
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:AGGF1-AP3B1 (FusionGDB2 ID:2881) |
Fusion Gene Summary for AGGF1-AP3B1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: AGGF1-AP3B1 | Fusion gene ID: 2881 | Hgene | Tgene | Gene symbol | AGGF1 | AP3B1 | Gene ID | 55109 | 8546 |
| Gene name | angiogenic factor with G-patch and FHA domains 1 | adaptor related protein complex 3 subunit beta 1 | |
| Synonyms | GPATC7|GPATCH7|HSU84971|HUS84971|VG5Q | ADTB3|ADTB3A|HPS|HPS2|PE | |
| Cytomap | 5q13.3 | 5q14.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | angiogenic factor with G patch and FHA domains 1G patch domain-containing protein 7angiogenic factor VG5Qvasculogenesis gene on 5q protein | AP-3 complex subunit beta-1AP-3 complex beta-3A subunitadaptor protein complex AP-3 subunit beta-1adaptor related protein complex 3 beta 1 subunitbeta-3A-adaptinclathrin assembly protein complex 3 beta-1 large chain | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8N302 | O00203 | |
| Ensembl transtripts involved in fusion gene | ENST00000312916, ENST00000503538, ENST00000506806, | ENST00000523204, ENST00000255194, ENST00000519295, | |
| Fusion gene scores | * DoF score | 3 X 2 X 3=18 | 11 X 11 X 6=726 |
| # samples | 3 | 12 | |
| ** MAII score | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(12/726*10)=-2.59693514238723 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: AGGF1 [Title/Abstract] AND AP3B1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | AGGF1(76335544)-AP3B1(77412058), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | AGGF1 | GO:0001938 | positive regulation of endothelial cell proliferation | 14961121 |
| Hgene | AGGF1 | GO:0007155 | cell adhesion | 14961121 |
| Hgene | AGGF1 | GO:0045766 | positive regulation of angiogenesis | 14961121 |
 Fusion gene breakpoints across AGGF1 (5'-gene) Fusion gene breakpoints across AGGF1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
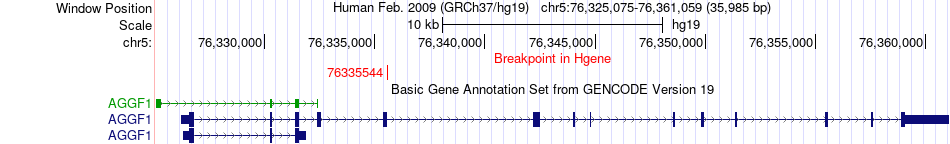 |
 Fusion gene breakpoints across AP3B1 (3'-gene) Fusion gene breakpoints across AP3B1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
