
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FAM155B-EDA (FusionGDB2 ID:28590) |
Fusion Gene Summary for FAM155B-EDA |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FAM155B-EDA | Fusion gene ID: 28590 | Hgene | Tgene | Gene symbol | FAM155B | EDA | Gene ID | 27112 | 1896 |
| Gene name | family with sequence similarity 155 member B | ectodysplasin A | |
| Synonyms | CXorf63|TED|TMEM28|bB57D9.1 | ECTD1|ED1|ED1-A1|ED1-A2|EDA-A1|EDA-A2|EDA1|EDA2|HED|HED1|ODT1|STHAGX1|TNLG7C|XHED|XLHED | |
| Cytomap | Xq13.1 | Xq13.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | transmembrane protein FAM155BbB57D9.1 (TED protein)transmembrane protein 28 | ectodysplasin-AX-linked anhidroitic ectodermal dysplasia proteinoligodontia 1tumor necrosis factor ligand 7C | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | O75949 | Q8WWZ3 | |
| Ensembl transtripts involved in fusion gene | ENST00000252338, | ENST00000502251, ENST00000338901, ENST00000525810, ENST00000527388, ENST00000374552, ENST00000374553, ENST00000524573, | |
| Fusion gene scores | * DoF score | 2 X 2 X 2=8 | 6 X 6 X 5=180 |
| # samples | 2 | 7 | |
| ** MAII score | log2(2/8*10)=1.32192809488736 | log2(7/180*10)=-1.36257007938471 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FAM155B [Title/Abstract] AND EDA [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FAM155B(68725986)-EDA(69176877), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | EDA | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 11039935 |
 Fusion gene breakpoints across FAM155B (5'-gene) Fusion gene breakpoints across FAM155B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
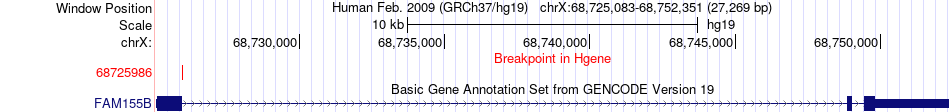 |
 Fusion gene breakpoints across EDA (3'-gene) Fusion gene breakpoints across EDA (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
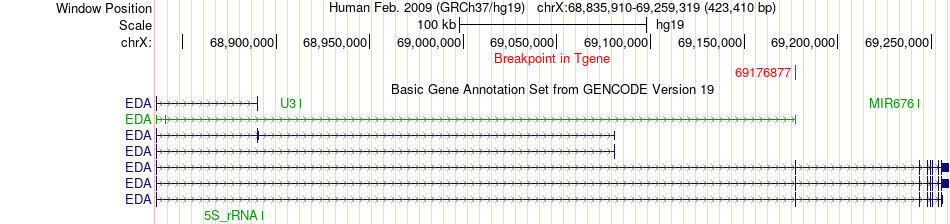 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | GBM | TCGA-06-0221-02A | FAM155B | chrX | 68725986 | + | EDA | chrX | 69176877 | + |
Top |
Fusion Gene ORF analysis for FAM155B-EDA |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000252338 | ENST00000502251 | FAM155B | chrX | 68725986 | + | EDA | chrX | 69176877 | + |
| 5CDS-intron | ENST00000252338 | ENST00000338901 | FAM155B | chrX | 68725986 | + | EDA | chrX | 69176877 | + |
| 5CDS-intron | ENST00000252338 | ENST00000525810 | FAM155B | chrX | 68725986 | + | EDA | chrX | 69176877 | + |
| 5CDS-intron | ENST00000252338 | ENST00000527388 | FAM155B | chrX | 68725986 | + | EDA | chrX | 69176877 | + |
| In-frame | ENST00000252338 | ENST00000374552 | FAM155B | chrX | 68725986 | + | EDA | chrX | 69176877 | + |
| In-frame | ENST00000252338 | ENST00000374553 | FAM155B | chrX | 68725986 | + | EDA | chrX | 69176877 | + |
| In-frame | ENST00000252338 | ENST00000524573 | FAM155B | chrX | 68725986 | + | EDA | chrX | 69176877 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000252338 | FAM155B | chrX | 68725986 | + | ENST00000374553 | EDA | chrX | 69176877 | + | 5537 | 903 | 42 | 1676 | 544 |
