
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FAM134B-SLC6A19 (FusionGDB2 ID:28497) |
Fusion Gene Summary for FAM134B-SLC6A19 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FAM134B-SLC6A19 | Fusion gene ID: 28497 | Hgene | Tgene | Gene symbol | FAM134B | SLC6A19 | Gene ID | 54463 | 340024 |
| Gene name | reticulophagy regulator 1 | solute carrier family 6 member 19 | |
| Synonyms | FAM134B|JK-1|JK1 | B0AT1|HND | |
| Cytomap | 5p15.1 | 5p15.33 | |
| Type of gene | protein-coding | protein-coding | |
| Description | reticulophagy regulator 1family with sequence similarity 134 member Bprotein FAM134Breticulophagy receptor 1reticulophagy receptor FAM134B | sodium-dependent neutral amino acid transporter B(0)AT1sodium-dependent amino acid transporter system B0solute carrier family 6 (neurotransmitter transporter), member 19solute carrier family 6 (neutral amino acid transporter), member 19system B(0) neu | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000306320, ENST00000399793, ENST00000509048, | ENST00000304460, | |
| Fusion gene scores | * DoF score | 7 X 6 X 5=210 | 6 X 5 X 6=180 |
| # samples | 8 | 7 | |
| ** MAII score | log2(8/210*10)=-1.39231742277876 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/180*10)=-1.36257007938471 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FAM134B [Title/Abstract] AND SLC6A19 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FAM134B(16481118)-SLC6A19(1208861), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across FAM134B (5'-gene) Fusion gene breakpoints across FAM134B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
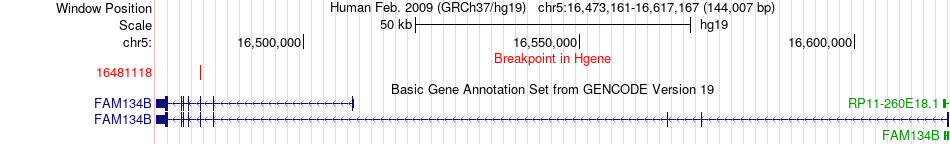 |
 Fusion gene breakpoints across SLC6A19 (3'-gene) Fusion gene breakpoints across SLC6A19 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
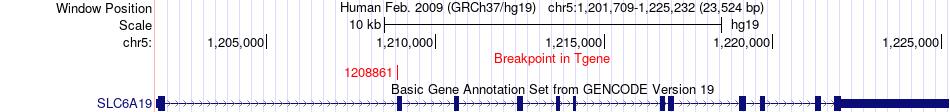 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-7959-01A | FAM134B | chr5 | 16481118 | - | SLC6A19 | chr5 | 1208861 | + |
Top |
Fusion Gene ORF analysis for FAM134B-SLC6A19 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000306320 | ENST00000304460 | FAM134B | chr5 | 16481118 | - | SLC6A19 | chr5 | 1208861 | + |
| In-frame | ENST00000399793 | ENST00000304460 | FAM134B | chr5 | 16481118 | - | SLC6A19 | chr5 | 1208861 | + |
| intron-3CDS | ENST00000509048 | ENST00000304460 | FAM134B | chr5 | 16481118 | - | SLC6A19 | chr5 | 1208861 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000306320 | FAM134B | chr5 | 16481118 | - | ENST00000304460 | SLC6A19 | chr5 | 1208861 | + | 5673 | 757 | 765 | 2459 | 564 |
| ENST00000399793 | FAM134B | chr5 | 16481118 | - | ENST00000304460 | SLC6A19 | chr5 | 1208861 | + | 5549 | 633 | 641 | 2335 | 564 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000306320 | ENST00000304460 | FAM134B | chr5 | 16481118 | - | SLC6A19 | chr5 | 1208861 | + | 0.001088485 | 0.9989115 |
