
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FAM20C-MKRN1 (FusionGDB2 ID:28892) |
Fusion Gene Summary for FAM20C-MKRN1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FAM20C-MKRN1 | Fusion gene ID: 28892 | Hgene | Tgene | Gene symbol | FAM20C | MKRN1 | Gene ID | 56975 | 23608 |
| Gene name | FAM20C golgi associated secretory pathway kinase | makorin ring finger protein 1 | |
| Synonyms | DMP-4|DMP4|G-CK|GEF-CK|RNS | RNF61 | |
| Cytomap | 7p22.3 | 7q34 | |
| Type of gene | protein-coding | protein-coding | |
| Description | extracellular serine/threonine protein kinase FAM20CGolgi casein kinasedentin matrix protein 4family with sequence similarity 20, member Cgolgi-enriched fraction casein kinase | E3 ubiquitin-protein ligase makorin-1RING finger protein 61RING-type E3 ubiquitin transferase makorin-1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8IXL6 | Q9UHC7 | |
| Ensembl transtripts involved in fusion gene | ENST00000313766, ENST00000471328, | ENST00000437223, ENST00000443720, ENST00000474576, ENST00000480552, ENST00000481705, ENST00000255977, | |
| Fusion gene scores | * DoF score | 4 X 1 X 4=16 | 5 X 4 X 3=60 |
| # samples | 4 | 5 | |
| ** MAII score | log2(4/16*10)=1.32192809488736 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(5/60*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FAM20C [Title/Abstract] AND MKRN1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FAM20C(208976)-MKRN1(140171811), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | FAM20C | GO:0006468 | protein phosphorylation | 23754375|25789606|26091039 |
| Tgene | MKRN1 | GO:0000209 | protein polyubiquitination | 19536131 |
 Fusion gene breakpoints across FAM20C (5'-gene) Fusion gene breakpoints across FAM20C (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
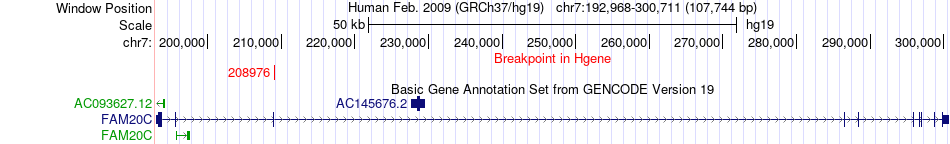 |
 Fusion gene breakpoints across MKRN1 (3'-gene) Fusion gene breakpoints across MKRN1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
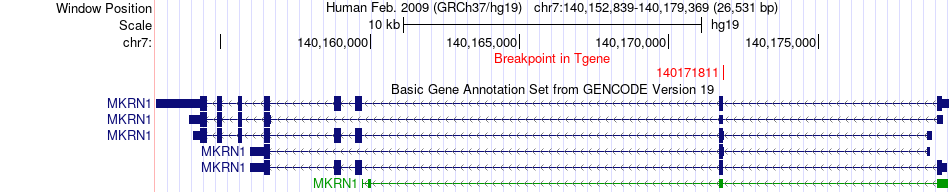 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUSC | TCGA-56-7223-01A | FAM20C | chr7 | 208976 | + | MKRN1 | chr7 | 140171811 | - |
Top |
Fusion Gene ORF analysis for FAM20C-MKRN1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000313766 | ENST00000437223 | FAM20C | chr7 | 208976 | + | MKRN1 | chr7 | 140171811 | - |
| 5CDS-5UTR | ENST00000313766 | ENST00000443720 | FAM20C | chr7 | 208976 | + | MKRN1 | chr7 | 140171811 | - |
| 5CDS-5UTR | ENST00000313766 | ENST00000474576 | FAM20C | chr7 | 208976 | + | MKRN1 | chr7 | 140171811 | - |
| 5CDS-5UTR | ENST00000313766 | ENST00000480552 | FAM20C | chr7 | 208976 | + | MKRN1 | chr7 | 140171811 | - |