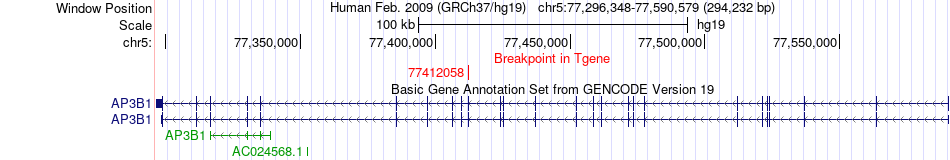 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-HU-A4GT-01A | AGGF1 | chr5 | 76335544 | + | AP3B1 | chr5 | 77412058 | - |
Top |
Fusion Gene ORF analysis for AGGF1-AP3B1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000312916 | ENST00000523204 | AGGF1 | chr5 | 76335544 | + | AP3B1 | chr5 | 77412058 | - |
| In-frame | ENST00000312916 | ENST00000255194 | AGGF1 | chr5 | 76335544 | + | AP3B1 | chr5 | 77412058 | - |
| In-frame | ENST00000312916 | ENST00000519295 | AGGF1 | chr5 | 76335544 | + | AP3B1 | chr5 | 77412058 | - |
| intron-3CDS | ENST00000503538 | ENST00000255194 | AGGF1 | chr5 | 76335544 | + | AP3B1 | chr5 | 77412058 | - |
| intron-3CDS | ENST00000503538 | ENST00000519295 | AGGF1 | chr5 | 76335544 | + | AP3B1 | chr5 | 77412058 | - |
| intron-3CDS | ENST00000506806 | ENST00000255194 | AGGF1 | chr5 | 76335544 | + | AP3B1 | chr5 | 77412058 | - |
| intron-3CDS | ENST00000506806 | ENST00000519295 | AGGF1 | chr5 | 76335544 | + | AP3B1 | chr5 | 77412058 | - |
| intron-intron | ENST00000503538 | ENST00000523204 | AGGF1 | chr5 | 76335544 | + | AP3B1 | chr5 | 77412058 | - |
| intron-intron | ENST00000506806 | ENST00000523204 | AGGF1 | chr5 | 76335544 | + | AP3B1 | chr5 | 77412058 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000312916 | AGGF1 | chr5 | 76335544 | + | ENST00000255194 | AP3B1 | chr5 | 77412058 | - | 4946 | 1252 | 286 | 2568 | 760 |
| ENST00000312916 | AGGF1 | chr5 | 76335544 | + | ENST00000519295 | AP3B1 | chr5 | 77412058 | - | 3145 | 1252 | 286 | 2568 | 760 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000312916 | ENST00000255194 | AGGF1 | chr5 | 76335544 | + | AP3B1 | chr5 | 77412058 | - | 0.000369278 | 0.9996307 |
| ENST00000312916 | ENST00000519295 | AGGF1 | chr5 | 76335544 | + | AP3B1 | chr5 | 77412058 | - | 0.002043796 | 0.99795616 |
Top |
Fusion Genomic Features for AGGF1-AP3B1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
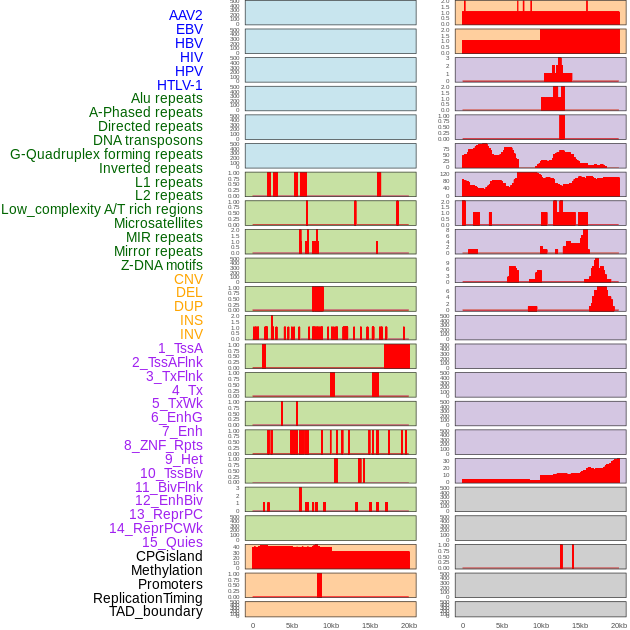 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for AGGF1-AP3B1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:76335544/chr5:77412058) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| AGGF1 | AP3B1 |