| ENST00000399793 | ENST00000304460 | FAM134B | chr5 | 16481118 | - | SLC6A19 | chr5 | 1208861 | + | 0.000886836 | 0.99911314 |
Top |
Fusion Genomic Features for FAM134B-SLC6A19 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| FAM134B | chr5 | 16481117 | - | SLC6A19 | chr5 | 1208860 | + | 1.19E-05 | 0.9999881 |
| FAM134B | chr5 | 16481117 | - | SLC6A19 | chr5 | 1208860 | + | 1.19E-05 | 0.9999881 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
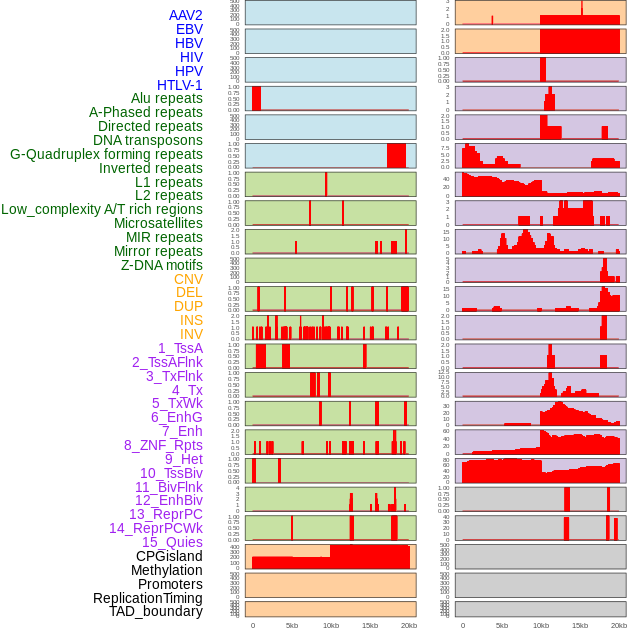 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
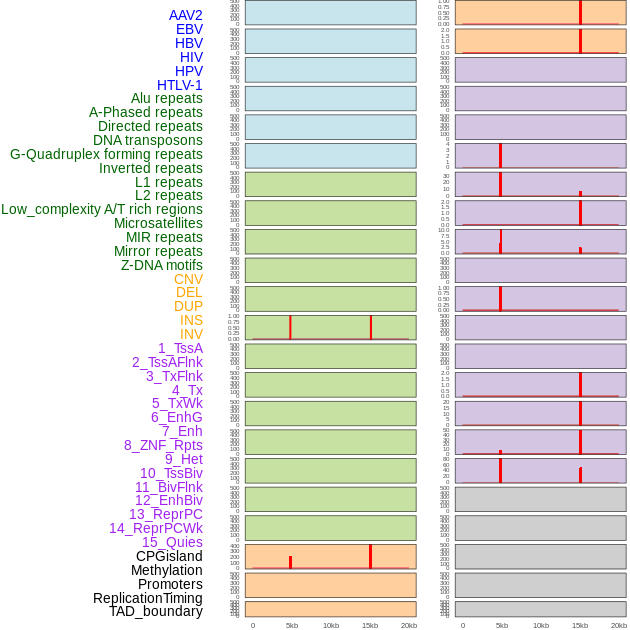 |
Top |
Fusion Protein Features for FAM134B-SLC6A19 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:16481118/chr5:1208861) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000306320 | - | 5 | 9 | 117_118 | 223 | 498.0 | Topological domain | Cytoplasmic |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000306320 | - | 5 | 9 | 140_208 | 223 | 498.0 | Topological domain | Lumenal |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000306320 | - | 5 | 9 | 1_59 | 223 | 498.0 | Topological domain | Cytoplasmic |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000306320 | - | 5 | 9 | 81_95 | 223 | 498.0 | Topological domain | Lumenal |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000399793 | - | 3 | 7 | 1_59 | 82 | 357.0 | Topological domain | Cytoplasmic |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000306320 | - | 5 | 9 | 119_139 | 223 | 498.0 | Transmembrane | Helical |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000306320 | - | 5 | 9 | 60_80 | 223 | 498.0 | Transmembrane | Helical |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000306320 | - | 5 | 9 | 96_116 | 223 | 498.0 | Transmembrane | Helical |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000399793 | - | 3 | 7 | 60_80 | 82 | 357.0 | Transmembrane | Helical |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 142_192 | 67 | 635.0 | Topological domain | Extracellular | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 214_221 | 67 | 635.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 243_268 | 67 | 635.0 | Topological domain | Extracellular | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 290_304 | 67 | 635.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 326_413 | 67 | 635.0 | Topological domain | Extracellular | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 435_456 | 67 | 635.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 478_490 | 67 | 635.0 | Topological domain | Extracellular | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 512_531 | 67 | 635.