| 5CDS-5UTR | ENST00000313766 | ENST00000481705 | FAM20C | chr7 | 208976 | + | MKRN1 | chr7 | 140171811 | - |
| In-frame | ENST00000313766 | ENST00000255977 | FAM20C | chr7 | 208976 | + | MKRN1 | chr7 | 140171811 | - |
| intron-3CDS | ENST00000471328 | ENST00000255977 | FAM20C | chr7 | 208976 | + | MKRN1 | chr7 | 140171811 | - |
| intron-5UTR | ENST00000471328 | ENST00000437223 | FAM20C | chr7 | 208976 | + | MKRN1 | chr7 | 140171811 | - |
| intron-5UTR | ENST00000471328 | ENST00000443720 | FAM20C | chr7 | 208976 | + | MKRN1 | chr7 | 140171811 | - |
| intron-5UTR | ENST00000471328 | ENST00000474576 | FAM20C | chr7 | 208976 | + | MKRN1 | chr7 | 140171811 | - |
| intron-5UTR | ENST00000471328 | ENST00000480552 | FAM20C | chr7 | 208976 | + | MKRN1 | chr7 | 140171811 | - |
| intron-5UTR | ENST00000471328 | ENST00000481705 | FAM20C | chr7 | 208976 | + | MKRN1 | chr7 | 140171811 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000313766 | FAM20C | chr7 | 208976 | + | ENST00000255977 | MKRN1 | chr7 | 140171811 | - | 3835 | 1094 | 171 | 2357 | 728 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000313766 | ENST00000255977 | FAM20C | chr7 | 208976 | + | MKRN1 | chr7 | 140171811 | - | 0.001502893 | 0.9984971 |
Top |
Fusion Genomic Features for FAM20C-MKRN1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
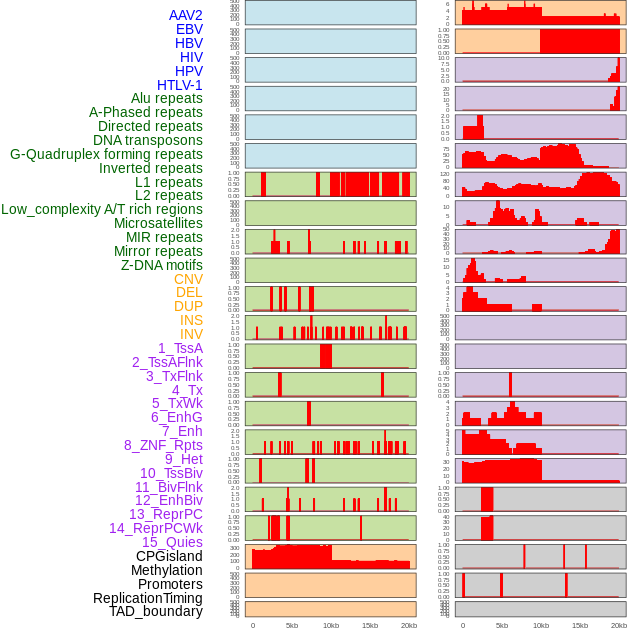 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for FAM20C-MKRN1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:208976/chr7:140171811) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FAM20C | MKRN1 |
| FUNCTION: Golgi serine/threonine protein kinase that phosphorylates secretory pathway proteins within Ser-x-Glu/pSer motifs and plays a key role in biomineralization of bones and teeth (PubMed:22582013, PubMed:23754375, PubMed:25789606). Constitutes the main protein kinase for extracellular proteins, generating the majority of the extracellular phosphoproteome (PubMed:26091039). Mainly phosphorylates proteins within the Ser-x-Glu/pSer motif, but also displays a broader substrate specificity (PubMed:26091039). Phosphorylates casein as well as a number of proteins involved in biomineralization such as AMELX, AMTN, ENAM and SPP1 (PubMed:22582013, PubMed:25789606). In addition to its role in biomineralization, also plays a role in lipid homeostasis, wound healing and cell migration and adhesion (PubMed:26091039). {ECO:0000269|PubMed:22582013, ECO:0000269|PubMed:23754375, ECO:0000269|PubMed:25789606, ECO:0000269|PubMed:26091039}. | FUNCTION: E3 ubiquitin ligase catalyzing the covalent attachment of ubiquitin moieties onto substrate proteins. These substrates include FILIP1, p53/TP53, CDKN1A