| ENST00000252338 | FAM155B | chrX | 68725986 | + | ENST00000374552 | EDA | chrX | 69176877 | + | 5543 | 903 | 42 | 1682 | 546 |
| ENST00000252338 | FAM155B | chrX | 68725986 | + | ENST00000524573 | EDA | chrX | 69176877 | + | 1696 | 903 | 42 | 1667 | 541 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000252338 | ENST00000374553 | FAM155B | chrX | 68725986 | + | EDA | chrX | 69176877 | + | 0.02609995 | 0.9739 |
| ENST00000252338 | ENST00000374552 | FAM155B | chrX | 68725986 | + | EDA | chrX | 69176877 | + | 0.025298415 | 0.9747015 |
| ENST00000252338 | ENST00000524573 | FAM155B | chrX | 68725986 | + | EDA | chrX | 69176877 | + | 0.07908973 | 0.9209103 |
Top |
Fusion Genomic Features for FAM155B-EDA |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| FAM155B | chrX | 68725986 | + | EDA | chrX | 69176876 | + | 0.11657933 | 0.8834207 |
| FAM155B | chrX | 68725986 | + | EDA | chrX | 69176876 | + | 0.11657933 | 0.8834207 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
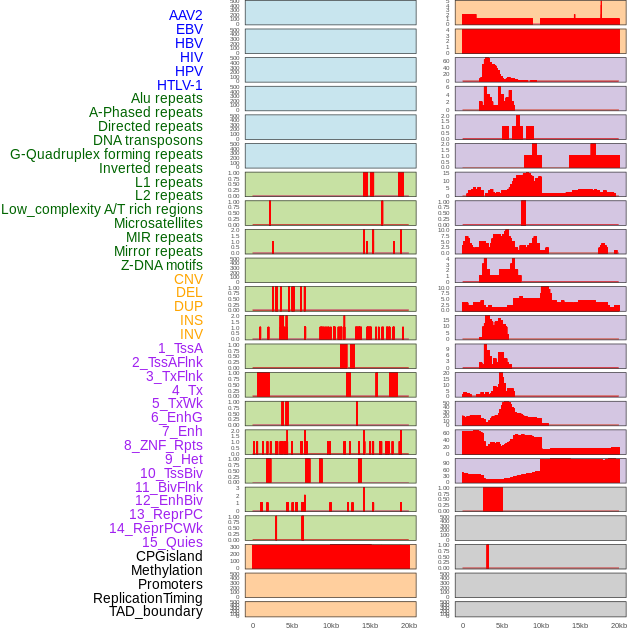 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
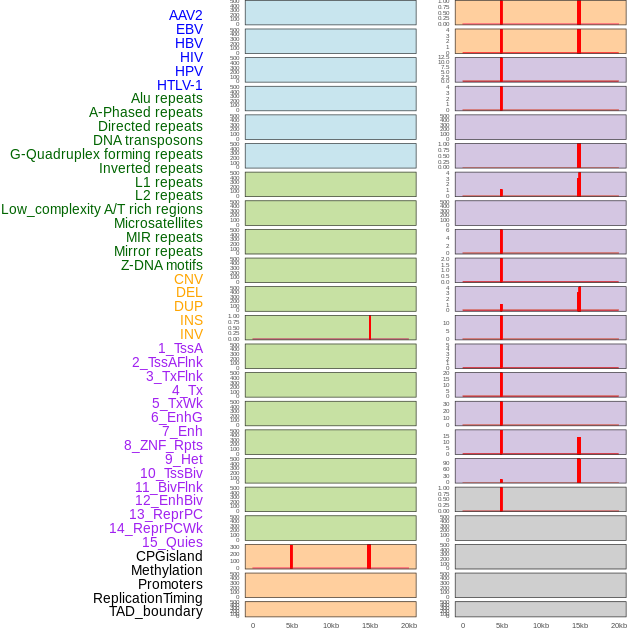 |
Top |
Fusion Protein Features for FAM155B-EDA |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chrX:68725986/chrX:69176877) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FAM155B | EDA |
| FUNCTION: Adapter protein that interacts with EDAR DEATH domain and couples the receptor to EDA signaling pathway during morphogenesis of ectodermal organs. Mediates the activation of NF-kappa-B. {ECO:0000269|PubMed:11882293}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FAM155B | chrX:68725986 | chrX:69176877 | ENST00000252338 | + | 1 | 3 | 80_180 | 287 | 473.0 | Compositional bias | Pro-rich |
| Hgene | FAM155B | chrX:68725986 | chrX:69176877 | ENST00000252338 | + | 1 | 3 | 47_67 | 287 | 473.0 | Transmembrane | Helical |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000338901 | 0 | 2 | 180_229 | 0 | 149.0 | Domain | Note=Collagen-like | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000374552 | 0 | 8 | 180_229 | 132 | 392.0 | Domain | Note=Collagen-like | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000374553 | 0 | 8 | 180_229 | 132 | 390.0 | Domain | Note=Collagen-like | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000524573 | 0 | 8 | 180_229 | 132 | 387.0 | Domain | Note=Collagen-like | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000525810 | 0 | 2 | 180_229 | 0 | 136.0 | Domain | Note=Collagen-like | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000527388 | 0 | 3 | 180_229 | 0 | 4.666666666666667 | Domain | Note=Collagen-like | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000338901 | 0 | 2 | 1_41 | 0 | 149.0 | Topological domain | Cytoplasmic | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000338901 | 0 | 2 | 63_391 | 0 | 149.0 | Topological domain | Extracellular | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000525810 | 0 | 2 | 1_41 | 0 | 136.0 | Topological domain | Cytoplasmic | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000525810 | 0 | 2 | 63_391 | 0 | 136.0 | Topological domain | Extracellular | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000527388 | 0 | 3 | 1_41 | 0 | 4.666666666666667 | Topological domain | Cytoplasmic | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000527388 | 0 | 3 | 63_391 | 0 | 4.666666666666667 | Topological domain | Extracellular | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000338901 | 0 | 2 | 42_62 | 0 | 149.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000525810 | 0 | 2 | 42_62 | 0 | 136.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000527388 | 0 | 3 | 42_62 | 0 | 4.666666666666667 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FAM155B | chrX:68725986 | chrX:69176877 | ENST00000252338 | + | 1 | 3 | 382_405 | 287 | 473.0 | Compositional bias | His-rich |
| Hgene | FAM155B | chrX:68725986 | chrX:69176877 | ENST00000252338 | + | 1 | 3 | 433_453 | 287 | 473.0 | Transmembrane | Helical |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000374552 | 0 | 8 | 1_41 | 132 | 392.0 | Topological domain | Cytoplasmic | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000374552 | 0 | 8 | 63_391 | 132 | 392.0 | Topological domain | Extracellular | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000374553 | 0 | 8 | 1_41 | 132 | 390.0 | Topological domain | Cytoplasmic | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000374553 | 0 | 8 | 63_391 | 132 | 390.0 | Topological domain | Extracellular | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000524573 | 0 | 8 | 1_41 | 132 | 387.0 | Topological domain | Cytoplasmic | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000524573 | 0 | 8 | 63_391 | 132 | 387.0 | Topological domain | Extracellular | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000374552 | 0 | 8 | 42_62 | 132 | 392.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000374553 | 0 | 8 | 42_62 | 132 | 390.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | EDA | chrX:68725986 | chrX:69176877 | ENST00000524573 | 0 | 8 | 42_62 | 132 | 387.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
Top |
Fusion Gene Sequence for FAM155B-EDA |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >28590_28590_1_FAM155B-EDA_FAM155B_chrX_68725986_ENST00000252338_EDA_chrX_69176877_ENST00000374552_length(transcript)=5543nt_BP=903nt CTCGCGCCTGCCTGTTCCCTCCAGCCCGGACCCCCCTGAAATATGTTCAGGGGCGCTTGGATGTGGCCCGGGAAAGACGCCGCCGCGCTG ACTATCTGCTGCTGCTGCTGCTGCTGGGCTCCCAGGCCGAGCGACAAACCTTGCGCCGACTCCGAGCGGGCGCAGCGATGGCGACTGTCC CTGGCGTCCCTGCTCTTCTTCACCGTGCTGCTCGCTGACCATCTGTGGCTGTGCGCGGGGGCCCGGCCCCGGGCCAGGGAGCTGAGCAGC GCCATGCGGCCCCCATGGGGGGCCGGCCGGGAGCGGCAGCCGGTGCCTCCTCGCGCGGTGCTGCCGCTGCCGCCGCCGCCGCCCGGCGAG CCCAGCGCGCCCCCAGGCACCTGCGGCCCCAGATACAGCAACCTGACCAAAGCCGCCCCCGCCGCCGGCTCTCGGCCGGTCTGCGGCGGC GTCCCAGAGCCCACGGGGCTGGACGCAGCTTGCACCAAATTGCAATCTTTGCAGAGACTTTTCGAACCGACTACTCCGGCCCCCCCTCTG