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 553_581 | 67 | 635.0 | Topological domain | Extracellular | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 603_634 | 67 | 635.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 89_120 | 67 | 635.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 121_141 | 67 | 635.0 | Transmembrane | Helical%3B Name%3D3 | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 193_213 | 67 | 635.0 | Transmembrane | Helical%3B Name%3D4 | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 222_242 | 67 | 635.0 | Transmembrane | Helical%3B Name%3D5 | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 269_289 | 67 | 635.0 | Transmembrane | Helical%3B Name%3D6 | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 305_325 | 67 | 635.0 | Transmembrane | Helical%3B Name%3D7 | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 414_434 | 67 | 635.0 | Transmembrane | Helical%3B Name%3D8 | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 457_477 | 67 | 635.0 | Transmembrane | Helical%3B Name%3D9 | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 491_511 | 67 | 635.0 | Transmembrane | Helical%3B Name%3D10 | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 532_552 | 67 | 635.0 | Transmembrane | Helical%3B Name%3D11 | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 582_602 | 67 | 635.0 | Transmembrane | Helical%3B Name%3D12 | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 68_88 | 67 | 635.0 | Transmembrane | Helical%3B Name%3D2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000306320 | - | 5 | 9 | 84_233 | 223 | 498.0 | Region | Reticulon homology domain |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000399793 | - | 3 | 7 | 84_233 | 82 | 357.0 | Region | Reticulon homology domain |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000306320 | - | 5 | 9 | 230_497 | 223 | 498.0 | Topological domain | Cytoplasmic |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000399793 | - | 3 | 7 | 117_118 | 82 | 357.0 | Topological domain | Cytoplasmic |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000399793 | - | 3 | 7 | 140_208 | 82 | 357.0 | Topological domain | Lumenal |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000399793 | - | 3 | 7 | 230_497 | 82 | 357.0 | Topological domain | Cytoplasmic |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000399793 | - | 3 | 7 | 81_95 | 82 | 357.0 | Topological domain | Lumenal |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000306320 | - | 5 | 9 | 209_229 | 223 | 498.0 | Transmembrane | Helical |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000399793 | - | 3 | 7 | 119_139 | 82 | 357.0 | Transmembrane | Helical |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000399793 | - | 3 | 7 | 209_229 | 82 | 357.0 | Transmembrane | Helical |