and TERT. Keeps cells alive by suppressing p53/TP53 under normal conditions, but stimulates apoptosis by repressing CDKN1A under stress conditions. Acts as a negative regulator of telomerase. Has negative and positive effects on RNA polymerase II-dependent transcription. {ECO:0000269|PubMed:16785614, ECO:0000269|PubMed:19536131}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000255977 | 0 | 8 | 236_263 | 61 | 483.0 | Region | Note=Makorin-type Cys-His | |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000443720 | 0 | 5 | 236_263 | 61 | 330.0 | Region | Note=Makorin-type Cys-His | |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000474576 | 0 | 8 | 236_263 | 0 | 419.0 | Region | Note=Makorin-type Cys-His | |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000255977 | 0 | 8 | 208_235 | 61 | 483.0 | Zinc finger | C3H1-type 3 | |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000255977 | 0 | 8 | 281_335 | 61 | 483.0 | Zinc finger | RING-type | |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000255977 | 0 | 8 | 364_393 | 61 | 483.0 | Zinc finger | C3H1-type 4 | |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000255977 | 0 | 8 | 84_111 | 61 | 483.0 | Zinc finger | C3H1-type 2 | |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000443720 | 0 | 5 | 208_235 | 61 | 330.0 | Zinc finger | C3H1-type 3 | |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000443720 | 0 | 5 | 281_335 | 61 | 330.0 | Zinc finger | RING-type | |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000443720 | 0 | 5 | 364_393 | 61 | 330.0 | Zinc finger | C3H1-type 4 | |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000443720 | 0 | 5 | 84_111 | 61 | 330.0 | Zinc finger | C3H1-type 2 | |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000474576 | 0 | 8 | 208_235 | 0 | 419.0 | Zinc finger | C3H1-type 3 | |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000474576 | 0 | 8 | 281_335 | 0 | 419.0 | Zinc finger | RING-type | |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000474576 | 0 | 8 | 364_393 | 0 | 419.0 | Zinc finger | C3H1-type 4 | |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000474576 | 0 | 8 | 55_82 | 0 | 419.0 | Zinc finger | C3H1-type 1 | |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000474576 | 0 | 8 | 84_111 | 0 | 419.0 | Zinc finger | C3H1-type 2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FAM20C | chr7:208976 | chr7:140171811 | ENST00000313766 | + | 3 | 10 | 389_392 | 287 | 585.0 | Nucleotide binding | ATP |
| Hgene | FAM20C | chr7:208976 | chr7:140171811 | ENST00000313766 | + | 3 | 10 | 354_565 | 287 | 585.0 | Region | Kinase domain |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000255977 | 0 | 8 | 55_82 | 61 | 483.0 | Zinc finger | C3H1-type 1 | |