CGGCCCCCTGACTCCCTTTCCCGTGCCCCGGCCGAGTTCCCCTCCGCCAAAAAAAACTTGCTCAAAGGCCACTTTCGGAACTTCACTCTC TCCTTTTGCGACACCTACACGGTCTGGGACTTGCTGCTGGGCATGGACCGCCCCGACAGCCTGGACTGTAGCCTGGACACCCTGATGGGG GACCTGCTGGCCGTGGTGGCCAGCCCGGGCTCCGGGGCCTGGGAGGCGTGTAGCAACTGTATCGAGGCGTACCAGCGGCTGGACCGACAC GCTCAGGAAAAATATGACGAGTTCGACCTCGTGCTGCATAAATACTTACAGGCGGAAGAGTACTCAATCCGGTCCTGCACGAAAGGCTGT AAGATGGCCCTATTGAATTTCTTCTTCCCTGATGAAAAGCCATACTCTGAAGAAGAAAGTAGGCGTGTTCGCCGCAATAAAAGAAGCAAA AGCAATGAAGGAGCAGATGGCCCAGTTAAAAACAAGAAAAAGGGAAAGAAAGCAGGACCTCCTGGACCCAATGGCCCTCCAGGACCCCCA GGACCTCCAGGACCCCAGGGACCCCCAGGAATTCCAGGGATTCCTGGAATTCCAGGAACAACTGTTATGGGACCACCTGGTCCTCCAGGT CCTCCTGGTCCTCAAGGACCCCCTGGCCTCCAGGGACCTTCTGGTGCTGCTGATAAAGCTGGAACTCGAGAAAACCAGCCAGCTGTGGTG CATCTACAGGGCCAAGGGTCAGCAATTCAAGTCAAGAATGATCTTTCAGGTGGAGTGCTCAATGACTGGTCTCGCATCACTATGAACCCC AAGGTGTTTAAGCTACATCCCCGCAGCGGGGAGCTGGAGGTACTGGTGGACGGCACCTACTTCATCTATAGTCAGGTAGAAGTATACTAC ATCAACTTCACTGACTTTGCCAGCTATGAGGTGGTGGTGGATGAGAAGCCCTTCCTGCAGTGCACACGCAGCATCGAGACGGGCAAGACC AACTACAACACTTGCTATACCGCAGGCGTCTGCCTCCTCAAGGCCCGGCAGAAGATCGCCGTCAAGATGGTGCACGCTGACATCTCCATC AACATGAGCAAGCACACCACGTTCTTTGGGGCCATCAGGCTGGGTGAAGCCCCTGCATCCTAGATTCCCCCCATTTTGCCTCTGTCCGTG CCCCTTCCCTGGGTTTGGGAGCCAGGACTCCCAGAACCTCTAAGTGCTGCTGTGGAGTGAGGTGTATTGGTGTTGCAGCCGCAGAGAAAT GCCCCAGTGTTATTTATTCCCCAGTGACTCCAGGGTGACAAGGCCTGCTTGACTTTCCAGAATGACCTTGAGTTAACAGGACAGTTGATG GAGCCCCAGGGTTTACATGAAGCAGAACCTTCTTTGGTTCCATGTTGACTGACTTATGGCATGACTCTTCAACCCCGAGGTCCCTGTTGT CAGATCTATTGTTTGTTGCACTAAAATGAGGATCCAGGGCAGCAGGCCAGAGAAAGCAAAGGTGCACTCCAGACTCTGGGGGTGGACATC TGACCCCAAGGGGGCTGCTGCTCCTCTCTTGGGTAGGGTAGTGGCTGGGGTGGAGTGGGAAGGGAGCATTGCAGCCTAAGAAGAAGGCCA GAGAGGGAAAAGGCAGGTGCTTTTGGCAGAGACCATAAGAGAAACCTGCCAAGGAGCATCCTTGGCAGTGGGAATGTTCTTTCTGCTCTA TACTGTGGCCTGCAGGAGGGTTGGAGTGCTCTTCCCACTCCAGCTGACAGCCACACCGTGGCAGCTTGCTGGGCTTTGGGAAGTTTGCTG TGCTTTGGAACAATCACAGGGAATGGCCACAAACCTGCCCGCCTAAGACCCTGAATCCGTACTTGGGTCACATGACTCTCATTTTATTTA CAGCTGTGCTCCACACTCAGAAAATTCCCTGGGGTCACCTTCTAGTTGCCCCCATTCCCAGCCTGACTAGAACTCCTGTCTTCTTTCTCC ATGGAGCCTACCTCTGTCTGAGACAGGTGCCTAACCTGGGACCTGTGGTCATGTGAGTCTGGGATATTCTTTAGCTTACCTGGGCACAAA CAGAATTTTCCATTTATTAAGCAGTACAAATGTTTTTCATCCATTCCTAATCAAATTCTGTCTGGGGACGAAGGGTTGGACGGGATGACC TCCAGAAGTCCCTTCAATTTCTAGTACCTGTGACTCTTAGCCCTCACCACAGCCTTCTAAATTCCCAAATCCTAGACTGCTCCTGGGCAT TAGCAAGGCAGAGCCTTTTTACCTGGCCTAGAAAGGGCAAGGGGTGAGGATAGGACAGAGGGATTTTGTTCAAGTTTGCTGCAACCCAAG TGGACGTTAGGCCAGGCCTTATCTGAAAGGCCAGCAGCTGATGCTGTACTAACCCAGTCTTTCTTCACTCTGGCTTCAAAAAGCCACAGC AGAGCATTGTCACCGCAGGTGCTCATGCTGCTCCCCTAAAGCCAGGCTCAGGAGAAGCCAGTGTCTAGGCACTGAGCAGGGATCTGCCCC CTAGTTCAGGTCCAAATTCACCTTCCCCTAAACCCCAAGCTTCCCAACAGATCATATGGTAGGACCCTCGAGAGCCTTACTTCAAAGTGC CTGGGCTCAGCCTGGTTTCTGGGTGCTAGATCCAGCCCAAACCTGGGAAGGCCAGCCTTGTACAGTCTGCTCCTCTTGTTCCTGAAATGT GTTTCCTTTTCAGGAGATGGGGAATAATTTCCTTCAGGCAGCTGAAATTCACCAAGAACAGCGGGTACTTATTTCTCAGCTGTGCCTTCC CTTTCTAAGCAACCACACTGCTTGGCCCTTCAAGGGTCAGGGTGAGACGTGATGGGCTAGGCCTCCGTTGTCTGGTTGCTAATGACAGCC