
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FAM118A-CYB5R3 (FusionGDB2 ID:28379) |
Fusion Gene Summary for FAM118A-CYB5R3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FAM118A-CYB5R3 | Fusion gene ID: 28379 | Hgene | Tgene | Gene symbol | FAM118A | CYB5R3 | Gene ID | 55007 | 1727 |
| Gene name | family with sequence similarity 118 member A | cytochrome b5 reductase 3 | |
| Synonyms | C22orf8 | B5R|DIA1 | |
| Cytomap | 22q13.31 | 22q13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein FAM118AbK268H5.C22.4 | NADH-cytochrome b5 reductase 3NADH-cytochrome b5 reductase 3 membrane-bound formNADH-cytochrome b5 reductase 3 soluble formdiaphorase-1mutant NADH-cytochrome b5 reductase | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | Q9NWS6 | P00387 | |
| Ensembl transtripts involved in fusion gene | ENST00000216214, ENST00000405673, ENST00000441876, ENST00000405548, ENST00000491671, | ENST00000352397, ENST00000396303, ENST00000402438, ENST00000407332, ENST00000407623, ENST00000361740, | |
| Fusion gene scores | * DoF score | 11 X 7 X 8=616 | 14 X 14 X 8=1568 |
| # samples | 11 | 18 | |
| ** MAII score | log2(11/616*10)=-2.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(18/1568*10)=-3.12285674778553 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FAM118A [Title/Abstract] AND CYB5R3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FAM118A(45719308)-CYB5R3(43032852), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across FAM118A (5'-gene) Fusion gene breakpoints across FAM118A (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
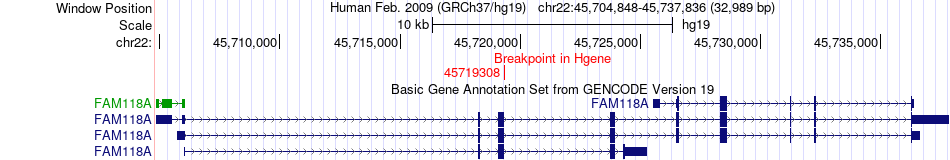 |
 Fusion gene breakpoints across CYB5R3 (3'-gene) Fusion gene breakpoints across CYB5R3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
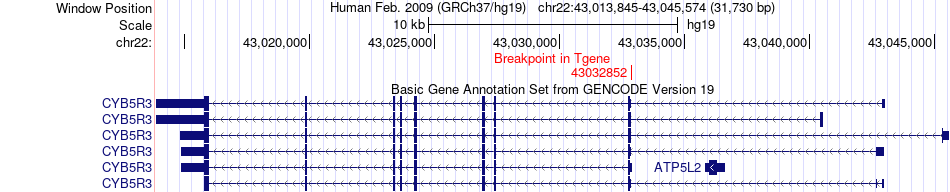 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-JY-A93D | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
Top |
Fusion Gene ORF analysis for FAM118A-CYB5R3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000216214 | ENST00000352397 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| 5CDS-5UTR | ENST00000216214 | ENST00000396303 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| 5CDS-5UTR | ENST00000216214 | ENST00000402438 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| 5CDS-5UTR | ENST00000216214 | ENST00000407332 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| 5CDS-5UTR | ENST00000216214 | ENST00000407623 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| 5CDS-5UTR | ENST00000405673 | ENST00000352397 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| 5CDS-5UTR | ENST00000405673 | ENST00000396303 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| 5CDS-5UTR | ENST00000405673 | ENST00000402438 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| 5CDS-5UTR | ENST00000405673 | ENST00000407332 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| 5CDS-5UTR | ENST00000405673 | ENST00000407623 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| 5CDS-5UTR | ENST00000441876 | ENST00000352397 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| 5CDS-5UTR | ENST00000441876 | ENST00000396303 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| 5CDS-5UTR | ENST00000441876 | ENST00000402438 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| 5CDS-5UTR | ENST00000441876 | ENST00000407332 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| 5CDS-5UTR | ENST00000441876 | ENST00000407623 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| In-frame | ENST00000216214 | ENST00000361740 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| In-frame | ENST00000405673 | ENST00000361740 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| In-frame | ENST00000441876 | ENST00000361740 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| intron-3CDS | ENST00000405548 | ENST00000361740 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| intron-3CDS | ENST00000491671 | ENST00000361740 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| intron-5UTR | ENST00000405548 | ENST00000352397 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| intron-5UTR | ENST00000405548 | ENST00000396303 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| intron-5UTR | ENST00000405548 | ENST00000402438 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| intron-5UTR | ENST00000405548 | ENST00000407332 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| intron-5UTR | ENST00000405548 | ENST00000407623 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| intron-5UTR | ENST00000491671 | ENST00000352397 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| intron-5UTR | ENST00000491671 | ENST00000396303 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| intron-5UTR | ENST00000491671 | ENST00000402438 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| intron-5UTR | ENST00000491671 | ENST00000407332 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
| intron-5UTR | ENST00000491671 | ENST00000407623 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000216214 | FAM118A | chr22 | 45719308 | + | ENST00000361740 | CYB5R3 | chr22 | 43032852 | - | 3952 | 1134 | 678 | 2018 | 446 |
| ENST00000441876 | FAM118A | chr22 | 45719308 | + | ENST00000361740 | CYB5R3 | chr22 | 43032852 | - | 3464 | 646 | 190 | 1530 | 446 |
| ENST00000405673 | FAM118A | chr22 | 45719308 | + | ENST00000361740 | CYB5R3 | chr22 | 43032852 | - | 3187 | 369 | 69 | 1253 | 394 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000216214 | ENST00000361740 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - | 0.00817803 | 0.99182194 |