| Tgene | MKRN1 | chr7:208976 | chr7:140171811 | ENST00000443720 | 0 | 5 | 55_82 | 61 | 330.0 | Zinc finger | C3H1-type 1 |
Top |
Fusion Gene Sequence for FAM20C-MKRN1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >28892_28892_1_FAM20C-MKRN1_FAM20C_chr7_208976_ENST00000313766_MKRN1_chr7_140171811_ENST00000255977_length(transcript)=3835nt_BP=1094nt GCGGCCGCTCCGGAGAGGCCGAGCGGGGACGGGCCCGAGATGGCGCGGGGACCCAGCGCTCCGGCGGCGGGCGCCCTGCACGCGGCCCGG GCCCGGGGACAGCCCCGGAGCTGGTAGCCGCCCGGCACCGATGGACCTTGACCCGCGAGGCGGCGCCGCGCTCGTGCCCAGCTGCAGCTA GAGGGGCGCGCGGGCAGAACGCGCTCCAGGCCCGGGCCGGCCCGCGCGGCCATGAAGATGATGCTGGTGCGCCGGTTCCGCGTGCTCATC CTGATGGTGTTCCTGGTGGCCTGCGCGCTGCACATCGCCCTGGACCTGCTGCCCAGGCTGGAGCGACGCGGCGCGCGGCCCTCGGGGGAG CCCGGCTGTTCGTGCGCGCAGCCCGCCGCCGAGGTGGCCGCGCCCGGCTGGGCCCAGGTTCGGGGCCGCCCCGGGGAGCCCCCGGCCGCC TCCTCCGCCGCCGGCGACGCGGGCTGGCCCAACAAGCACACGCTCCGCATCCTGCAGGACTTCAGCTCCGACCCCTCCTCCAACCTCTCG TCCCACTCGCTGGAGAAACTGCCGCCCGCGGCCGAGCCGGCCGAGCGCGCCTTGCGGGGGCGGGATCCCGGCGCCCTAAGACCCCACGAC CCCGCGCACCGGCCGCTGCTGCGAGACCCCGGCCCGCGTCGGTCCGAGTCGCCCCCCGGCCCCGGCGGAGACGCCTCCCTCCTGGCCAGG CTGTTCGAGCACCCGCTTTACCGGGTGGCGGTTCCGCCGCTCACGGAGGAGGACGTCCTGTTCAATGTGAACAGCGACACCAGGCTCAGC CCCAAAGCGGCGGAGAACCCGGACTGGCCGCATGCGGGTGCTGAAGGTGCAGAATTCCTCTCCCCCGGGGAGGCGGCCGTGGACTCCTAT CCCAACTGGCTCAAGTTCCACATTGGTATCAACCGGTACGAGCTGTACTCCAGACACAACCCGGCCATCGAGGCCCTGCTGCACGACCTC AGCTCCCAGAGGATCACCAGCGTGGCCATGAAGTCGGGGGGCACGCAGCTGAAGCTCATCATGACCTTCCAGAATTACGGGCAAGCGCTG TTCAAACCCATGAAGTATTTTATGCATGGGGTTTGTAAGGAAGGAGACAACTGTCGCTACTCGCATGACCTCTCTGACAGTCCGTATAGT GTAGTGTGCAAGTATTTTCAGCGAGGGTACTGTATTTATGGAGACCGCTGCAGATATGAACATAGCAAACCATTGAAACAGGAAGAAGCA ACTGCTACAGAGCTAACTACAAAGTCATCCCTTGCTGCTTCCTCAAGTCTCTCATCGATAGTTGGACCACTTGTTGAAATGAATACAGGC GAAGCTGAGTCAAGAAATTCAAACTTTGCAACTGTAGGAGCAGGTTCAGAGGACTGGGTGAATGCTATTGAGTTTGTTCCTGGGCAACCC TACTGTGGCCGTACTGCGCCTTCCTGCACTGAAGCACCCCTGCAGGGCTCAGTGACCAAGGAAGAATCAGAGAAAGAGCAAACCGCCGTG GAGACAAAGAAGCAGCTGTGCCCCTATGCTGCAGTGGGAGAGTGCCGATACGGGGAGAACTGTGTGTATCTCCACGGAGATTCTTGTGAC ATGTGTGGGCTGCAGGTCCTGCATCCAATGGATGCTGCCCAGAGATCGCAGCATATCAAATCGTGCATTGAGGCCCATGAGAAGGACATG GAGCTCTCATTTGCCGTGCAGCGCAGCAAGGACATGGTGTGTGGGATCTGCATGGAGGTGGTCTATGAGAAAGCCAACCCCAGTGAGCGC CGCTTCGGGATCCTCTCCAACTGCAACCACACCTACTGTCTCAAGTGCATTCGCAAGTGGAGGAGTGCTAAGCAATTTGAGAGCAAGATC ATAAAGTCCTGCCCAGAATGCCGGATCACATCTAACTTTGTCATTCCAAGTGAGTACTGGGTGGAGGAGAAAGAAGAGAAGCAGAAACTC ATTCTGAAATACAAGGAGGCAATGAGCAACAAGGCGTGCAGGTATTTTGATGAAGGACGTGGGAGCTGCCCATTTGGAGGGAACTGTTTT TACAAGCATGCGTACCCTGATGGCCGTAGAGAGGAGCCACAGAGACAGAAAGTGGGAACATCAAGCAGATACCGGGCCCAACGAAGGAAC CACTTCTGGGAACTCATTGAGGAAAGAGAGAACAGCAACCCCTTTGACAACGATGAAGAAGAGGTTGTCACCTTTGAGCTGGGCGAGATG