TTGCAACCCAAGGTGAGGTGAACTCCAGGCATGTGTCTGGCCCTAACTCCTATAAAGTGCCTCGGACAGTCCGCAGTTGTAGCAGAAACC AACAAGAACCACTCCTTCATGTTTGGAAAATAATTTCTCTTGTATTATCTCCTTTGAAGAAGGCAAGGCTGATAATATGACAAACATCAT TGTTTAGATGAGGCTCAGAGAGGTAGCACTCTCAGAGTGTTTTGACCAGTTTAAGCCGCAGACCTGGAGCTTCAGCCAGGTCTGACTCCA AAGCTGTTCCATTACACCACAGCATTGTGTGGAATTTGAGGTCTAGAGAGAACCAATAAAAGTGGTAATTGGGAACTGAAATCCTTGAGA GTTCCGGGGAGAAACCCAGAGATGCCTGATTTCATTCCTCGATGGTAATACCCGTCCTCTCGGCTGCCAGGGGCTCTGTGGCAAAAAGAG TCAGACATTTCTTTGGAAAACAGCGAACAGCCTTAGAGCTCTTGTGTTCAGAAGAATCTTCCTGGCACAATGTTGGAGCAGCAGGCCTCT GGGACCCACAGAACTTGTGGCCTTTATGTTCTTTCACCCATCCTAGGAACCAGCCAACCATCATGTGTAGAGCCCCTACTGTGGGCAAAG TCCTCCTTTCATTACCCTACAGACAGCTTACAGGAGCCAGCCTGCTTCCCACAACTACTAGTGTGACTCCTTATCTCTTTCCACCATACC TTAGAGACTTTGATACTACCAGGGTCTCTCAGGGATGGAGGGAAGACCTGAAAGAGAGGACTGGTTCTGAGGCCAGAAAGGTGTGAGGAG AGAGGAGGAAAAGTCTTCCTAATTGTGCCCCTAAAGAGCATCCTGATACCATTCTATTCTCCAGACATGGAGGGGATGATAAAGGAAATA GGATCTCCACTGGACCCTTGATTCATTCTGAACCCTCCAAAGGAACTCTAGAGGGCGAGGGATGATGAGGGAAGCAATAGGTAGCTGGGG AGCCCTATTGCTGCTAAGTCATTGGCAAAGTGACAAAGCAATTTACTGATGAGAGAATGTGGAAATAGATGTGCAGTTTGGAATTATGTT GGTGTGAATTTGCCAGAGGACCAATGCTTGCATGGAGAATGGGACGAGGACATTTGTGGGCAAGCAGATGACAGAGGTTTGAAGGAGAAT GGCATGGCAGGAGTCTCTGCCAGTTACTTGGGCTTCAACAGCCAAGCTGGCACAAAAGACAGCTGGCGGAGGCTGCTCGGCTACTGGTTA CCTGGAGAAGTAGTATTTGCCTATTTCCCCCTTCATCCATCCTGAGCCAAATTTCTTTTGCTGAACAGGAAAGAGCTAGGAACCCTGGAG GTAAACAAAGACTTTGATCCATGTATGAGTGTATGTGTTTATGTAACTTCCTGTGGATGCAAATAGATTCAGAGAAATTTAGAGCTAAAA AGGCCCTTAGAGGGAATCTAGCCCAACCTACATTCCACCCTGTTACTTATGTAGAAACTGAGGCCCAGAGAGGGAAGATGACCTGCCCCA AGTGGTGAGCAAGCACCAACCTCCAGACTCAGCAGAGTGAGGGGGTAAAGCAGTTCCTGTCCCACATGGCCATCTTCTTTCTTCCACCCA CAAACTCCAGGCTGGAAGTACTTGGCCCCCTTCAGGAGCCTGGCCAGGCAGGGAGAGAGTAGCTGCAGCCTTCATCAGAACTCTTCCTCC TCCCAAGGCATTCTCCCAGCTCTAGCCTCTGGACTGGAAAGCACAAGACTGGCCCAGTGCCAGCAAGTCCTTAGGCTACTGTAATGCTGC CTCAGGACCCATCCCTGCCTGGAGGCTCCTCTAGGCCCTGTGAGCACAAAGAAGAAAGCTGATTTTTGTCTTTTAATCCATTTCAGGACT CTCTCCAGGAGGGCTCGGGGTGTGTCATTTCTATATTCCTCCAGCTGGGATTGGGGGGTGGGCTTTGTTGTGAGAATGGCCTGGAGCAGG CCCAATGCTGCTTTTGGGGGTCAGCATCCAGTGTGAGATACTGTGTATATAAACTATATATAATGTATATAAACTGGGATGTAAGTTTGT >28590_28590_1_FAM155B-EDA_FAM155B_chrX_68725986_ENST00000252338_EDA_chrX_69176877_ENST00000374552_length(amino acids)=546AA_BP=0 MFRGAWMWPGKDAAALTICCCCCCWAPRPSDKPCADSERAQRWRLSLASLLFFTVLLADHLWLCAGARPRARELSSAMRPPWGAGRERQP VPPRAVLPLPPPPPGEPSAPPGTCGPRYSNLTKAAPAAGSRPVCGGVPEPTGLDAACTKLQSLQRLFEPTTPAPPLRPPDSLSRAPAEFP SAKKNLLKGHFRNFTLSFCDTYTVWDLLLGMDRPDSLDCSLDTLMGDLLAVVASPGSGAWEACSNCIEAYQRLDRHAQEKYDEFDLVLHK YLQAEEYSIRSCTKGCKMALLNFFFPDEKPYSEEESRRVRRNKRSKSNEGADGPVKNKKKGKKAGPPGPNGPPGPPGPPGPQGPPGIPGI PGIPGTTVMGPPGPPGPPGPQGPPGLQGPSGAADKAGTRENQPAVVHLQGQGSAIQVKNDLSGGVLNDWSRITMNPKVFKLHPRSGELEV LVDGTYFIYSQVEVYYINFTDFASYEVVVDEKPFLQCTRSIETGKTNYNTCYTAGVCLLKARQKIAVKMVHADISINMSKHTTFFGAIRL -------------------------------------------------------------- >28590_28590_2_FAM155B-EDA_FAM155B_chrX_68725986_ENST00000252338_EDA_chrX_69176877_ENST00000374553_length(transcript)=5537nt_BP=903nt CTCGCGCCTGCCTGTTCCCTCCAGCCCGGACCCCCCTGAAATATGTTCAGGGGCGCTTGGATGTGGCCCGGGAAAGACGCCGCCGCGCTG