| FUNCTION: Promotes angiogenesis and the proliferation of endothelial cells. Able to bind to endothelial cells and promote cell proliferation, suggesting that it may act in an autocrine fashion. {ECO:0000269|PubMed:14961121}. | FUNCTION: Subunit of non-clathrin- and clathrin-associated adaptor protein complex 3 (AP-3) that plays a role in protein sorting in the late-Golgi/trans-Golgi network (TGN) and/or endosomes. The AP complexes mediate both the recruitment of clathrin to membranes and the recognition of sorting signals within the cytosolic tails of transmembrane cargo molecules. AP-3 appears to be involved in the sorting of a subset of transmembrane proteins targeted to lysosomes and lysosome-related organelles. In concert with the BLOC-1 complex, AP-3 is required to target cargos into vesicles assembled at cell bodies for delivery into neurites and nerve terminals. {ECO:0000305|PubMed:9151686}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AGGF1 | chr5:76335544 | chr5:77412058 | ENST00000312916 | + | 5 | 14 | 18_88 | 290 | 715.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | AGGF1 | chr5:76335544 | chr5:77412058 | ENST00000312916 | + | 5 | 14 | 12_15 | 290 | 715.0 | Compositional bias | Note=Poly-Pro |
| Tgene | AP3B1 | chr5:76335544 | chr5:77412058 | ENST00000255194 | 16 | 27 | 677_802 | 656 | 1095.0 | Compositional bias | Note=Glu/Ser-rich |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AGGF1 | chr5:76335544 | chr5:77412058 | ENST00000506806 | + | 1 | 3 | 18_88 | 0 | 177.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | AGGF1 | chr5:76335544 | chr5:77412058 | ENST00000506806 | + | 1 | 3 | 12_15 | 0 | 177.0 | Compositional bias | Note=Poly-Pro |
| Hgene | AGGF1 | chr5:76335544 | chr5:77412058 | ENST00000312916 | + | 5 | 14 | 434_487 | 290 | 715.0 | Domain | FHA |
| Hgene | AGGF1 | chr5:76335544 | chr5:77412058 | ENST00000312916 | + | 5 | 14 | 619_665 | 290 | 715.0 | Domain | G-patch |
| Hgene | AGGF1 | chr5:76335544 | chr5:77412058 | ENST00000506806 | + | 1 | 3 | 434_487 | 0 | 177.0 | Domain | FHA |