| Hgene | FAM134B | chr5:16481118 | chr5:1208861 | ENST00000399793 | - | 3 | 7 | 96_116 | 82 | 357.0 | Transmembrane | Helical |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 1_41 | 67 | 635.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 63_67 | 67 | 635.0 | Topological domain | Extracellular | |
| Tgene | SLC6A19 | chr5:16481118 | chr5:1208861 | ENST00000304460 | 0 | 12 | 42_62 | 67 | 635.0 | Transmembrane | Helical%3B Name%3D1 |
Top |
Fusion Gene Sequence for FAM134B-SLC6A19 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >28497_28497_1_FAM134B-SLC6A19_FAM134B_chr5_16481118_ENST00000306320_SLC6A19_chr5_1208861_ENST00000304460_length(transcript)=5673nt_BP=757nt ACCGCCGCCCCGCCCTCGGCGGCACCCACACCCAGGCGCGCCCGCGCGCGCGCCCGGCCCCGTCCCTGTCTGGAAGCACAGCTGAAGATG GCGAGCCCGGCGCCTCCGGAGCACGCCGAGGAGGGATGCCCGGCTCCTGCCGCCGAGGAGCAGGCGCCGCCGTCGCCGCCACCGCCCCAG GCATCCCCCGCAGAGCGGCAGCAGCAGGAGGAGGAAGCGCAGGAAGCTGGGGCGGCGGAGGGCGCGGGGTTGCAGGTGGAGGAGGCCGCG GGCCGGGCGGCGGCCGCGGTAACCTGGCTGCTCGGGGAGCCGGTGCTGTGGCTGGGCTGCCGCGCCGACGAGCTGCTGAGCTGGAAGAGG CCGCTGCGGAGCCTGCTCGGCTTCGTCGCTGCCAACCTGCTGTTCTGGTTCCTTGCATTGACTCCATGGAGAGTATATCACCTGATTTCC GTCATGATACTTGGGCGTGTTATTATGCAAATAATAAAGGATATGGTTTTGTCTAGAACAAGAGGTGCACAGTTGTGGAGAAGCCTCAGT GAAAGCTGGGAAGTTATCAATTCCAAACCAGATGAAAGACCCAGGCTCAGCCACTGTATTGCAGAATCATGGATGAATTTCAGCATATTT CTTCAAGAAATGTCTCTTTTTAAACAGCAGAGCCCTGGCAAGTTTTGTCTCCTGGTCTGTAGTGTGTGCACATTTTTTACGATCTTGGGA AGTTACATTCCTGGGGTTATACTCAGCTATCTACTGTGAGCCTTCATGATCCCGTTCCTCATCCTGCTGGTCCTGGAGGGCATCCCCCTG CTGTACCTGGAGTTCGCCATCGGGCAGCGGCTGCGGCGGGGCAGCCTGGGTGTGTGGAGCTCCATCCACCCGGCCCTGAAGGGCCTAGGC CTGGCCTCCATGCTCACGTCCTTCATGGTGGGACTGTATTACAACACCATCATCTCCTGGATCATGTGGTACTTATTCAACTCCTTCCAG GAGCCTCTGCCCTGGAGCGACTGCCCGCTCAACGAGAACCAGACAGGGTATGTGGACGAGTGCGCCAGGAGCTCCCCTGTGGACTACTTC TGGTACCGAGAGACGCTCAACATCTCCACGTCCATCAGCGACTCGGGCTCCATCCAGTGGTGGATGCTGCTGTGCCTGGCCTGCGCATGG AGCGTCCTGTACATGTGCACCATCCGCGGCATCGAGACCACCGGGAAGGCCGTGTACATCACCTCCACGCTGCCCTATGTCGTCCTGACC ATCTTCCTCATCCGAGGCCTGACGCTGAAGGGCGCCACCAATGGCATCGTCTTCCTCTTCACGCCCAACGTCACGGAGCTGGCCCAGCCG GACACCTGGCTGGACGCGGGCGCACAGGTCTTCTTCTCCTTCTCCCTGGCCTTCGGGGGCCTCATCTCCTTCTCCAGCTACAACTCTGTG CACAACAACTGCGAGAAGGACTCGGTGATTGTGTCCATCATCAACGGCTTCACATCGGTGTATGTGGCCATCGTGGTCTACTCCGTCATT GGGTTCCGCGCCACACAGCGCTACGACGACTGCTTCAGCACGAACATCCTGACCCTCATCAACGGGTTCGACCTGCCTGAAGGCAACGTG ACCCAGGAGAACTTTGTGGACATGCAGCAGCGGTGCAACGCCTCCGACCCCGCGGCCTACGCGCAGCTGGTGTTCCAGACCTGCGACATC AACGCCTTCCTCTCAGAGGCCGTGGAGGGCACAGGCCTGGCCTTCATCGTCTTCACCGAGGCCATCACCAAGATGCCGTTGTCCCCACTG TGGTCTGTGCTCTTCTTCATTATGCTCTTCTGCCTGGGGCTGTCATCTATGTTTGGGAACATGGAGGGCGTCGTTGTGCCCCTGCAGGAC CTCAGAGTCATCCCCCCGAAGTGGCCCAAGGAGGTGCTCACAGGCCTCATCTGCCTGGGGACATTCCTCATTGGCTTCATCTTCACGCTG AACTCCGGCCAGTACTGGCTCTCCCTGCTGGACAGCTATGCCGGCTCCATTCCCCTGCTCATCATCGCCTTCTGCGAGATGTTCTCTGTG