ACTATCTGCTGCTGCTGCTGCTGCTGGGCTCCCAGGCCGAGCGACAAACCTTGCGCCGACTCCGAGCGGGCGCAGCGATGGCGACTGTCC CTGGCGTCCCTGCTCTTCTTCACCGTGCTGCTCGCTGACCATCTGTGGCTGTGCGCGGGGGCCCGGCCCCGGGCCAGGGAGCTGAGCAGC GCCATGCGGCCCCCATGGGGGGCCGGCCGGGAGCGGCAGCCGGTGCCTCCTCGCGCGGTGCTGCCGCTGCCGCCGCCGCCGCCCGGCGAG CCCAGCGCGCCCCCAGGCACCTGCGGCCCCAGATACAGCAACCTGACCAAAGCCGCCCCCGCCGCCGGCTCTCGGCCGGTCTGCGGCGGC GTCCCAGAGCCCACGGGGCTGGACGCAGCTTGCACCAAATTGCAATCTTTGCAGAGACTTTTCGAACCGACTACTCCGGCCCCCCCTCTG CGGCCCCCTGACTCCCTTTCCCGTGCCCCGGCCGAGTTCCCCTCCGCCAAAAAAAACTTGCTCAAAGGCCACTTTCGGAACTTCACTCTC TCCTTTTGCGACACCTACACGGTCTGGGACTTGCTGCTGGGCATGGACCGCCCCGACAGCCTGGACTGTAGCCTGGACACCCTGATGGGG GACCTGCTGGCCGTGGTGGCCAGCCCGGGCTCCGGGGCCTGGGAGGCGTGTAGCAACTGTATCGAGGCGTACCAGCGGCTGGACCGACAC GCTCAGGAAAAATATGACGAGTTCGACCTCGTGCTGCATAAATACTTACAGGCGGAAGAGTACTCAATCCGGTCCTGCACGAAAGGCTGT AAGATGGCCCTATTGAATTTCTTCTTCCCTGATGAAAAGCCATACTCTGAAGAAGAAAGTAGGCGTGTTCGCCGCAATAAAAGAAGCAAA AGCAATGAAGGAGCAGATGGCCCAGTTAAAAACAAGAAAAAGGGAAAGAAAGCAGGACCTCCTGGACCCAATGGCCCTCCAGGACCCCCA GGACCTCCAGGACCCCAGGGACCCCCAGGAATTCCAGGGATTCCTGGAATTCCAGGAACAACTGTTATGGGACCACCTGGTCCTCCAGGT CCTCCTGGTCCTCAAGGACCCCCTGGCCTCCAGGGACCTTCTGGTGCTGCTGATAAAGCTGGAACTCGAGAAAACCAGCCAGCTGTGGTG CATCTACAGGGCCAAGGGTCAGCAATTCAAGTCAAGAATGATCTTTCAGGTGGAGTGCTCAATGACTGGTCTCGCATCACTATGAACCCC AAGGTGTTTAAGCTACATCCCCGCAGCGGGGAGCTGGAGGTACTGGTGGACGGCACCTACTTCATCTATAGTCAGGTATACTACATCAAC TTCACTGACTTTGCCAGCTATGAGGTGGTGGTGGATGAGAAGCCCTTCCTGCAGTGCACACGCAGCATCGAGACGGGCAAGACCAACTAC AACACTTGCTATACCGCAGGCGTCTGCCTCCTCAAGGCCCGGCAGAAGATCGCCGTCAAGATGGTGCACGCTGACATCTCCATCAACATG AGCAAGCACACCACGTTCTTTGGGGCCATCAGGCTGGGTGAAGCCCCTGCATCCTAGATTCCCCCCATTTTGCCTCTGTCCGTGCCCCTT CCCTGGGTTTGGGAGCCAGGACTCCCAGAACCTCTAAGTGCTGCTGTGGAGTGAGGTGTATTGGTGTTGCAGCCGCAGAGAAATGCCCCA GTGTTATTTATTCCCCAGTGACTCCAGGGTGACAAGGCCTGCTTGACTTTCCAGAATGACCTTGAGTTAACAGGACAGTTGATGGAGCCC CAGGGTTTACATGAAGCAGAACCTTCTTTGGTTCCATGTTGACTGACTTATGGCATGACTCTTCAACCCCGAGGTCCCTGTTGTCAGATC TATTGTTTGTTGCACTAAAATGAGGATCCAGGGCAGCAGGCCAGAGAAAGCAAAGGTGCACTCCAGACTCTGGGGGTGGACATCTGACCC CAAGGGGGCTGCTGCTCCTCTCTTGGGTAGGGTAGTGGCTGGGGTGGAGTGGGAAGGGAGCATTGCAGCCTAAGAAGAAGGCCAGAGAGG GAAAAGGCAGGTGCTTTTGGCAGAGACCATAAGAGAAACCTGCCAAGGAGCATCCTTGGCAGTGGGAATGTTCTTTCTGCTCTATACTGT GGCCTGCAGGAGGGTTGGAGTGCTCTTCCCACTCCAGCTGACAGCCACACCGTGGCAGCTTGCTGGGCTTTGGGAAGTTTGCTGTGCTTT GGAACAATCACAGGGAATGGCCACAAACCTGCCCGCCTAAGACCCTGAATCCGTACTTGGGTCACATGACTCTCATTTTATTTACAGCTG TGCTCCACACTCAGAAAATTCCCTGGGGTCACCTTCTAGTTGCCCCCATTCCCAGCCTGACTAGAACTCCTGTCTTCTTTCTCCATGGAG CCTACCTCTGTCTGAGACAGGTGCCTAACCTGGGACCTGTGGTCATGTGAGTCTGGGATATTCTTTAGCTTACCTGGGCACAAACAGAAT TTTCCATTTATTAAGCAGTACAAATGTTTTTCATCCATTCCTAATCAAATTCTGTCTGGGGACGAAGGGTTGGACGGGATGACCTCCAGA AGTCCCTTCAATTTCTAGTACCTGTGACTCTTAGCCCTCACCACAGCCTTCTAAATTCCCAAATCCTAGACTGCTCCTGGGCATTAGCAA GGCAGAGCCTTTTTACCTGGCCTAGAAAGGGCAAGGGGTGAGGATAGGACAGAGGGATTTTGTTCAAGTTTGCTGCAACCCAAGTGGACG TTAGGCCAGGCCTTATCTGAAAGGCCAGCAGCTGATGCTGTACTAACCCAGTCTTTCTTCACTCTGGCTTCAAAAAGCCACAGCAGAGCA