TTGCTTATGCTTTTGGCTGCAGGTGGGGACGACGAACTAACAGACTCTGAAGATGAGTGGGACTTGTTTCATGATGAGCTGGAAGATTTT TATGACTTGGATCTATAGCAACCTTGCGTGGCGTGTGAACTGGTCTGCTGACCTCAGACAGCAGCTGTCCCCTGTGGTGGTGTGGCAGTG CCTGTGTTCTCTCCTAGGCAGGCCTCTCAACTCCAGGTGCTGTCCTAAGAATTTTTACCCAGGGCCTGTCTTCTCAACCCCTCACCTTTC CCTGAGGAGTGTGTTGTTTTCCCTGTTGAAAAAAGTTACAAAAATAAATCTTAAAGTTAGTTTTTTGTAACACGAATTTAACTGTCAGAC AGTTAGTGTAGGTGTGTTGCGTCATCTGTTTTCAACCAGATTGCATTTATGGACTTTTCACACACTCATTTTGAGGACCCCAGGTTCAAA AGTAAAAGCAGTGGCCCTGCTTTGGGGTCCAAGAATAGGAGTGATGGGTGAAGGGACCTAAGCTGGCCAATAGCCCTCTGCCCCAGACAT GGGATGTGGATCCTTGAGGTTTCTGGTGAAATCTGCACATCTGTGTTTTTATATCTGTTCCCTACCCTGTAATCCCTACCACGTGCACTT GTTCTGTGGTTTTGGTCTCTTGTTTAATTGCACACAAGTAATACTACTGGGTAACCAGAATCAGGTGTGAATGTGTTGAGATTTTTTACT GTTTTGCATGATAGGAAAATTGAGAAAGAATACGTATAAAAGATAGAGAGGCATAACATCAATGCAGAGTTGGAAGTTGGCTCCCAAGGG CTGACATGGTGTGAGTGTGTGGGTGTGTGATAAGCTTCTCATCCCTGCATAGATGCAGTATTCTTAGCCTTAGTAGAAAAACCTGGTTTA GTGGTTTAAGCCTTGTGTGGCAGATAGATCTTAAAGGGCAAAGCAGTATATTGGTAGTTGTCAATATAGCAGTGCTAGCTCTGTCTATAT AAATAGAGAAATGGGGTTAGCCATAGAGGTTAAAACTACCTGGTTATCCCATATAATAACACAAACTGGGTCTTGGATACACAGTTGTAT TTAATGTTTTACGATCTAGCCTTTCCAGTACAGGCACTTTCTGAGAAACCTTTGTCCTCACTTGAGGCATTTTGTTGTCGGGTTTTTGTG TTTGTTTTTGTGGGTATTTGCCTCATTCCACCCCTGAGCTTTCAGGTAGACAGACGTGATTCAAAACTCTGTTCTAAGGTGTTTATTGTA GTGGAGTAATGGGTTTGCAGTGATAAGTCATACTTTTCCACCGAAAGGGAGGGCTTGGGAATCCCTGAGATTAGCTAAAGTTAAGTTGTT GGAAGAATTCCTTGATTGGAAATTGTACCTTTGTGTTTTGTTGCTCTGTTTCCTGAAAATAACTCGGGGATGCTCCTGGTTTGTCCATCT ACTGCTTTGATTCCTTGGATCCCACCCATTCTTTCACTTTAAGAAAAAACAAATAATTGTTGCAGAGGTCTCTGTATTTTGCAGCTGCCC >28892_28892_1_FAM20C-MKRN1_FAM20C_chr7_208976_ENST00000313766_MKRN1_chr7_140171811_ENST00000255977_length(amino acids)=728AA_BP=308 MQLEGRAGRTRSRPGPARAAMKMMLVRRFRVLILMVFLVACALHIALDLLPRLERRGARPSGEPGCSCAQPAAEVAAPGWAQVRGRPGEP PAASSAAGDAGWPNKHTLRILQDFSSDPSSNLSSHSLEKLPPAAEPAERALRGRDPGALRPHDPAHRPLLRDPGPRRSESPPGPGGDASL LARLFEHPLYRVAVPPLTEEDVLFNVNSDTRLSPKAAENPDWPHAGAEGAEFLSPGEAAVDSYPNWLKFHIGINRYELYSRHNPAIEALL HDLSSQRITSVAMKSGGTQLKLIMTFQNYGQALFKPMKYFMHGVCKEGDNCRYSHDLSDSPYSVVCKYFQRGYCIYGDRCRYEHSKPLKQ EEATATELTTKSSLAASSSLSSIVGPLVEMNTGEAESRNSNFATVGAGSEDWVNAIEFVPGQPYCGRTAPSCTEAPLQGSVTKEESEKEQ TAVETKKQLCPYAAVGECRYGENCVYLHGDSCDMCGLQVLHPMDAAQRSQHIKSCIEAHEKDMELSFAVQRSKDMVCGICMEVVYEKANP SERRFGILSNCNHTYCLKCIRKWRSAKQFESKIIKSCPECRITSNFVIPSEYWVEEKEEKQKLILKYKEAMSNKACRYFDEGRGSCPFGG NCFYKHAYPDGRREEPQRQKVGTSSRYRAQRRNHFWELIEERENSNPFDNDEEEVVTFELGEMLLMLLAAGGDDELTDSEDEWDLFHDEL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FAM20C-MKRN1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FAM20C-MKRN1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FAM20C-MKRN1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