GTCTACGTGTACGGTGTGGACAGGTTCAATAAGGACATCGAGTTCATGATCGGCCACAAGCCCAACATCTTCTGGCAAGTCACGTGGCGC GTGGTCAGCCCCCTGCTCATGCTGATCATCTTCCTCTTCTTCTTCGTGGTAGAGGTCAGTCAGGAGCTGACCTACAGCATCTGGGACCCT GGCTACGAGGAATTTCCCAAATCCCAGAAGATCTCCTACCCGAACTGGGTGTATGTGGTGGTGGTGATTGTGGCTGGAGTGCCCTCCCTC ACCATCCCTGGCTATGCCATCTACAAGCTCATCAGGAACCACTGCCAGAAGCCAGGGGACCATCAGGGGCTGGTGAGCACACTGTCCACA GCCTCCATGAACGGGGACCTGAAGTACTGAGAAGGCCCATCCCACGGCGTGCCATACACTGGTGTCAGGGAAGGAGGAACCAGCAAGACC TGTGGGGTGGGGGCCGGGCTGCACCTGCATGTGTGTAAGCGTGAGTGTATGCTCGTGTGTGAGTGTGTGTATTGTACACGCATGTGCCAT GTGTGCAGATATGTATCGTGTGTGCATGTACATGCATGGGCACTGTGTGAGTGTGCACGTGTATGCACACATATACATGTGTGTGGGTGT GTGTATTGTATGTGCATGTGCCATGTGTGCAGATGTGTCATGTTGTGTGTGTGCATGTACATGTATGGACATTGTGTGAGTGTGCAAGTG TGCATGCATATACATGTGTGCGATATTTGCTGCCCGTGTGTGTGCATGTATATATAGACATACATGCCTATGTTGTGTGTGGTGTGCATA TGTGTGAACACACACGTGTATACATGCATGCACATGTGCTCGTACAATGGGTGTCCACATGCACGTGTATATGTATATCTGTGAGTGTAT ATACATGCATGCAATTGTGTGTATGTGTGTTCTGTGTGTGCGTTTGCAAGTATATATGCACATGTGTATATGTACATGTATGCCTGTGTG ACGTGTGTATATGTGAGCATGTGTACGTGTGTGTATACGTGTGTTGTGTATATGTGTGTGTCTGTACCTGTTTGTGTATATGTGTGTGAT GTGTGCTCGTGTGTGTGCATATTCAGGCAGGTGTGCATTTGTGCATGCCAGTGTGTATGTATGTGCGCATATGGACACGCATGGACACGC ATATGGACACATATGGACACACATATGGACACGTGTGGATATGTGTGCGTACACGTCGCTGGGACACATGCCTGCCACTCGGGGCCCAGC TGCCCTCTGTGTTTGTCCTTGCCACAGTCACGGGGTGCATGTGCAGAGGGGAGCAGACCACTGGGGACGTGCTGTGCCCTGCACGTGCCC GGGGGAAGCGGAAGCTGCAGCTGGGGTGGGGGCAGCACCTCTATGCTTCATCTCTGTGGGTGGCAGGAGACAAAAGCACAGGGTACTATC TTGGCTCCTGGGAGCGACTCTTGCTACCCACCCCCACCCATCCCCTTCCCCTTGGTGTTGACCTTTGACCTGGGGGTTCCCAGAGCCCTG TAGCCCTCGACCCGGAGCAGCCTCTCGGAAGCCGGAGTGGGCAGTTGCTGGCGATTCTGAGAAAACTTGGCCGCATCCACCGGGGCCCTG CCTCCAGTCGGCCGCTGCCGAGTCTCTGCGTTCTGGCCGCTTCCCGGCTTAATGAATGCCAGCCATTTAATCATTGCTCCTGCCACCACA AATAGATGAGCAGTTAAATAAAACTCAACTTGGCATAATTCAAGGCAAATACCACTCTGTGCATTTTCTTAAGAGGACATGAGCTGTGTG AATTTTTAGCCAGCCTTTGGAAAAGATGGGTTACAGGGTAACTCAACCCTGGCTGCCATCCTTGGGCACTGTGTGTGTCCAGGGCACCTT GGAGGACCGTGCAGCCCCCAGAAGCTTCCAGCTCCCGCACCACTCAGTGAAGCCCAGCCTGGCGCCTGCCCTGCCCCCGTCACGGGATGG GCCCCCATTGGGGTTCAACATTCCATCGCAGCCAAAGGCAGTCGGCACTTGGGACATCTGCTTCCACGGACAGGTCACCTCCGCTTTGCA CGGAAGAATCTGGATGCTTACATTAAACTGGTGTTCTGAGAGTTCCTACGGACAGGTCACCTCCGCTTTGCATGGAAGAATCTGGATGCT TACATTAAACTGGTGTTCTGAGAGTTCCTACGGACAGGTCACCTCTGCTTTCCATAGAAGAATCTGGACGCTTACATTAAACTGATGTTC TGAGAATTCCTACAGGCAGGACTGAAAGCCTGGTGTGTGCCAGTATGATGTTCCACCCACAGAAACCTGGTCACAATCGTCCCTTCCAGC ACCCCATCCAGCAGTGACTGCACACACTGAGTCCCCTACCAGCCCCTTTCACCCTGCTGACTGTCACTGGGCCCTGGGATGCGCAAGACT CCACAGCAGCAGAGGTGGGGGGACATATCACAGCCTCTGCCCCCGGCTGTGATGCCACCGAGGGGCTCGCCTGCTGATGGCTTCAACAGG GTCTCACCTCATCTTTTCCTGCTCTTTGGCCCTGGATCGAGAAAATTTCCATCAGTGCCCCATTAATATGCTGCCCTGTGGCATCTGCCC AGGAGGCCCTGCCAGGCGTGCACAGGTGTGCATTGGTGTACCCTGGCATGCACAGGTGTGCACTGATGTGCCCTGGCATCCATTGGTGTA