| ENST00000441876 | ENST00000361740 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - | 0.008921691 | 0.9910783 |
| ENST00000405673 | ENST00000361740 | FAM118A | chr22 | 45719308 | + | CYB5R3 | chr22 | 43032852 | - | 0.008767543 | 0.99123245 |
Top |
Fusion Genomic Features for FAM118A-CYB5R3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
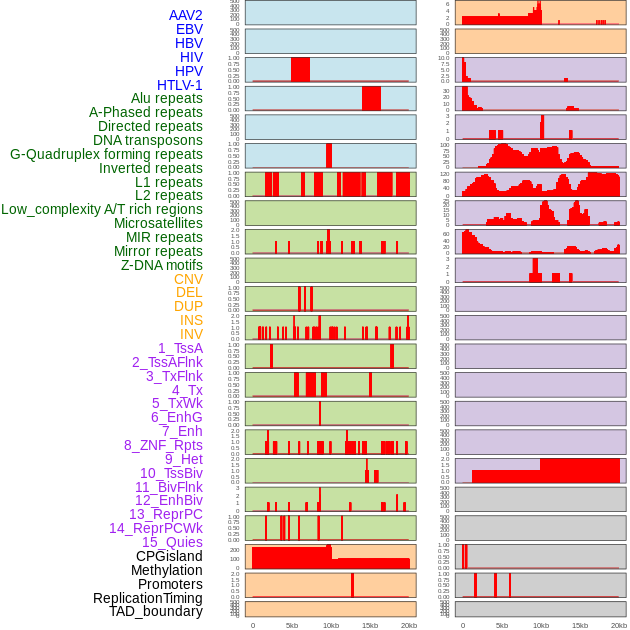 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for FAM118A-CYB5R3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr22:45719308/chr22:43032852) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FAM118A | CYB5R3 |
| FUNCTION: Desaturation and elongation of fatty acids, cholesterol biosynthesis, drug metabolism, and, in erythrocyte, methemoglobin reduction. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FAM118A | chr22:45719308 | chr22:43032852 | ENST00000216214 | + | 4 | 10 | 30_50 | 100 | 358.0 | Transmembrane | Helical |
| Hgene | FAM118A | chr22:45719308 | chr22:43032852 | ENST00000441876 | + | 3 | 9 | 30_50 | 100 | 358.0 | Transmembrane | Helical |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000352397 | 0 | 9 | 40_152 | 7 | 302.0 | Domain | FAD-binding FR-type | |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000361740 | 0 | 9 | 40_152 | 40 | 335.0 | Domain | FAD-binding FR-type | |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000396303 | 0 | 9 | 40_152 | 0 | 279.0 | Domain | FAD-binding FR-type | |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000402438 | 1 | 10 | 40_152 | 0 | 279.0 | Domain | FAD-binding FR-type | |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000407332 | 0 | 8 | 40_152 | 0 | 279.0 | Domain | FAD-binding FR-type | |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000407623 | 0 | 9 | 40_152 | 0 | 279.0 | Domain | FAD-binding FR-type | |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000352397 | 0 | 9 | 132_147 | 7 | 302.0 | Nucleotide binding | FAD | |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000352397 | 0 | 9 | 171_206 | 7 | 302.0 | Nucleotide binding | FAD | |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000361740 | 0 | 9 | 132_147 | 40 | 335.0 | Nucleotide binding | FAD | |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000361740 | 0 | 9 | 171_206 | 40 | 335.0 | Nucleotide binding | FAD | |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000396303 | 0 | 9 | 132_147 | 0 | 279.0 | Nucleotide binding | FAD | |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000396303 | 0 | 9 | 171_206 | 0 | 279.0 | Nucleotide binding | FAD | |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000402438 | 1 | 10 | 132_147 | 0 | 279.0 | Nucleotide binding | FAD | |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000402438 | 1 | 10 | 171_206 | 0 | 279.0 | Nucleotide binding | FAD | |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000407332 | 0 | 8 | 132_147 | 0 | 279.0 | Nucleotide binding | FAD | |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000407332 | 0 | 8 | 171_206 | 0 | 279.0 | Nucleotide binding | FAD | |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000407623 | 0 | 9 | 132_147 | 0 | 279.0 | Nucleotide binding | FAD | |
| Tgene | CYB5R3 | chr22:45719308 | chr22:43032852 | ENST00000407623 | 0 | 9 | 171_206 | 0 | 279.0 | Nucleotide binding | FAD |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Gene Sequence for FAM118A-CYB5R3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >28379_28379_1_FAM118A-CYB5R3_FAM118A_chr22_45719308_ENST00000216214_CYB5R3_chr22_43032852_ENST00000361740_length(transcript)=3952nt_BP=1134nt AAAAGACTTCAGTGGCAGACAAAGGAGGAGTAATAAGATCGCTAGGGGGCCCGTGCCCAGCCCACCCACGCACAATCTCAGTCCTCGCAA TACCCACAAGGTAGGTGCTAGGATCACACCCTTTACGGACGCGGCACCTGCGACAGGGATGCGCGAGGAGTCAGGGGGCCTCGCCGGATC GAACCTAAGCTGGGGAAGAGTATTTCTTGTATTTTTAGGAGAAATTCTCAGCCTCGGGGAAGAGTATTTCTTGATGAGGGAAGAGCGCGG GGAAGACACTCACGCACGCACAAACATGTGGGCGGCCATGGTGTGCCCAGCGCCGTGCTGGCTTCTGGGAACCCCCAGTGGACAAGACGG ACAAGGTACCGGCTCTCAGGGGAAGTGGGAGCCAGTCACAAGCGTACCTAATTTCGGAGAGTGACAAGTACTCTGAAAAAGAAAGAAGGT AGGGCTGGTGACTGGCCAATTTAAGCGGGCAGGAGTCTGCTGGGGGACGGAGACCAGCCTCAGGTCTGGGTTGGGGACAGAAGCTGTGCC TAAGTGTGGTGCAGGATGCAGTTGCAAAGGAGCGCTTCCGATCGCACTTGATGCTCGCCACGTCCCTGCAAAGTGCTCCCGCCCCCTTTC TGCAAATGAGGAAACGGGACGCGCGGCTCGCCGGGCCAGCCCGCGTGCCTGCGCAGTCCCCTCCCCGAGAACCATCCCCTTGCCCCGCCC AGCGTCAGGGGTGCGCGGCCGCCGAGAGACCCCGGAGGCGTAGCCGGCTGCGGAGGCGAAGAGGTGGCAGCGCGAGCTGGGACCAGCGTC TCGGAGGCGCCGCAGAATTCACAGATGGATTCAGTGGAAAAGACAACAAATAGAAGTGAACAAAAATCCAGAAAGTTTTTAAAAAGCCTC ATCCGGAAACAGCCCCAGGAACTGCTCCTGGTTATCGGGACTGGCGTCAGCGCAGCAGTGGCCCCCGGAATCCCTGCCCTTTGCTCGTGG AGAAGCTGCATCGAGGCCGTCATCGAGGCTGCAGAGCAGCTGGAGGTGCTGCACCCCGGAGACGTCGCCGAGTTCCGGAGGAAAGTGACA AAGGACCGGGACCTGTTGGTTGTCGCCCATGATCTGATCCGGAAGATGTCACCTTTGGGCCATATGGTGCTCTTCCCAGTCTGGTTCCTG TACAGTCTGCTCATGAAGCTGTTCCAGCGCTCCACGCCAGCCATCACCCTCGAGAGCCCGGACATCAAGTACCCGCTGCGGCTCATCGAC CGGGAGATCATCAGCCATGACACCCGGCGCTTCCGCTTTGCCCTGCCGTCACCCCAGCACATCCTGGGCCTCCCTGTCGGCCAGCACATC TACCTCTCGGCTCGAATTGATGGAAACCTGGTCGTCCGGCCCTATACACCCATCTCCAGCGATGATGACAAGGGCTTCGTGGACCTGGTC ATCAAGGTTTACTTCAAGGACACCCATCCCAAGTTTCCCGCTGGAGGGAAGATGTCTCAGTACCTGGAGAGCATGCAGATTGGAGACACC ATTGAGTTCCGGGGCCCCAGTGGGCTGCTGGTCTACCAGGGCAAAGGGAAGTTCGCCATCCGACCTGACAAAAAGTCCAACCCTATCATC AGGACAGTGAAGTCTGTGGGCATGATCGCGGGAGGGACAGGCATCACCCCGATGCTGCAGGTGATCCGCGCCATCATGAAGGACCCTGAT GACCACACTGTGTGCCACCTGCTCTTTGCCAACCAGACCGAGAAGGACATCCTGCTGCGACCTGAGCTGGAGGAACTCAGGAACAAACAT