATGATGTGCTGACAGCCACCTCACAGCCCTCAGATGACAGAAGCAGCAGCACTGAACCACCTCCTCC TGTTCGCCAGGAGCCATCTCCCAAGCCCAACAACAAGACCCCTGCAATTCTGTATACCTACAGTGGCCTGCGTAATAGACGAGCTGCCGT TTATGTGGGCAGCTTCTCCTGGTGGACCACAGACCAGCAGCTGATCCAGGTTATTCGCTCTATAGGAGTCTATGATGTGGTGGAGTTGAA ATTTGCAGAGAATCGAGCAAATGGCCAGTCCAAAGGGTATGCTGAGGTGGTGGTAGCCTCTGAAAACTCTGTCCACAAATTGTTGGAACT CCTACCAGGGAAAGTTCTTAATGGAGAAAAAGTGGACGTGAGGCCGGCCACCCGGCAGAACCTGTCACAGTTTGAGGCACAGGCTCGGAA ACGTGAGTGTGTCCGAGTCCCAAGAGGGGGAATACCTCCACGGGCCCATTCCCGAGATTCTAGTGATTCTGCTGATGGACGGGCCACACC CTCTGAGAACCTTGTACCCTCATCTGCTCGTGTGGATAAGCCCCCCAGTGTGCTGCCCTACTTCAATCGTCCTCCTTCGGCCCTTCCCCT GATGGGTCTGCCCCCACCACCAATTCCACCCCCACCACCTCTCTCCTCAAGCTTTGGGGTCCCTCCTCCTCCTCCTGGTATCCACTACCA GCATCTCATGCCCCCACCTCCTCGATTACCTCCTCATCTTGCTGTACCTCCCCCTGGGGCCATCCCACCTGCCCTTCACCTCAATCCAGC CTTCTTCCCCCCACCAAACGCTACAGTGGGGCCTCCACCAGATACTTACATGAAGGCCTCTGCCCCCTATAACCACCATGGCAGCCGAGA TTCGGGCCCTCCACCCTCTACAGTGAGTGAAGCCGAATTTGAAGATATCATGAAGCGAAACAGAGCAATTTCCAGCAGTGCCATTTCCAA AGCAGTATCTGGAGCCAGTGCAGGGGATTACAGTGACGCAATTGAGACGCTGCTCACAGCCATTGCGGTTATCAAACAGTCCCGGGTTGC CAATGATGAGCGTTGCCGTGTCCTCATCTCCTCTCTTAAGGACTGTCTTCATGGCATCGAAGCCAAGTCCTACAGTGTGGGTGCCAGTGG GAGCTCTTCCAGGAAAAGACATCGCTCCCGGGAAAGGTCACCTAGCCGGTCCCGGGAGAGCAGCAGGAGGCACCGGGATCTGCTTCATAA TGAAGATCGGCATGATGATTATTTCCAAGAAAGGAACCGGGAGCATGAGAGACACCGGGATAGAGAACGGGACCGGCACCACTGAGAAAG GAGTCTGGTTGGAAGCAAATGTTTTTTTAATGGACTTGCATCTCCTCACCTTGATCAGGACTAAAGGACGGAGGCCGCCCCACCCCCTTC CCTTTCCTCCAAACCCCTAACTCCCTCCAGACACCCAGGGAATACCCTCTGCCCCACAGGATTGAAGACTGCTTGGCAGTCCTCCCAATC CCACACCTCCTGTTTGCCAGGGGAAAGAACCTAAAGACTTCGTGTGATTGGGAGGGGTGGCAGACAGGAAGAAAACATGTCCAGGCCCCT GGTCTCCATAGAGAATGGTGCTTTGTCCAAGAAAACGTATGAGTTTCTGATTCTCCGGGAGCCGTTCAATGGTGAGGTTGATGGGAAGAC TTCCTTCCCAAAGAAAATAGATCCTCCATGCAGGATCTAGGAGAGTGACTGGGTGTGCCAAAATATGCCCAGGGTCCTGCCCTCAGCACT AGATTTAATGGGGCCAAGAGGGTCCAAACCCCTTGCTAACATACCACTTCTTTGTTTAACTCCTTTACCTTTCCAGCCCTTTGAGGAGGG ACCATGAGAACAGAAATTACCTTATGAAAAGCTACTTCTGTTCCTGCTTTCCCTCTCACGTATTGACGGTTTATTTCTTTGACCTCCCAG AGGGCTGAACTCTTTCAACTCTGCGCTGCCCAGCCTTCTCAGTGGACTTGCCCCTCCTAAGCAGAGAAGGCCTATGAGGTTGCTTGCTGC