CCCTGGTGTGCCTGCCATAGGACCCTGGGCGGGAGCTCCCATCTCATCTACATCTCCTGATTCATGCGTTGTTTCATAGGTTTCAATGTC TCTGTAAATGTGGTAGAAATGCAGGCTTTATGGGCATAAAGTGTACATTTCTAAATAAATCCCTTCTATTGAGTATGCTCACCCTAGAAG TTACTGTTGTCCAGACGTAGAGGGATGAGTGAGCCAGTGACCTCAGACGGGATGGTGGGGACGGCAGGTCCAGCTCCTGCCTCCTCCTGG GGGGTCTGGCTTTGGGGGCTTGCTCCGAAGAGGCCATGGCCCAGGCCTGTGGCCTCACAATGGGGACCAACCAGCTCTTCTCATCTTCTT CCCTCACACTTCCTCTCACTCAAATAAGAACCTTCCAAAAATGTGTCCACCTGGGCCCCTGCCCTGGGACTCATGGATTTGGAGTTGTGG CCACACGGTTGAGGGGTGCAGTGTCCAGTGGAATGGGGCAATTGCGGGCCTGGGGGCCCTTGGCCTGTCCGTGGCGGGAGCATCTGCAAG GAGGAGCCCCAGAGTCCAGGGAGCACTGTGGGGAGCTCCTTAGAGCTGAACTCACCCGGCGTCAACTCATCAACCCTCCACCCATGGACA GGGGTGCCCCCAGCACAGGAGAGGACTCAGCCCTCTGCCCCCACGCACGGTGGGTGCCTGTCACCCTGTCCTGCCCAGCGGCCCGAGGGC AGCAGTGGGTGTGAGGGCAGCCCCCGGCCTCCCAAGAGCAGCTGAGAGGATCCCTGCGGGAATCCGGGCTTCGGGTGCATGCGATCTGAT CTGAGTTGTTTCTGACAGTGACAGAGTGACAATCTATAAGTATCTCAAGATCAAATGGTTAAATAAAACATAAGAAATTTAAAACGATTA >28497_28497_1_FAM134B-SLC6A19_FAM134B_chr5_16481118_ENST00000306320_SLC6A19_chr5_1208861_ENST00000304460_length(amino acids)=564AA_BP=0 MIPFLILLVLEGIPLLYLEFAIGQRLRRGSLGVWSSIHPALKGLGLASMLTSFMVGLYYNTIISWIMWYLFNSFQEPLPWSDCPLNENQT GYVDECARSSPVDYFWYRETLNISTSISDSGSIQWWMLLCLACAWSVLYMCTIRGIETTGKAVYITSTLPYVVLTIFLIRGLTLKGATNG IVFLFTPNVTELAQPDTWLDAGAQVFFSFSLAFGGLISFSSYNSVHNNCEKDSVIVSIINGFTSVYVAIVVYSVIGFRATQRYDDCFSTN ILTLINGFDLPEGNVTQENFVDMQQRCNASDPAAYAQLVFQTCDINAFLSEAVEGTGLAFIVFTEAITKMPLSPLWSVLFFIMLFCLGLS SMFGNMEGVVVPLQDLRVIPPKWPKEVLTGLICLGTFLIGFIFTLNSGQYWLSLLDSYAGSIPLLIIAFCEMFSVVYVYGVDRFNKDIEF MIGHKPNIFWQVTWRVVSPLLMLIIFLFFFVVEVSQELTYSIWDPGYEEFPKSQKISYPNWVYVVVVIVAGVPSLTIPGYAIYKLIRNHC -------------------------------------------------------------- >28497_28497_2_FAM134B-SLC6A19_FAM134B_chr5_16481118_ENST00000399793_SLC6A19_chr5_1208861_ENST00000304460_length(transcript)=5549nt_BP=633nt ATCCAATGAAGGTTACTGGCTGAGCGGGGACACCTTCTCACAGGACTGGAGAGAGAATGCGGGGCAGCTGGGCAGGGCTCACTTCCAGCC GCCTGTCACAGTACTGGGAGTAAGAGGTGACCTATTTATTTTTAGAAGGGGGCAGTGATAATAACCCAGCTCCTAGCTTCATTCAAGGGA GGCAGGCGCTTTGGAAGTTTGTAAACACCAACTTTCTGAGTAAGGGAGGAGCACTTTTTTTCCAAAAAGGAAAGAACGTCTCTACTGGGT TTTTTTCCTCCTGATATTCAGCATTAGAGTAGAAAAGAAACTATTGTTTGGCCACATTAGCCGTGGTTAGCAGGTGCTGCAGCCTTTGCC ACTGTTATTATTTTTAAAGGGCAGAAATGCCTGAAGGTGAAGACTTTGGACCAGGCAAAAGCTGGGAAGTTATCAATTCCAAACCAGATG AAAGACCCAGGCTCAGCCACTGTATTGCAGAATCATGGATGAATTTCAGCATATTTCTTCAAGAAATGTCTCTTTTTAAACAGCAGAGCC CTGGCAAGTTTTGTCTCCTGGTCTGTAGTGTGTGCACATTTTTTACGATCTTGGGAAGTTACATTCCTGGGGTTATACTCAGCTATCTAC TGTGAGCCTTCATGATCCCGTTCCTCATCCTGCTGGTCCTGGAGGGCATCCCCCTGCTGTACCTGGAGTTCGCCATCGGGCAGCGGCTGC GGCGGGGCAGCCTGGGTGTGTGGAGCTCCATCCACCCGGCCCTGAAGGGCCTAGGCCTGGCCTCCATGCTCACGTCCTTCATGGTGGGAC TGTATTACAACACCATCATCTCCTGGATCATGTGGTACTTATTCAACTCCTTCCAGGAGCCTCTGCCCTGGAGCGACTGCCCGCTCAACG AGAACCAGACAGGGTATGTGGACGAGTGCGCCAGGAGCTCCCCTGTGGACTACTTCTGGTACCGAGAGACGCTCAACATCTCCACGTCCA