| Hgene | AGGF1 | chr5:76335544 | chr5:77412058 | ENST00000506806 | + | 1 | 3 | 619_665 | 0 | 177.0 | Domain | G-patch |
Top |
Fusion Gene Sequence for AGGF1-AP3B1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >2881_2881_1_AGGF1-AP3B1_AGGF1_chr5_76335544_ENST00000312916_AP3B1_chr5_77412058_ENST00000255194_length(transcript)=4946nt_BP=1252nt GGTCCCGCCTGCCGCTGGCGCCGTTGTTTCCGGCTCAACTGGGGAGCTGCTGGAGCTCTTCTGGCCTCTGGTTTTCCGACTGCTTATCCG ACGCTCCTCCCTCTGTCTCTGTAGCTGGAGAAGGTAGTTTCCAGGAAAGTTTTCCGGTTTGCAGGCCGCGCACATCGGGCAGGGGCCATC CTCGGTCCCCTTGCTCGTTGCTCGCAGCCCCGTTCGGCTACAAGTGAGTTTCAGGGCGTCATGGCCAGGGGCCACCGCGGCCAGCCGGGT GTGAGGCTGCCTTTCGCTGCCCGCGCGCTCCAGTGGTCTCTGGGTCCGCCGGCGTCCGTTTCGGCCTGAACGCAGCCCCTCCGCGGCGAC GAGCAGTCTCGCGCCGGAGCTCATGGCCTCGGAGGCGCCGTCCCCGCCGCGGTCGCCGCCGCCGCCCACCTCCCCCGAGCCTGAGCTGGC CCAGCTAAGGCGGAAGGTGGAGAAGTTGGAACGTGAACTGCGGAGCTGCAAGCGGCAGGTGCGGGAGATCGAGAAGCTGCTGCATCACAC AGAACGGCTGTACCAGAACGCAGAAAGCAACAACCAGGAGCTCCGCACGCAGGTGGAAGAACTCAGTAAAATACTCCAACGTGGGAGAAA TGAAGATAATAAAAAGTCTGATGTAGAAGTACAAACAGAGAACCATGCTCCTTGGTCAATCTCAGATTATTTTTATCAGACGTACTACAA TGACGTTAGTCTTCCAAATAAAGTGACTGAACTGTCAGATCAACAAGATCAAGCTATCGAAACTTCTATTTTGAATTCTAAAGACCATTT ACAAGTAGAAAATGATGCTTACCCTGGTACCGATAGAACAGAAAATGTTAAATATAGACAAGTGGACCATTTTGCCTCAAATTCACAGGA GCCAGCATCTGCATTAGCAACAGAAGATACCTCCTTAGAAGGCTCATCATTAGCTGAAAGTTTGAGAGCTGCAGCAGAAGCGGCTGTATC ACAGACTGGATTTAGTTATGATGAAAATACTGGACTGTATTTTGACCACAGCACTGGTTTCTATTATGATTCTGAAAATCAACTCTATTA TGATCCTTCCACTGGAATTTATTACTATTGTGATGTGGAAAGTGGTCGTTATCAGTTTCATTCTCGAGTAGATTTGCAACCTTATCCGAC TTCTAGCACAAAACAAAGTAAAGATAAAAAATTGAAGAAGAAAAGAAAAGATCCAGATTCTTCTGCAACAAATGAGGAAAAGGCAAAAGA ATGGACCCCAGCAGGAAAAGCAAAGCAAGAGAATTCTGCTAAGAAGTTTTATTCTGAATCTGAGGAAGAGGAGGACTCTTCTGATAGTAG CAGTGACAGTGAGAGTGAATCTGGAAGTGAAAGTGGAGAACAAGGCGAAAGTGGGGAGGAAGGAGACAGCAATGAGGACAGCAGTGAGGA CTCCTCCAGTGAGCAGGACAGTGAGAGTGGACGGGAGTCAGGCCTAGAAAACAAAAGAACAGCCAAGAGGAACTCAAAAGCCAAAGGAAA AAGTGATTCTGAAGATGGGGAGAAGGAAAATGAAAAATCTAAAACTTCAGATTCTTCAAATGACGAATCTAGTTCAATAGAAGACAGTTC TTCCGATTCTGAATCAGAGTCAGAACCTGAAAGTGAATCTGAATCCAGAAGAGTCACTAAGGAGAAAGAAAAGAAAACAAAGCAAGATAG