TCTGCACGCTTCAAGCTCTGGTACACGCTGGACAGAGCCCCTGAAGCCTGGGACTACGGCCAGGGCTTCGTGAATGAGGAGATGATCCGG GACCACCTTCCACCCCCAGAGGAGGAGCCGCTGGTGCTGATGTGTGGCCCCCCACCCATGATCCAGTACGCCTGCCTTCCCAACCTGGAC CACGTGGGCCACCCCACGGAGCGCTGCTTCGTCTTCTGAGGGCCGGGCACGGTCACACGGCCACCCGCCCCGCGCACCCCACGCCCTGTT CACGCTCACCCAGTCACCTCCCCACATCGCACACTGGGGCCCCGGGTTCAGCCTGGCCTGCCCGTGCCCTGGTGAATCACCTGGCTGAGC AGTTCCCCTGGAGCCCCTTCGGGAGCAGGGCTGTGTCCCAGATGGGCCACGGCTGAGCCTTCAGAGTACGTCCTGCCTGGCACTTACTGG TCCTTACCAGAGACGCCCAGCCCCATCCCTGTCCTCATGACCCCTCGTCCACCCCCCACACACACTATAAGGCTGAGGGCTGCCAGCAGC CCCGTCTGCCCACCATTCCCGGCCGTGGACCATAGTCGGGATGTCAGCAGACACACATGGGCAGCCCAAAGCTGCAGGTGCCAGGGCCCA CCCCAGCCTCGCCTGTCACCCCCACTCCCGCCTCAGGGCCAGGCCCAGGCCTCACCACCTGACGCTGCATGAGACATTGACACCAGAAAG CCCTCTTGGGGGCACTGCTCCCTACCCCAGGGCCCTGGCCAGCCGGGAGCTTGGCTCTCCTCTGGCTAGAGTGGGAAGAGGGGGCTGGCC ATGGGGCCCTCCCAGAACCTCAGCATTTCCTTCCAGCCCATCCAAACACTGAGGCAGCCTTGGGGAACCCCGAGCTGGGGGGTTGGCAGC CCACTGCACCGCCTCAGGGTTTTGGGGTCCTGGGCTGGGGCCACCATCCCTGATGGCAGAACTCCCACAACCACATGTATTTATTCCTCT GTCCTAAACCGTCCCCTCCTTCCCTCACCCCCAGCACAGGGGGATTCTGAGCAGTGCCTCTTGTCTGAGGGACATATCAGTGACCTCGAC GTTGCCTTTAGACTACAGTTGTGTTAGCCTCTTGCGTATTGGCTTTTTCAGAGTCATTTATGAGCAGAAAAAAAAAAAGTAAAACTTTGC TAATATTAACCCTTCTCTAGCTCCTCGAGGGTCTGTGACCTGCAACACAAGGGGTGGGGTCAGGAAAGGGCTGGGGAAGACCTAGCATTT TTTTTTTCTTTTTTTTTTTTTTTTGAGACGGAGTCTCCCTTTGTCACCCAGGCTGGAGTGGCATGATCTCAGCTCACTACAACCTCCACC TCTCGGGTTCAAGCGATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGACTAAAAGTGCCCACCACCACACCCAGCTAGTTTTTGTATTTT TTTTTTTTTTTTTTTGAGACGGAGTCTCGCTCTGTCGCCCAGGCTGGAGTGCAGTGGCGGGATCTCGGCTCACTGCAAGCTCCGCCTCCC GGGTTCACGCCATTCTCCTGCCTCAGCCTCCCAAGTAGCTGGGACTACAGGCGCCCGCCACTACGCCCGGCTAATTTTTTGTATTTTTAG TAGAGACGGGGTTTCACCGTTTTAGCCGGGATGGTCTCGATCTCCTGACCTCGTGATCCGCCCGCCTCGGCCTCCCAAAGTGCTGGGATT ACAGGCGTGAGCCACTGCGCCCGGCTCCAGCATTTATTTCTGATGTATCTTTGTGGTAGAAAATTTGGAAAGTGCAGAGAAGTATACACA GGAAGAAAAATTCCCAACCCCCAGAGGCAAACCAGCTGAAACCACGCAACCCCAGTCACCCCAATGCACCGCGAGGCTGCTGCCTCCTGT CAGGGTCAGATGAGCCTCGAGGCTCAGGAAAGTCAGAGGATGCCATCTGCATGGTGGTAAATTACAGAGGTGATGAGGCAAGGTGGGTGT GGGGCTGTTCTTAAAACGGGGCAGCAGGAAGGCCCCAAGGAGATGGATTTGGGCTGGGACGGGAAGAGAGAGCTGGCCATGCTGGGGTGG >28379_28379_1_FAM118A-CYB5R3_FAM118A_chr22_45719308_ENST00000216214_CYB5R3_chr22_43032852_ENST00000361740_length(amino acids)=446AA_BP=152 MRSPLPENHPLAPPSVRGARPPRDPGGVAGCGGEEVAARAGTSVSEAPQNSQMDSVEKTTNRSEQKSRKFLKSLIRKQPQELLLVIGTGV SAAVAPGIPALCSWRSCIEAVIEAAEQLEVLHPGDVAEFRRKVTKDRDLLVVAHDLIRKMSPLGHMVLFPVWFLYSLLMKLFQRSTPAIT LESPDIKYPLRLIDREIISHDTRRFRFALPSPQHILGLPVGQHIYLSARIDGNLVVRPYTPISSDDDKGFVDLVIKVYFKDTHPKFPAGG KMSQYLESMQIGDTIEFRGPSGLLVYQGKGKFAIRPDKKSNPIIRTVKSVGMIAGGTGITPMLQVIRAIMKDPDDHTVCHLLFANQTEKD -------------------------------------------------------------- >28379_28379_2_FAM118A-CYB5R3_FAM118A_chr22_45719308_ENST00000405673_CYB5R3_chr22_43032852_ENST00000361740_length(transcript)=3187nt_BP=369nt