TTGTCACCGCAGGTGCTCATGCTGCTCCCCTAAAGCCAGGCTCAGGAGAAGCCAGTGTCTAGGCACTGAGCAGGGATCTGCCCCCTAGTT CAGGTCCAAATTCACCTTCCCCTAAACCCCAAGCTTCCCAACAGATCATATGGTAGGACCCTCGAGAGCCTTACTTCAAAGTGCCTGGGC TCAGCCTGGTTTCTGGGTGCTAGATCCAGCCCAAACCTGGGAAGGCCAGCCTTGTACAGTCTGCTCCTCTTGTTCCTGAAATGTGTTTCC TTTTCAGGAGATGGGGAATAATTTCCTTCAGGCAGCTGAAATTCACCAAGAACAGCGGGTACTTATTTCTCAGCTGTGCCTTCCCTTTCT AAGCAACCACACTGCTTGGCCCTTCAAGGGTCAGGGTGAGACGTGATGGGCTAGGCCTCCGTTGTCTGGTTGCTAATGACAGCCTTGCAA CCCAAGGTGAGGTGAACTCCAGGCATGTGTCTGGCCCTAACTCCTATAAAGTGCCTCGGACAGTCCGCAGTTGTAGCAGAAACCAACAAG AACCACTCCTTCATGTTTGGAAAATAATTTCTCTTGTATTATCTCCTTTGAAGAAGGCAAGGCTGATAATATGACAAACATCATTGTTTA GATGAGGCTCAGAGAGGTAGCACTCTCAGAGTGTTTTGACCAGTTTAAGCCGCAGACCTGGAGCTTCAGCCAGGTCTGACTCCAAAGCTG TTCCATTACACCACAGCATTGTGTGGAATTTGAGGTCTAGAGAGAACCAATAAAAGTGGTAATTGGGAACTGAAATCCTTGAGAGTTCCG GGGAGAAACCCAGAGATGCCTGATTTCATTCCTCGATGGTAATACCCGTCCTCTCGGCTGCCAGGGGCTCTGTGGCAAAAAGAGTCAGAC ATTTCTTTGGAAAACAGCGAACAGCCTTAGAGCTCTTGTGTTCAGAAGAATCTTCCTGGCACAATGTTGGAGCAGCAGGCCTCTGGGACC CACAGAACTTGTGGCCTTTATGTTCTTTCACCCATCCTAGGAACCAGCCAACCATCATGTGTAGAGCCCCTACTGTGGGCAAAGTCCTCC TTTCATTACCCTACAGACAGCTTACAGGAGCCAGCCTGCTTCCCACAACTACTAGTGTGACTCCTTATCTCTTTCCACCATACCTTAGAG ACTTTGATACTACCAGGGTCTCTCAGGGATGGAGGGAAGACCTGAAAGAGAGGACTGGTTCTGAGGCCAGAAAGGTGTGAGGAGAGAGGA GGAAAAGTCTTCCTAATTGTGCCCCTAAAGAGCATCCTGATACCATTCTATTCTCCAGACATGGAGGGGATGATAAAGGAAATAGGATCT CCACTGGACCCTTGATTCATTCTGAACCCTCCAAAGGAACTCTAGAGGGCGAGGGATGATGAGGGAAGCAATAGGTAGCTGGGGAGCCCT ATTGCTGCTAAGTCATTGGCAAAGTGACAAAGCAATTTACTGATGAGAGAATGTGGAAATAGATGTGCAGTTTGGAATTATGTTGGTGTG AATTTGCCAGAGGACCAATGCTTGCATGGAGAATGGGACGAGGACATTTGTGGGCAAGCAGATGACAGAGGTTTGAAGGAGAATGGCATG GCAGGAGTCTCTGCCAGTTACTTGGGCTTCAACAGCCAAGCTGGCACAAAAGACAGCTGGCGGAGGCTGCTCGGCTACTGGTTACCTGGA GAAGTAGTATTTGCCTATTTCCCCCTTCATCCATCCTGAGCCAAATTTCTTTTGCTGAACAGGAAAGAGCTAGGAACCCTGGAGGTAAAC AAAGACTTTGATCCATGTATGAGTGTATGTGTTTATGTAACTTCCTGTGGATGCAAATAGATTCAGAGAAATTTAGAGCTAAAAAGGCCC TTAGAGGGAATCTAGCCCAACCTACATTCCACCCTGTTACTTATGTAGAAACTGAGGCCCAGAGAGGGAAGATGACCTGCCCCAAGTGGT GAGCAAGCACCAACCTCCAGACTCAGCAGAGTGAGGGGGTAAAGCAGTTCCTGTCCCACATGGCCATCTTCTTTCTTCCACCCACAAACT CCAGGCTGGAAGTACTTGGCCCCCTTCAGGAGCCTGGCCAGGCAGGGAGAGAGTAGCTGCAGCCTTCATCAGAACTCTTCCTCCTCCCAA GGCATTCTCCCAGCTCTAGCCTCTGGACTGGAAAGCACAAGACTGGCCCAGTGCCAGCAAGTCCTTAGGCTACTGTAATGCTGCCTCAGG ACCCATCCCTGCCTGGAGGCTCCTCTAGGCCCTGTGAGCACAAAGAAGAAAGCTGATTTTTGTCTTTTAATCCATTTCAGGACTCTCTCC AGGAGGGCTCGGGGTGTGTCATTTCTATATTCCTCCAGCTGGGATTGGGGGGTGGGCTTTGTTGTGAGAATGGCCTGGAGCAGGCCCAAT GCTGCTTTTGGGGGTCAGCATCCAGTGTGAGATACTGTGTATATAAACTATATATAATGTATATAAACTGGGATGTAAGTTTGTGTAAAT >28590_28590_2_FAM155B-EDA_FAM155B_chrX_68725986_ENST00000252338_EDA_chrX_69176877_ENST00000374553_length(amino acids)=544AA_BP=0 MFRGAWMWPGKDAAALTICCCCCCWAPRPSDKPCADSERAQRWRLSLASLLFFTVLLADHLWLCAGARPRARELSSAMRPPWGAGRERQP VPPRAVLPLPPPPPGEPSAPPGTCGPRYSNLTKAAPAAGSRPVCGGVPEPTGLDAACTKLQSLQRLFEPTTPAPPLRPPDSLSRAPAEFP SAKKNLLKGHFRNFTLSFCDTYTVWDLLLGMDRPDSLDCSLDTLMGDLLAVVASPGSGAWEACSNCIEAYQRLDRHAQEKYDEFDLVLHK YLQAEEYSIRSCTKGCKMALLNFFFPDEKPYSEEESRRVRRNKRSKSNEGADGPVKNKKKGKKAGPPGPNGPPGPPGPPGPQGPPGIPGI