AACTCCTCTTACCAAAGATGTTTCACTTCTAGATCTGGATGATTTTAACCCAGTATCCACTCCAGTTGCACTTCCCACACCAGCTCTTTC TCCAAGTTTGATGGCTGATCTTGAAGGTTTACACTTGTCAACTTCCTCTTCAGTCATCAGTGTCAGTACTCCTGCATTTGTACCAACGAA AACTCACGTGCTGCTTCATCGAATGAGTGGAAAAGGACTAGCTGCCCATTATTTCTTTCCAAGACAGCCTTGCATTTTTGGTGATAAGAT GGTCTCTATACAAATAACACTGAATAACACTACTGATCGAAAGATAGAAAATATCCACATAGGGGAAAAAAAACTTCCTATAGGCATGAA AATGCATGTTTTTAATCCAATAGACTCTCTTGAGCCTGAGGGATCCATTACAGTTTCAATGGGTATTGACTTTTGTGATTCTACTCAGAC TGCCAGTTTCCAGTTGTGTACCAAGGATGATTGCTTCAATGTTAATATTCAGCCACCTGTTGGAGAACTGCTTTTACCTGTGGCCATGTC AGAGAAAGATTTTAAGAAAGAGCAAGGAGTGCTAACAGGAATGAATGAAACTTCTGCTGTAATCATTGCTGCACCACAGAATTTCACTCC CTCTGTGATCTTTCAGAAGGTTGTAAATGTAGCCAATGTAGGTGCAGTCCCTTCTGGCCAGGATAATATACACAGGTTTGCAGCTAAAAC TGTGCACAGTGGGTCATTGATGCTAGTCACAGTGGAACTGAAGGAAGGCTCTACAGCCCAGCTTATCATAAACACTGAGAAAACTGTGAT TGGCTCTGTTCTGCTGCGGGAACTGAAGCCTGTCCTGTCTCAGGGGTAACCTGCTTACATCTGGACTTTAGAATCTGGCACACAACAAAA GTGCCTGGCATCCACTACTGCTGCCTTTCATTTATAATAATAGCCCTTCCATCTGGCAGTGGGGGTAGAATACACTCTTGACATTCTTGT CTCCTGCTTTAGAATGCTAGTGTGTATCTATCATGTATGCAATACTTTCCCCCTTTTTGCTTTGCTAACCAAAGAGCATATATTTTACTG TCAGTTGTCTCAACTCTTGAATCCATGTGGCGTTTTCTCTGTCCTGCTGCTTCTTTTGGCCTCCTCGTTTTCCTTCTCTTTTTCGACAAT GGTAGACATGAATGAGATATTTAAAGTTCATTGGAAATCTTCTTCCCTACAGCAGTAAGCAAAAATTAGCAAAGAGATAGTCTAAATGGC CTCTCAGCTTGGTATGTGAAAATGAGATCACATACTTTTTAAATCCAAATACAAAAGCATAGTCTCTGCAAGATTTTGTTCTTTGAATTT CTTGATATTGTAATTGATTATTGATAACTGTCATCATGAAATTATCTCTCAATAATAAGATAAATAAACTAGCATATGAATCATATTTGT CTTTGCATTTTTAATGAGAAACTATTAATAATCCTGAAGTTCATATGCTAAAGTTTTCTTATTTTAAAGCAAGTATTTTGAGTGTTACAT TATGCAGAGAAGCAATGTGGTTTAGTTAGCGAAAGAAAAACCACTTCCAAACCAAGTAAATTTTCTACTTATAGTTTACTCCAAGGAAAA AAAATTGGTGTATTTAATAACAAAAACAAAAATCTTCATATTGCAAAATAAACATCTTCTAGGTAGTTTTGATCTATTAGTAGATATCTG ATTATGTGGCCTGCTGAATAAATTCTTTGAATCTCATTTTTCCTTATGCCCAACTTTGGAAACTAACTAGCATAAACTTATAGAAATGTT TAAAACTAGTTATATGATCAACCTATTAGCATCTTTTCCTAGAGTATTTGTTTTATTTGCTACCTTAAGTTACTGTTTTCAAGGTTGTTC TATAATCAGAGAACTTTTCTAATACTTTCGTACCTTTTTAAAACCAGCTGAAATTTTACTTCCTTTTTGAAGCAAGGTAATGGACTTTCA GGTAGTATAAGCTACTCAGTTCTTTTTGTATAGAGCCACATACATAATGATTTTTAAAAAGTAAAAAGGAAGTGACTGTACATTGCTTGA