GGCTGCGGAGGCGAAGAGGTGGCAGCGCGAGCTGGGACCAGCGTCTCGGAGGCGCCGCAGAATTCACAGATGGATTCAGTGGAAAAGACA ACAAATAGAAGTGAACAAAAATCCAGAAAGTTTTTAAAAAGCCTCATCCGGAAACAGCCCCAGGAACTGCTCCTGGTTATCGGGACTGGC GTCAGCGCAGCAGTGGCCCCCGGAATCCCTGCCCTTTGCTCGTGGAGAAGCTGCATCGAGGCCGTCATCGAGGCTGCAGAGCAGCTGGAG GTGCTGCACCCCGGAGACGTCGCCGAGTTCCGGAGGAAAGTGACAAAGGACCGGGACCTGTTGGTTGTCGCCCATGATCTGATCCGGAAG ATGTCACCTTTGGGCCATATGGTGCTCTTCCCAGTCTGGTTCCTGTACAGTCTGCTCATGAAGCTGTTCCAGCGCTCCACGCCAGCCATC ACCCTCGAGAGCCCGGACATCAAGTACCCGCTGCGGCTCATCGACCGGGAGATCATCAGCCATGACACCCGGCGCTTCCGCTTTGCCCTG CCGTCACCCCAGCACATCCTGGGCCTCCCTGTCGGCCAGCACATCTACCTCTCGGCTCGAATTGATGGAAACCTGGTCGTCCGGCCCTAT ACACCCATCTCCAGCGATGATGACAAGGGCTTCGTGGACCTGGTCATCAAGGTTTACTTCAAGGACACCCATCCCAAGTTTCCCGCTGGA GGGAAGATGTCTCAGTACCTGGAGAGCATGCAGATTGGAGACACCATTGAGTTCCGGGGCCCCAGTGGGCTGCTGGTCTACCAGGGCAAA GGGAAGTTCGCCATCCGACCTGACAAAAAGTCCAACCCTATCATCAGGACAGTGAAGTCTGTGGGCATGATCGCGGGAGGGACAGGCATC ACCCCGATGCTGCAGGTGATCCGCGCCATCATGAAGGACCCTGATGACCACACTGTGTGCCACCTGCTCTTTGCCAACCAGACCGAGAAG GACATCCTGCTGCGACCTGAGCTGGAGGAACTCAGGAACAAACATTCTGCACGCTTCAAGCTCTGGTACACGCTGGACAGAGCCCCTGAA GCCTGGGACTACGGCCAGGGCTTCGTGAATGAGGAGATGATCCGGGACCACCTTCCACCCCCAGAGGAGGAGCCGCTGGTGCTGATGTGT GGCCCCCCACCCATGATCCAGTACGCCTGCCTTCCCAACCTGGACCACGTGGGCCACCCCACGGAGCGCTGCTTCGTCTTCTGAGGGCCG GGCACGGTCACACGGCCACCCGCCCCGCGCACCCCACGCCCTGTTCACGCTCACCCAGTCACCTCCCCACATCGCACACTGGGGCCCCGG GTTCAGCCTGGCCTGCCCGTGCCCTGGTGAATCACCTGGCTGAGCAGTTCCCCTGGAGCCCCTTCGGGAGCAGGGCTGTGTCCCAGATGG GCCACGGCTGAGCCTTCAGAGTACGTCCTGCCTGGCACTTACTGGTCCTTACCAGAGACGCCCAGCCCCATCCCTGTCCTCATGACCCCT CGTCCACCCCCCACACACACTATAAGGCTGAGGGCTGCCAGCAGCCCCGTCTGCCCACCATTCCCGGCCGTGGACCATAGTCGGGATGTC AGCAGACACACATGGGCAGCCCAAAGCTGCAGGTGCCAGGGCCCACCCCAGCCTCGCCTGTCACCCCCACTCCCGCCTCAGGGCCAGGCC CAGGCCTCACCACCTGACGCTGCATGAGACATTGACACCAGAAAGCCCTCTTGGGGGCACTGCTCCCTACCCCAGGGCCCTGGCCAGCCG GGAGCTTGGCTCTCCTCTGGCTAGAGTGGGAAGAGGGGGCTGGCCATGGGGCCCTCCCAGAACCTCAGCATTTCCTTCCAGCCCATCCAA ACACTGAGGCAGCCTTGGGGAACCCCGAGCTGGGGGGTTGGCAGCCCACTGCACCGCCTCAGGGTTTTGGGGTCCTGGGCTGGGGCCACC ATCCCTGATGGCAGAACTCCCACAACCACATGTATTTATTCCTCTGTCCTAAACCGTCCCCTCCTTCCCTCACCCCCAGCACAGGGGGAT TCTGAGCAGTGCCTCTTGTCTGAGGGACATATCAGTGACCTCGACGTTGCCTTTAGACTACAGTTGTGTTAGCCTCTTGCGTATTGGCTT TTTCAGAGTCATTTATGAGCAGAAAAAAAAAAAGTAAAACTTTGCTAATATTAACCCTTCTCTAGCTCCTCGAGGGTCTGTGACCTGCAA CACAAGGGGTGGGGTCAGGAAAGGGCTGGGGAAGACCTAGCATTTTTTTTTTCTTTTTTTTTTTTTTTTGAGACGGAGTCTCCCTTTGTC ACCCAGGCTGGAGTGGCATGATCTCAGCTCACTACAACCTCCACCTCTCGGGTTCAAGCGATTCTCCTGCCTCAGCCTCCCGAGTAGCTG GGACTAAAAGTGCCCACCACCACACCCAGCTAGTTTTTGTATTTTTTTTTTTTTTTTTTTGAGACGGAGTCTCGCTCTGTCGCCCAGGCT GGAGTGCAGTGGCGGGATCTCGGCTCACTGCAAGCTCCGCCTCCCGGGTTCACGCCATTCTCCTGCCTCAGCCTCCCAAGTAGCTGGGAC