PGIPGTTVMGPPGPPGPPGPQGPPGLQGPSGAADKAGTRENQPAVVHLQGQGSAIQVKNDLSGGVLNDWSRITMNPKVFKLHPRSGELEV LVDGTYFIYSQVYYINFTDFASYEVVVDEKPFLQCTRSIETGKTNYNTCYTAGVCLLKARQKIAVKMVHADISINMSKHTTFFGAIRLGE -------------------------------------------------------------- >28590_28590_3_FAM155B-EDA_FAM155B_chrX_68725986_ENST00000252338_EDA_chrX_69176877_ENST00000524573_length(transcript)=1696nt_BP=903nt CTCGCGCCTGCCTGTTCCCTCCAGCCCGGACCCCCCTGAAATATGTTCAGGGGCGCTTGGATGTGGCCCGGGAAAGACGCCGCCGCGCTG ACTATCTGCTGCTGCTGCTGCTGCTGGGCTCCCAGGCCGAGCGACAAACCTTGCGCCGACTCCGAGCGGGCGCAGCGATGGCGACTGTCC CTGGCGTCCCTGCTCTTCTTCACCGTGCTGCTCGCTGACCATCTGTGGCTGTGCGCGGGGGCCCGGCCCCGGGCCAGGGAGCTGAGCAGC GCCATGCGGCCCCCATGGGGGGCCGGCCGGGAGCGGCAGCCGGTGCCTCCTCGCGCGGTGCTGCCGCTGCCGCCGCCGCCGCCCGGCGAG CCCAGCGCGCCCCCAGGCACCTGCGGCCCCAGATACAGCAACCTGACCAAAGCCGCCCCCGCCGCCGGCTCTCGGCCGGTCTGCGGCGGC GTCCCAGAGCCCACGGGGCTGGACGCAGCTTGCACCAAATTGCAATCTTTGCAGAGACTTTTCGAACCGACTACTCCGGCCCCCCCTCTG CGGCCCCCTGACTCCCTTTCCCGTGCCCCGGCCGAGTTCCCCTCCGCCAAAAAAAACTTGCTCAAAGGCCACTTTCGGAACTTCACTCTC TCCTTTTGCGACACCTACACGGTCTGGGACTTGCTGCTGGGCATGGACCGCCCCGACAGCCTGGACTGTAGCCTGGACACCCTGATGGGG GACCTGCTGGCCGTGGTGGCCAGCCCGGGCTCCGGGGCCTGGGAGGCGTGTAGCAACTGTATCGAGGCGTACCAGCGGCTGGACCGACAC GCTCAGGAAAAATATGACGAGTTCGACCTCGTGCTGCATAAATACTTACAGGCGGAAGAGTACTCAATCCGGTCCTGCACGAAAGGCTGT AAGATGGCCCTATTGAATTTCTTCTTCCCTGATGAAAAGCCATACTCTGAAGAAGAAAGTAGGCGTGTTCGCCGCAATAAAAGAAGCAAA AGCAATGAAGGAGCAGATGGCCCAGTTAAAAACAAGAAAAAGGGAAAGAAAGCAGGACCTCCTGGACCCAATGGCCCTCCAGGACCCCCA GGACCTCCAGGACCCCAGGGACCCCCAGGAATTCCAGGGATTCCTGGAATTCCAGGAACAACTGTTATGGGACCACCTGGTCCTCCAGGT CCTCCTGGTCCTCAAGGACCCCCTGGCCTCCAGGGACCTTCTGGTGCTGCTGATAAAGCTGGAACTCGAGAAAACCAGCCAGCTGTGGTG CATCTACAGGGCCAAGGGTCAGCAATTCAAGTCAAGAATGGTGGAGTGCTCAATGACTGGTCTCGCATCACTATGAACCCCAAGGTGTTT AAGCTACATCCCCGCAGCGGGGAGCTGGAGGTACTGGTGGACGGCACCTACTTCATCTATAGTCAGGTATACTACATCAACTTCACTGAC TTTGCCAGCTATGAGGTGGTGGTGGATGAGAAGCCCTTCCTGCAGTGCACACGCAGCATCGAGACGGGCAAGACCAACTACAACACTTGC TATACCGCAGGCGTCTGCCTCCTCAAGGCCCGGCAGAAGATCGCCGTCAAGATGGTGCACGCTGACATCTCCATCAACATGAGCAAGCAC >28590_28590_3_FAM155B-EDA_FAM155B_chrX_68725986_ENST00000252338_EDA_chrX_69176877_ENST00000524573_length(amino acids)=541AA_BP=0 MFRGAWMWPGKDAAALTICCCCCCWAPRPSDKPCADSERAQRWRLSLASLLFFTVLLADHLWLCAGARPRARELSSAMRPPWGAGRERQP VPPRAVLPLPPPPPGEPSAPPGTCGPRYSNLTKAAPAAGSRPVCGGVPEPTGLDAACTKLQSLQRLFEPTTPAPPLRPPDSLSRAPAEFP SAKKNLLKGHFRNFTLSFCDTYTVWDLLLGMDRPDSLDCSLDTLMGDLLAVVASPGSGAWEACSNCIEAYQRLDRHAQEKYDEFDLVLHK YLQAEEYSIRSCTKGCKMALLNFFFPDEKPYSEEESRRVRRNKRSKSNEGADGPVKNKKKGKKAGPPGPNGPPGPPGPPGPQGPPGIPGI PGIPGTTVMGPPGPPGPPGPQGPPGLQGPSGAADKAGTRENQPAVVHLQGQGSAIQVKNGGVLNDWSRITMNPKVFKLHPRSGELEVLVD GTYFIYSQVYYINFTDFASYEVVVDEKPFLQCTRSIETGKTNYNTCYTAGVCLLKARQKIAVKMVHADISINMSKHTTFFGAIRLGEAPA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FAM155B-EDA |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FAM155B-EDA |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FAM155B-EDA |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