TCAGCGACTCGGGCTCCATCCAGTGGTGGATGCTGCTGTGCCTGGCCTGCGCATGGAGCGTCCTGTACATGTGCACCATCCGCGGCATCG AGACCACCGGGAAGGCCGTGTACATCACCTCCACGCTGCCCTATGTCGTCCTGACCATCTTCCTCATCCGAGGCCTGACGCTGAAGGGCG CCACCAATGGCATCGTCTTCCTCTTCACGCCCAACGTCACGGAGCTGGCCCAGCCGGACACCTGGCTGGACGCGGGCGCACAGGTCTTCT TCTCCTTCTCCCTGGCCTTCGGGGGCCTCATCTCCTTCTCCAGCTACAACTCTGTGCACAACAACTGCGAGAAGGACTCGGTGATTGTGT CCATCATCAACGGCTTCACATCGGTGTATGTGGCCATCGTGGTCTACTCCGTCATTGGGTTCCGCGCCACACAGCGCTACGACGACTGCT TCAGCACGAACATCCTGACCCTCATCAACGGGTTCGACCTGCCTGAAGGCAACGTGACCCAGGAGAACTTTGTGGACATGCAGCAGCGGT GCAACGCCTCCGACCCCGCGGCCTACGCGCAGCTGGTGTTCCAGACCTGCGACATCAACGCCTTCCTCTCAGAGGCCGTGGAGGGCACAG GCCTGGCCTTCATCGTCTTCACCGAGGCCATCACCAAGATGCCGTTGTCCCCACTGTGGTCTGTGCTCTTCTTCATTATGCTCTTCTGCC TGGGGCTGTCATCTATGTTTGGGAACATGGAGGGCGTCGTTGTGCCCCTGCAGGACCTCAGAGTCATCCCCCCGAAGTGGCCCAAGGAGG TGCTCACAGGCCTCATCTGCCTGGGGACATTCCTCATTGGCTTCATCTTCACGCTGAACTCCGGCCAGTACTGGCTCTCCCTGCTGGACA GCTATGCCGGCTCCATTCCCCTGCTCATCATCGCCTTCTGCGAGATGTTCTCTGTGGTCTACGTGTACGGTGTGGACAGGTTCAATAAGG ACATCGAGTTCATGATCGGCCACAAGCCCAACATCTTCTGGCAAGTCACGTGGCGCGTGGTCAGCCCCCTGCTCATGCTGATCATCTTCC TCTTCTTCTTCGTGGTAGAGGTCAGTCAGGAGCTGACCTACAGCATCTGGGACCCTGGCTACGAGGAATTTCCCAAATCCCAGAAGATCT CCTACCCGAACTGGGTGTATGTGGTGGTGGTGATTGTGGCTGGAGTGCCCTCCCTCACCATCCCTGGCTATGCCATCTACAAGCTCATCA GGAACCACTGCCAGAAGCCAGGGGACCATCAGGGGCTGGTGAGCACACTGTCCACAGCCTCCATGAACGGGGACCTGAAGTACTGAGAAG GCCCATCCCACGGCGTGCCATACACTGGTGTCAGGGAAGGAGGAACCAGCAAGACCTGTGGGGTGGGGGCCGGGCTGCACCTGCATGTGT GTAAGCGTGAGTGTATGCTCGTGTGTGAGTGTGTGTATTGTACACGCATGTGCCATGTGTGCAGATATGTATCGTGTGTGCATGTACATG CATGGGCACTGTGTGAGTGTGCACGTGTATGCACACATATACATGTGTGTGGGTGTGTGTATTGTATGTGCATGTGCCATGTGTGCAGAT GTGTCATGTTGTGTGTGTGCATGTACATGTATGGACATTGTGTGAGTGTGCAAGTGTGCATGCATATACATGTGTGCGATATTTGCTGCC CGTGTGTGTGCATGTATATATAGACATACATGCCTATGTTGTGTGTGGTGTGCATATGTGTGAACACACACGTGTATACATGCATGCACA TGTGCTCGTACAATGGGTGTCCACATGCACGTGTATATGTATATCTGTGAGTGTATATACATGCATGCAATTGTGTGTATGTGTGTTCTG TGTGTGCGTTTGCAAGTATATATGCACATGTGTATATGTACATGTATGCCTGTGTGACGTGTGTATATGTGAGCATGTGTACGTGTGTGT ATACGTGTGTTGTGTATATGTGTGTGTCTGTACCTGTTTGTGTATATGTGTGTGATGTGTGCTCGTGTGTGTGCATATTCAGGCAGGTGT GCATTTGTGCATGCCAGTGTGTATGTATGTGCGCATATGGACACGCATGGACACGCATATGGACACATATGGACACACATATGGACACGT GTGGATATGTGTGCGTACACGTCGCTGGGACACATGCCTGCCACTCGGGGCCCAGCTGCCCTCTGTGTTTGTCCTTGCCACAGTCACGGG GTGCATGTGCAGAGGGGAGCAGACCACTGGGGACGTGCTGTGCCCTGCACGTGCCCGGGGGAAGCGGAAGCTGCAGCTGGGGTGGGGGCA GCACCTCTATGCTTCATCTCTGTGGGTGGCAGGAGACAAAAGCACAGGGTACTATCTTGGCTCCTGGGAGCGACTCTTGCTACCCACCCC CACCCATCCCCTTCCCCTTGGTGTTGACCTTTGACCTGGGGGTTCCCAGAGCCCTGTAGCCCTCGACCCGGAGCAGCCTCTCGGAAGCCG GAGTGGGCAGTTGCTGGCGATTCTGAGAAAACTTGGCCGCATCCACCGGGGCCCTGCCTCCAGTCGGCCGCTGCCGAGTCTCTGCGTTCT GGCCGCTTCCCGGCTTAATGAATGCCAGCCATTTAATCATTGCTCCTGCCACCACAAATAGATGAGCAGTTAAATAAAACTCAACTTGGC