TGGGAAGCCTGGCAGAGCCAATTACCACCCTCTGCTGCTTAGTGCTTGGGTACCTCTTGCAATAACCAGCTCTTAGTTGTTCCCTTTCCC TGGGGCTTTTCCATTTAACACATGGAGCCCTTCCCCCAGAAGGCTACTTCCTTGTTTTAGAGGAAGGTACTGCCCATTGGGAGATGGGGA CATTGGGACCTCAGCAATGAAGAACCCTTGTGAAGTAACCAGGAGGAATGGGGAAAGAAGCAAGTTGGGCAGGATATGGCCTACTTCCAT AGGCTTTTCTTTTTTCAGGTTTGATGTAAGCATGGGCTTACATCCCCCAGGTACATACTTTTACTTATTGTGGGATAACCTGGCACTAGT AGGCAGGTAAAGTCACAAATTTGGTGTCTTTTCACCTTTTGACTGTTGACTTAATAGCTCCTCTCACTCTGCCTGGAGATACTTCCTGCC TCAGATGAGGAGCCAGAAGAAACAGAGCCCGACTTGAATGAACTCAGCTCAGAGTTCTAAGGACCAGCATTCTGGGGGCCATTTTCTCTA CAGGCAAATGGAATTGCTTTTCCATAACATCCAAATTGTAATGTGGTTGCTGCTGAAGGAGGAGGCAGCAGCGAGGTCCTGCGGTACCCA TGGGGTGATGCTACTTCTGCATGCATCTACAGGGCATCTGACACCTAACATGAGACGTGGCATGTGAGATGAGACTTGGCATGTGAGACA TAGGGTCACTAGAGACCCTTCTGGGTCAGAGGAGAGAGACTGAATTGGACTAAACCCGTCCTCTGTTCCCAGCACGTTTCTCATATAGCC CTCAGTCACTGAGGGAGTCCCCCGCAGGATTGGAGAGGCACATTCCCTTGGGACAGAGGCTACAGGTTGGAGCTTTTTTTCCCCTGTCCC CCAACCCCATCCCCACCTCCACTTCAGAACATGGCACCCCACCCAACTGGCCAAGTGTTAAGTGATGTGCTTATTGAGAGCAACTCCGGG TGTCTTTTAAAATGTAGAGAAAAGGTGACAGTTTAAGGAAAAATATATATAGAATACCAGAAATGCCGTTTACCCGGAGAATTTTTTTCT CCCCATTTGTTTTGTTTTTACTCAATGACACCATTTTTAGTTTTATTTCCTGATAGCAAAAGGAAAAAAAACACCCATCCCTCAAAAAGG CCAAGGTCCCGTCCCCCTGTTGTCGGTGATTTGTTTGTCTTTCTGATAGGTTGAAAATTGTGTAATAAACTTGATGACGCTGTCAATCTT >27868_27868_3_EXD3-CPSF7_EXD3_chr9_140289754_ENST00000479452_CPSF7_chr11_61196762_ENST00000340437_length(amino acids)=484AA_BP= MSILRTRDLPSGAMSEGVDLIDIYADEEFNQDPEFNNTDQIDLYDDVLTATSQPSDDRSSSTEPPPPVRQEPSPKPNNKTPAILYTYSGL RNRRAAVYVGSFSWWTTDQQLIQVIRSIGVYDVVELKFAENRANGQSKGYAEVVVASENSVHKLLELLPGKVLNGEKVDVRPATRQNLSQ FEAQARKRECVRVPRGGIPPRAHSRDSSDSADGRATPSENLVPSSARVDKPPSVLPYFNRPPSALPLMGLPPPPIPPPPPLSSSFGVPPP PPGIHYQHLMPPPPRLPPHLAVPPPGAIPPALHLNPAFFPPPNATVGPPPDTYMKASAPYNHHGSRDSGPPPSTVSEAEFEDIMKRNRAI SSSAISKAVSGASAGDYSDAIETLLTAIAVIKQSRVANDERCRVLISSLKDCLHGIEAKSYSVGASGSSSRKRHRSRERSPSRSRESSRR -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for EXD3-CPSF7 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for EXD3-CPSF7 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for EXD3-CPSF7 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