AGCATTATAGGAAGGCTGAAAAGAAAAGAATGGAATTTTCTTGGAACATACATATTGAAAAGAACAAAACAGAGTCGAAAAATTATTTAA ATTTTCAAGCTGGTGGGATGACAAGAGTAAACAAAATTGTTAAATTTTCCTGCTCCAGAAAGAAATATAGCCCTGAATCTCCGGGAAGAG AATGTACAGAAGTGCAATGTACAGAAGCCACGTGTACATTTTGTGCCAATTCCGTACCATGTGTACATTCTGTACCATTCTGTACCAATG TATGGAAGCCACGAGTTCCACTTCCCTCTGGAGCCTGGGCGCTGTGGTAACACACCCAAAAGCCCCTTTGTAATCGCAGGTCGTTGGCAT TTCCATATTAGCCTTATTGCTCAGAGCTTCATAACTTTGAATTTTAAGTCAATAACTTTTTGAAATGTCAGACAGAATTTGACAGGTTTA ATACCTGTTTTCTGAGGAAGAAAAAAAAATTATATCCATGCTGTCTTCAGGCAGAGGGAAAGTGCATGTTGTGATTGAGAAAGGTCTTAT TTTTCCTTAAATTAGGACACTCATCTTTAAAAGATAGTTCACACATTTGTATTTTTTTAAACTAAGGAAATTTAATTAGGGCTTTACTTT TGACTAAGTTATTTTTGGCTGCTAACTTTTAAAATTATGGTAAATCAATAATATAATGTAGTATGAATCAGTGAACAGTATGTATCGATT GTAATGTGCGCTACCAGTTGAAACCATTTTAACACTAGTAGCTTAATATCATTAAAATGTTTTTCACGTAAAACAGAAAAACGACCAAAG CCCCAAGTTAAAGGTTTGGTTTTTTTTCTTTTGTGGTGTGGTGATTCAGAGTTCCTTGCCTCATATTTCCTTTAACTTTGTCATAATCCA TGATTATCTTTAAATTGTTCCATTGAGTCGAGTTAGTCAATGTTGAATTCTTAATTCTAGCTATATTTTTTCATTTTAATGTTCACTTAA TGGGACATTTTATGTACACCAATCTTGAAGAATGGCTCTCATAGTATATTTCAGACTAAGCCTCATTTTTGATATCACAATACATTTGCT >2881_2881_1_AGGF1-AP3B1_AGGF1_chr5_76335544_ENST00000312916_AP3B1_chr5_77412058_ENST00000255194_length(amino acids)=760AA_BP=322 MPARSSGLWVRRRPFRPERSPSAATSSLAPELMASEAPSPPRSPPPPTSPEPELAQLRRKVEKLERELRSCKRQVREIEKLLHHTERLYQ NAESNNQELRTQVEELSKILQRGRNEDNKKSDVEVQTENHAPWSISDYFYQTYYNDVSLPNKVTELSDQQDQAIETSILNSKDHLQVEND AYPGTDRTENVKYRQVDHFASNSQEPASALATEDTSLEGSSLAESLRAAAEAAVSQTGFSYDENTGLYFDHSTGFYYDSENQLYYDPSTG IYYYCDVESGRYQFHSRVDLQPYPTSSTKQSKDKKLKKKRKDPDSSATNEEKAKEWTPAGKAKQENSAKKFYSESEEEEDSSDSSSDSES ESGSESGEQGESGEEGDSNEDSSEDSSSEQDSESGRESGLENKRTAKRNSKAKGKSDSEDGEKENEKSKTSDSSNDESSSIEDSSSDSES ESEPESESESRRVTKEKEKKTKQDRTPLTKDVSLLDLDDFNPVSTPVALPTPALSPSLMADLEGLHLSTSSSVISVSTPAFVPTKTHVLL HRMSGKGLAAHYFFPRQPCIFGDKMVSIQITLNNTTDRKIENIHIGEKKLPIGMKMHVFNPIDSLEPEGSITVSMGIDFCDSTQTASFQL CTKDDCFNVNIQPPVGELLLPVAMSEKDFKKEQGVLTGMNETSAVIIAAPQNFTPSVIFQKVVNVANVGAVPSGQDNIHRFAAKTVHSGS -------------------------------------------------------------- >2881_2881_2_AGGF1-AP3B1_AGGF1_chr5_76335544_ENST00000312916_AP3B1_chr5_77412058_ENST00000519295_length(transcript)=3145nt_BP=1252nt