TACAGGCGCCCGCCACTACGCCCGGCTAATTTTTTGTATTTTTAGTAGAGACGGGGTTTCACCGTTTTAGCCGGGATGGTCTCGATCTCC TGACCTCGTGATCCGCCCGCCTCGGCCTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACTGCGCCCGGCTCCAGCATTTATTTCTGATG TATCTTTGTGGTAGAAAATTTGGAAAGTGCAGAGAAGTATACACAGGAAGAAAAATTCCCAACCCCCAGAGGCAAACCAGCTGAAACCAC GCAACCCCAGTCACCCCAATGCACCGCGAGGCTGCTGCCTCCTGTCAGGGTCAGATGAGCCTCGAGGCTCAGGAAAGTCAGAGGATGCCA TCTGCATGGTGGTAAATTACAGAGGTGATGAGGCAAGGTGGGTGTGGGGCTGTTCTTAAAACGGGGCAGCAGGAAGGCCCCAAGGAGATG GATTTGGGCTGGGACGGGAAGAGAGAGCTGGCCATGCTGGGGTGGGTGGGTGTTCAAATGGTGGAAACAGCAGACGCAAAGGCCCTGCCG >28379_28379_2_FAM118A-CYB5R3_FAM118A_chr22_45719308_ENST00000405673_CYB5R3_chr22_43032852_ENST00000361740_length(amino acids)=394AA_BP=100 MDSVEKTTNRSEQKSRKFLKSLIRKQPQELLLVIGTGVSAAVAPGIPALCSWRSCIEAVIEAAEQLEVLHPGDVAEFRRKVTKDRDLLVV AHDLIRKMSPLGHMVLFPVWFLYSLLMKLFQRSTPAITLESPDIKYPLRLIDREIISHDTRRFRFALPSPQHILGLPVGQHIYLSARIDG NLVVRPYTPISSDDDKGFVDLVIKVYFKDTHPKFPAGGKMSQYLESMQIGDTIEFRGPSGLLVYQGKGKFAIRPDKKSNPIIRTVKSVGM IAGGTGITPMLQVIRAIMKDPDDHTVCHLLFANQTEKDILLRPELEELRNKHSARFKLWYTLDRAPEAWDYGQGFVNEEMIRDHLPPPEE -------------------------------------------------------------- >28379_28379_3_FAM118A-CYB5R3_FAM118A_chr22_45719308_ENST00000441876_CYB5R3_chr22_43032852_ENST00000361740_length(transcript)=3464nt_BP=646nt CGGGGCGGGGCCTCCGGGGACCGCGGGGCCGTTGGTTTCGGGACGGAACGTTCACGCGGCTGGGGCGGGCGCGCGGGGGAAGGGTTTGCG GCGGCGCCGCTGCCGGCTAACGCGGAGGGGCGCCTGGAGGCGGCGTGGCGTCCGCTCTGGCTCCGACTCCGGCTCTCGCTCTCGCTTCTA GCCCGCGTGCCTGCGCAGTCCCCTCCCCGAGAACCATCCCCTTGCCCCGCCCAGCGTCAGGGGTGCGCGGCCGCCGAGAGACCCCGGAGG CGTAGCCGGCTGCGGAGGCGAAGAGGTGGCAGCGCGAGCTGGGACCAGCGTCTCGGAGGCGCCGCAGAATTCACAGATGGATTCAGTGGA AAAGACAACAAATAGAAGTGAACAAAAATCCAGAAAGTTTTTAAAAAGCCTCATCCGGAAACAGCCCCAGGAACTGCTCCTGGTTATCGG GACTGGCGTCAGCGCAGCAGTGGCCCCCGGAATCCCTGCCCTTTGCTCGTGGAGAAGCTGCATCGAGGCCGTCATCGAGGCTGCAGAGCA GCTGGAGGTGCTGCACCCCGGAGACGTCGCCGAGTTCCGGAGGAAAGTGACAAAGGACCGGGACCTGTTGGTTGTCGCCCATGATCTGAT CCGGAAGATGTCACCTTTGGGCCATATGGTGCTCTTCCCAGTCTGGTTCCTGTACAGTCTGCTCATGAAGCTGTTCCAGCGCTCCACGCC AGCCATCACCCTCGAGAGCCCGGACATCAAGTACCCGCTGCGGCTCATCGACCGGGAGATCATCAGCCATGACACCCGGCGCTTCCGCTT TGCCCTGCCGTCACCCCAGCACATCCTGGGCCTCCCTGTCGGCCAGCACATCTACCTCTCGGCTCGAATTGATGGAAACCTGGTCGTCCG GCCCTATACACCCATCTCCAGCGATGATGACAAGGGCTTCGTGGACCTGGTCATCAAGGTTTACTTCAAGGACACCCATCCCAAGTTTCC CGCTGGAGGGAAGATGTCTCAGTACCTGGAGAGCATGCAGATTGGAGACACCATTGAGTTCCGGGGCCCCAGTGGGCTGCTGGTCTACCA GGGCAAAGGGAAGTTCGCCATCCGACCTGACAAAAAGTCCAACCCTATCATCAGGACAGTGAAGTCTGTGGGCATGATCGCGGGAGGGAC AGGCATCACCCCGATGCTGCAGGTGATCCGCGCCATCATGAAGGACCCTGATGACCACACTGTGTGCCACCTGCTCTTTGCCAACCAGAC CGAGAAGGACATCCTGCTGCGACCTGAGCTGGAGGAACTCAGGAACAAACATTCTGCACGCTTCAAGCTCTGGTACACGCTGGACAGAGC CCCTGAAGCCTGGGACTACGGCCAGGGCTTCGTGAATGAGGAGATGATCCGGGACCACCTTCCACCCCCAGAGGAGGAGCCGCTGGTGCT