ATAATTCAAGGCAAATACCACTCTGTGCATTTTCTTAAGAGGACATGAGCTGTGTGAATTTTTAGCCAGCCTTTGGAAAAGATGGGTTAC AGGGTAACTCAACCCTGGCTGCCATCCTTGGGCACTGTGTGTGTCCAGGGCACCTTGGAGGACCGTGCAGCCCCCAGAAGCTTCCAGCTC CCGCACCACTCAGTGAAGCCCAGCCTGGCGCCTGCCCTGCCCCCGTCACGGGATGGGCCCCCATTGGGGTTCAACATTCCATCGCAGCCA AAGGCAGTCGGCACTTGGGACATCTGCTTCCACGGACAGGTCACCTCCGCTTTGCACGGAAGAATCTGGATGCTTACATTAAACTGGTGT TCTGAGAGTTCCTACGGACAGGTCACCTCCGCTTTGCATGGAAGAATCTGGATGCTTACATTAAACTGGTGTTCTGAGAGTTCCTACGGA CAGGTCACCTCTGCTTTCCATAGAAGAATCTGGACGCTTACATTAAACTGATGTTCTGAGAATTCCTACAGGCAGGACTGAAAGCCTGGT GTGTGCCAGTATGATGTTCCACCCACAGAAACCTGGTCACAATCGTCCCTTCCAGCACCCCATCCAGCAGTGACTGCACACACTGAGTCC CCTACCAGCCCCTTTCACCCTGCTGACTGTCACTGGGCCCTGGGATGCGCAAGACTCCACAGCAGCAGAGGTGGGGGGACATATCACAGC CTCTGCCCCCGGCTGTGATGCCACCGAGGGGCTCGCCTGCTGATGGCTTCAACAGGGTCTCACCTCATCTTTTCCTGCTCTTTGGCCCTG GATCGAGAAAATTTCCATCAGTGCCCCATTAATATGCTGCCCTGTGGCATCTGCCCAGGAGGCCCTGCCAGGCGTGCACAGGTGTGCATT GGTGTACCCTGGCATGCACAGGTGTGCACTGATGTGCCCTGGCATCCATTGGTGTACCCTGGTGTGCCTGCCATAGGACCCTGGGCGGGA GCTCCCATCTCATCTACATCTCCTGATTCATGCGTTGTTTCATAGGTTTCAATGTCTCTGTAAATGTGGTAGAAATGCAGGCTTTATGGG CATAAAGTGTACATTTCTAAATAAATCCCTTCTATTGAGTATGCTCACCCTAGAAGTTACTGTTGTCCAGACGTAGAGGGATGAGTGAGC CAGTGACCTCAGACGGGATGGTGGGGACGGCAGGTCCAGCTCCTGCCTCCTCCTGGGGGGTCTGGCTTTGGGGGCTTGCTCCGAAGAGGC CATGGCCCAGGCCTGTGGCCTCACAATGGGGACCAACCAGCTCTTCTCATCTTCTTCCCTCACACTTCCTCTCACTCAAATAAGAACCTT CCAAAAATGTGTCCACCTGGGCCCCTGCCCTGGGACTCATGGATTTGGAGTTGTGGCCACACGGTTGAGGGGTGCAGTGTCCAGTGGAAT GGGGCAATTGCGGGCCTGGGGGCCCTTGGCCTGTCCGTGGCGGGAGCATCTGCAAGGAGGAGCCCCAGAGTCCAGGGAGCACTGTGGGGA GCTCCTTAGAGCTGAACTCACCCGGCGTCAACTCATCAACCCTCCACCCATGGACAGGGGTGCCCCCAGCACAGGAGAGGACTCAGCCCT CTGCCCCCACGCACGGTGGGTGCCTGTCACCCTGTCCTGCCCAGCGGCCCGAGGGCAGCAGTGGGTGTGAGGGCAGCCCCCGGCCTCCCA AGAGCAGCTGAGAGGATCCCTGCGGGAATCCGGGCTTCGGGTGCATGCGATCTGATCTGAGTTGTTTCTGACAGTGACAGAGTGACAATC >28497_28497_2_FAM134B-SLC6A19_FAM134B_chr5_16481118_ENST00000399793_SLC6A19_chr5_1208861_ENST00000304460_length(amino acids)=564AA_BP=0 MIPFLILLVLEGIPLLYLEFAIGQRLRRGSLGVWSSIHPALKGLGLASMLTSFMVGLYYNTIISWIMWYLFNSFQEPLPWSDCPLNENQT GYVDECARSSPVDYFWYRETLNISTSISDSGSIQWWMLLCLACAWSVLYMCTIRGIETTGKAVYITSTLPYVVLTIFLIRGLTLKGATNG IVFLFTPNVTELAQPDTWLDAGAQVFFSFSLAFGGLISFSSYNSVHNNCEKDSVIVSIINGFTSVYVAIVVYSVIGFRATQRYDDCFSTN ILTLINGFDLPEGNVTQENFVDMQQRCNASDPAAYAQLVFQTCDINAFLSEAVEGTGLAFIVFTEAITKMPLSPLWSVLFFIMLFCLGLS SMFGNMEGVVVPLQDLRVIPPKWPKEVLTGLICLGTFLIGFIFTLNSGQYWLSLLDSYAGSIPLLIIAFCEMFSVVYVYGVDRFNKDIEF MIGHKPNIFWQVTWRVVSPLLMLIIFLFFFVVEVSQELTYSIWDPGYEEFPKSQKISYPNWVYVVVVIVAGVPSLTIPGYAIYKLIRNHC -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FAM134B-SLC6A19 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FAM134B-SLC6A19 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FAM134B-SLC6A19 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