GGTCCCGCCTGCCGCTGGCGCCGTTGTTTCCGGCTCAACTGGGGAGCTGCTGGAGCTCTTCTGGCCTCTGGTTTTCCGACTGCTTATCCG ACGCTCCTCCCTCTGTCTCTGTAGCTGGAGAAGGTAGTTTCCAGGAAAGTTTTCCGGTTTGCAGGCCGCGCACATCGGGCAGGGGCCATC CTCGGTCCCCTTGCTCGTTGCTCGCAGCCCCGTTCGGCTACAAGTGAGTTTCAGGGCGTCATGGCCAGGGGCCACCGCGGCCAGCCGGGT GTGAGGCTGCCTTTCGCTGCCCGCGCGCTCCAGTGGTCTCTGGGTCCGCCGGCGTCCGTTTCGGCCTGAACGCAGCCCCTCCGCGGCGAC GAGCAGTCTCGCGCCGGAGCTCATGGCCTCGGAGGCGCCGTCCCCGCCGCGGTCGCCGCCGCCGCCCACCTCCCCCGAGCCTGAGCTGGC CCAGCTAAGGCGGAAGGTGGAGAAGTTGGAACGTGAACTGCGGAGCTGCAAGCGGCAGGTGCGGGAGATCGAGAAGCTGCTGCATCACAC AGAACGGCTGTACCAGAACGCAGAAAGCAACAACCAGGAGCTCCGCACGCAGGTGGAAGAACTCAGTAAAATACTCCAACGTGGGAGAAA TGAAGATAATAAAAAGTCTGATGTAGAAGTACAAACAGAGAACCATGCTCCTTGGTCAATCTCAGATTATTTTTATCAGACGTACTACAA TGACGTTAGTCTTCCAAATAAAGTGACTGAACTGTCAGATCAACAAGATCAAGCTATCGAAACTTCTATTTTGAATTCTAAAGACCATTT ACAAGTAGAAAATGATGCTTACCCTGGTACCGATAGAACAGAAAATGTTAAATATAGACAAGTGGACCATTTTGCCTCAAATTCACAGGA GCCAGCATCTGCATTAGCAACAGAAGATACCTCCTTAGAAGGCTCATCATTAGCTGAAAGTTTGAGAGCTGCAGCAGAAGCGGCTGTATC ACAGACTGGATTTAGTTATGATGAAAATACTGGACTGTATTTTGACCACAGCACTGGTTTCTATTATGATTCTGAAAATCAACTCTATTA TGATCCTTCCACTGGAATTTATTACTATTGTGATGTGGAAAGTGGTCGTTATCAGTTTCATTCTCGAGTAGATTTGCAACCTTATCCGAC TTCTAGCACAAAACAAAGTAAAGATAAAAAATTGAAGAAGAAAAGAAAAGATCCAGATTCTTCTGCAACAAATGAGGAAAAGGCAAAAGA ATGGACCCCAGCAGGAAAAGCAAAGCAAGAGAATTCTGCTAAGAAGTTTTATTCTGAATCTGAGGAAGAGGAGGACTCTTCTGATAGTAG CAGTGACAGTGAGAGTGAATCTGGAAGTGAAAGTGGAGAACAAGGCGAAAGTGGGGAGGAAGGAGACAGCAATGAGGACAGCAGTGAGGA CTCCTCCAGTGAGCAGGACAGTGAGAGTGGACGGGAGTCAGGCCTAGAAAACAAAAGAACAGCCAAGAGGAACTCAAAAGCCAAAGGAAA AAGTGATTCTGAAGATGGGGAGAAGGAAAATGAAAAATCTAAAACTTCAGATTCTTCAAATGACGAATCTAGTTCAATAGAAGACAGTTC TTCCGATTCTGAATCAGAGTCAGAACCTGAAAGTGAATCTGAATCCAGAAGAGTCACTAAGGAGAAAGAAAAGAAAACAAAGCAAGATAG AACTCCTCTTACCAAAGATGTTTCACTTCTAGATCTGGATGATTTTAACCCAGTATCCACTCCAGTTGCACTTCCCACACCAGCTCTTTC TCCAAGTTTGATGGCTGATCTTGAAGGTTTACACTTGTCAACTTCCTCTTCAGTCATCAGTGTCAGTACTCCTGCATTTGTACCAACGAA AACTCACGTGCTGCTTCATCGAATGAGTGGAAAAGGACTAGCTGCCCATTATTTCTTTCCAAGACAGCCTTGCATTTTTGGTGATAAGAT GGTCTCTATACAAATAACACTGAATAACACTACTGATCGAAAGATAGAAAATATCCACATAGGGGAAAAAAAACTTCCTATAGGCATGAA