GATGTGTGGCCCCCCACCCATGATCCAGTACGCCTGCCTTCCCAACCTGGACCACGTGGGCCACCCCACGGAGCGCTGCTTCGTCTTCTG AGGGCCGGGCACGGTCACACGGCCACCCGCCCCGCGCACCCCACGCCCTGTTCACGCTCACCCAGTCACCTCCCCACATCGCACACTGGG GCCCCGGGTTCAGCCTGGCCTGCCCGTGCCCTGGTGAATCACCTGGCTGAGCAGTTCCCCTGGAGCCCCTTCGGGAGCAGGGCTGTGTCC CAGATGGGCCACGGCTGAGCCTTCAGAGTACGTCCTGCCTGGCACTTACTGGTCCTTACCAGAGACGCCCAGCCCCATCCCTGTCCTCAT GACCCCTCGTCCACCCCCCACACACACTATAAGGCTGAGGGCTGCCAGCAGCCCCGTCTGCCCACCATTCCCGGCCGTGGACCATAGTCG GGATGTCAGCAGACACACATGGGCAGCCCAAAGCTGCAGGTGCCAGGGCCCACCCCAGCCTCGCCTGTCACCCCCACTCCCGCCTCAGGG CCAGGCCCAGGCCTCACCACCTGACGCTGCATGAGACATTGACACCAGAAAGCCCTCTTGGGGGCACTGCTCCCTACCCCAGGGCCCTGG CCAGCCGGGAGCTTGGCTCTCCTCTGGCTAGAGTGGGAAGAGGGGGCTGGCCATGGGGCCCTCCCAGAACCTCAGCATTTCCTTCCAGCC CATCCAAACACTGAGGCAGCCTTGGGGAACCCCGAGCTGGGGGGTTGGCAGCCCACTGCACCGCCTCAGGGTTTTGGGGTCCTGGGCTGG GGCCACCATCCCTGATGGCAGAACTCCCACAACCACATGTATTTATTCCTCTGTCCTAAACCGTCCCCTCCTTCCCTCACCCCCAGCACA GGGGGATTCTGAGCAGTGCCTCTTGTCTGAGGGACATATCAGTGACCTCGACGTTGCCTTTAGACTACAGTTGTGTTAGCCTCTTGCGTA TTGGCTTTTTCAGAGTCATTTATGAGCAGAAAAAAAAAAAGTAAAACTTTGCTAATATTAACCCTTCTCTAGCTCCTCGAGGGTCTGTGA CCTGCAACACAAGGGGTGGGGTCAGGAAAGGGCTGGGGAAGACCTAGCATTTTTTTTTTCTTTTTTTTTTTTTTTTGAGACGGAGTCTCC CTTTGTCACCCAGGCTGGAGTGGCATGATCTCAGCTCACTACAACCTCCACCTCTCGGGTTCAAGCGATTCTCCTGCCTCAGCCTCCCGA GTAGCTGGGACTAAAAGTGCCCACCACCACACCCAGCTAGTTTTTGTATTTTTTTTTTTTTTTTTTTGAGACGGAGTCTCGCTCTGTCGC CCAGGCTGGAGTGCAGTGGCGGGATCTCGGCTCACTGCAAGCTCCGCCTCCCGGGTTCACGCCATTCTCCTGCCTCAGCCTCCCAAGTAG CTGGGACTACAGGCGCCCGCCACTACGCCCGGCTAATTTTTTGTATTTTTAGTAGAGACGGGGTTTCACCGTTTTAGCCGGGATGGTCTC GATCTCCTGACCTCGTGATCCGCCCGCCTCGGCCTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACTGCGCCCGGCTCCAGCATTTATT TCTGATGTATCTTTGTGGTAGAAAATTTGGAAAGTGCAGAGAAGTATACACAGGAAGAAAAATTCCCAACCCCCAGAGGCAAACCAGCTG AAACCACGCAACCCCAGTCACCCCAATGCACCGCGAGGCTGCTGCCTCCTGTCAGGGTCAGATGAGCCTCGAGGCTCAGGAAAGTCAGAG GATGCCATCTGCATGGTGGTAAATTACAGAGGTGATGAGGCAAGGTGGGTGTGGGGCTGTTCTTAAAACGGGGCAGCAGGAAGGCCCCAA GGAGATGGATTTGGGCTGGGACGGGAAGAGAGAGCTGGCCATGCTGGGGTGGGTGGGTGTTCAAATGGTGGAAACAGCAGACGCAAAGGC >28379_28379_3_FAM118A-CYB5R3_FAM118A_chr22_45719308_ENST00000441876_CYB5R3_chr22_43032852_ENST00000361740_length(amino acids)=446AA_BP=152 MRSPLPENHPLAPPSVRGARPPRDPGGVAGCGGEEVAARAGTSVSEAPQNSQMDSVEKTTNRSEQKSRKFLKSLIRKQPQELLLVIGTGV SAAVAPGIPALCSWRSCIEAVIEAAEQLEVLHPGDVAEFRRKVTKDRDLLVVAHDLIRKMSPLGHMVLFPVWFLYSLLMKLFQRSTPAIT LESPDIKYPLRLIDREIISHDTRRFRFALPSPQHILGLPVGQHIYLSARIDGNLVVRPYTPISSDDDKGFVDLVIKVYFKDTHPKFPAGG KMSQYLESMQIGDTIEFRGPSGLLVYQGKGKFAIRPDKKSNPIIRTVKSVGMIAGGTGITPMLQVIRAIMKDPDDHTVCHLLFANQTEKD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FAM118A-CYB5R3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FAM118A-CYB5R3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FAM118A-CYB5R3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