AATGCATGTTTTTAATCCAATAGACTCTCTTGAGCCTGAGGGATCCATTACAGTTTCAATGGGTATTGACTTTTGTGATTCTACTCAGAC TGCCAGTTTCCAGTTGTGTACCAAGGATGATTGCTTCAATGTTAATATTCAGCCACCTGTTGGAGAACTGCTTTTACCTGTGGCCATGTC AGAGAAAGATTTTAAGAAAGAGCAAGGAGTGCTAACAGGAATGAATGAAACTTCTGCTGTAATCATTGCTGCACCACAGAATTTCACTCC CTCTGTGATCTTTCAGAAGGTTGTAAATGTAGCCAATGTAGGTGCAGTCCCTTCTGGCCAGGATAATATACACAGGTTTGCAGCTAAAAC TGTGCACAGTGGGTCATTGATGCTAGTCACAGTGGAACTGAAGGAAGGCTCTACAGCCCAGCTTATCATAAACACTGAGAAAACTGTGAT TGGCTCTGTTCTGCTGCGGGAACTGAAGCCTGTCCTGTCTCAGGGGTAACCTGCTTACATCTGGACTTTAGAATCTGGCACACAACAAAA GTGCCTGGCATCCACTACTGCTGCCTTTCATTTATAATAATAGCCCTTCCATCTGGCAGTGGGGGTAGAATACACTCTTGACATTCTTGT CTCCTGCTTTAGAATGCTAGTGTGTATCTATCATGTATGCAATACTTTCCCCCTTTTTGCTTTGCTAACCAAAGAGCATATATTTTACTG TCAGTTGTCTCAACTCTTGAATCCATGTGGCGTTTTCTCTGTCCTGCTGCTTCTTTTGGCCTCCTCGTTTTCCTTCTCTTTTTCGACAAT GGTAGACATGAATGAGATATTTAAAGTTCATTGGAAATCTTCTTCCCTACAGCAGTAAGCAAAAATTAGCAAAGAGATAGTCTAAATGGC CTCTCAGCTTGGTATGTGAAAATGAGATCACATACTTTTTAAATCCAAATACAAAAGCATAGTCTCTGCAAGATTTTGTTCTTTGAATTT >2881_2881_2_AGGF1-AP3B1_AGGF1_chr5_76335544_ENST00000312916_AP3B1_chr5_77412058_ENST00000519295_length(amino acids)=760AA_BP=322 MPARSSGLWVRRRPFRPERSPSAATSSLAPELMASEAPSPPRSPPPPTSPEPELAQLRRKVEKLERELRSCKRQVREIEKLLHHTERLYQ NAESNNQELRTQVEELSKILQRGRNEDNKKSDVEVQTENHAPWSISDYFYQTYYNDVSLPNKVTELSDQQDQAIETSILNSKDHLQVEND AYPGTDRTENVKYRQVDHFASNSQEPASALATEDTSLEGSSLAESLRAAAEAAVSQTGFSYDENTGLYFDHSTGFYYDSENQLYYDPSTG IYYYCDVESGRYQFHSRVDLQPYPTSSTKQSKDKKLKKKRKDPDSSATNEEKAKEWTPAGKAKQENSAKKFYSESEEEEDSSDSSSDSES ESGSESGEQGESGEEGDSNEDSSEDSSSEQDSESGRESGLENKRTAKRNSKAKGKSDSEDGEKENEKSKTSDSSNDESSSIEDSSSDSES ESEPESESESRRVTKEKEKKTKQDRTPLTKDVSLLDLDDFNPVSTPVALPTPALSPSLMADLEGLHLSTSSSVISVSTPAFVPTKTHVLL HRMSGKGLAAHYFFPRQPCIFGDKMVSIQITLNNTTDRKIENIHIGEKKLPIGMKMHVFNPIDSLEPEGSITVSMGIDFCDSTQTASFQL CTKDDCFNVNIQPPVGELLLPVAMSEKDFKKEQGVLTGMNETSAVIIAAPQNFTPSVIFQKVVNVANVGAVPSGQDNIHRFAAKTVHSGS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for AGGF1-AP3B1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for AGGF1-AP3B1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for AGGF1-AP3B1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
