
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FAM53B-KCNN1 (FusionGDB2 ID:29097) |
Fusion Gene Summary for FAM53B-KCNN1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FAM53B-KCNN1 | Fusion gene ID: 29097 | Hgene | Tgene | Gene symbol | FAM53B | KCNN1 | Gene ID | 9679 | 3780 |
| Gene name | family with sequence similarity 53 member B | potassium calcium-activated channel subfamily N member 1 | |
| Synonyms | KIAA0140|bA12J10.2|smp | KCa2.1|SK1|SKCA1|hSK1 | |
| Cytomap | 10q26.13 | 19p13.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein FAM53Bprotein simpletsimplet homolog | small conductance calcium-activated potassium channel protein 1potassium channel, calcium activated intermediate/small conductance subfamily N alpha, member 1potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1small | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q14153 | Q92952 | |
| Ensembl transtripts involved in fusion gene | ENST00000280780, ENST00000337318, ENST00000392754, | ENST00000594192, ENST00000222249, | |
| Fusion gene scores | * DoF score | 9 X 5 X 6=270 | 4 X 3 X 4=48 |
| # samples | 11 | 4 | |
| ** MAII score | log2(11/270*10)=-1.29545588352617 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/48*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FAM53B [Title/Abstract] AND KCNN1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FAM53B(126370176)-KCNN1(18099224), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | FAM53B-KCNN1 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. FAM53B-KCNN1 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across FAM53B (5'-gene) Fusion gene breakpoints across FAM53B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
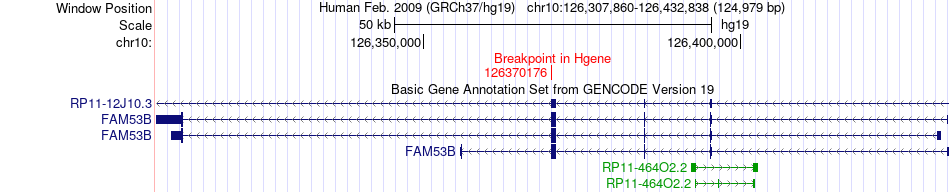 |
 Fusion gene breakpoints across KCNN1 (3'-gene) Fusion gene breakpoints across KCNN1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
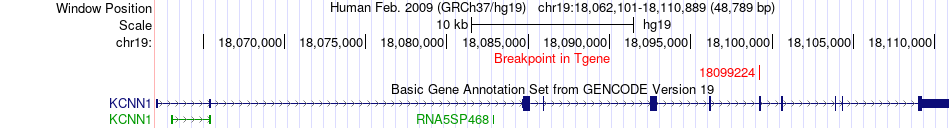 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-44-7670-01A | FAM53B | chr10 | 126370176 | - | KCNN1 | chr19 | 18099224 | + |
Top |
Fusion Gene ORF analysis for FAM53B-KCNN1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000280780 | ENST00000594192 | FAM53B | chr10 | 126370176 | - | KCNN1 | chr19 | 18099224 | + |
| 5CDS-intron | ENST00000337318 | ENST00000594192 | FAM53B | chr10 | 126370176 | - | KCNN1 | chr19 | 18099224 | + |
| 5CDS-intron | ENST00000392754 | ENST00000594192 | FAM53B | chr10 | 126370176 | - | KCNN1 | chr19 | 18099224 | + |
| Frame-shift | ENST00000392754 | ENST00000222249 | FAM53B | chr10 | 126370176 | - | KCNN1 | chr19 | 18099224 | + |
| In-frame | ENST00000280780 | ENST00000222249 | FAM53B | chr10 | 126370176 | - | KCNN1 | chr19 | 18099224 | + |
| In-frame | ENST00000337318 | ENST00000222249 | FAM53B | chr10 | 126370176 | - | KCNN1 | chr19 | 18099224 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000337318 | FAM53B | chr10 | 126370176 | - | ENST00000222249 | KCNN1 | chr19 | 18099224 | + | 3365 | 1118 | 167 | 1690 | 507 |
| ENST00000280780 | FAM53B | chr10 | 126370176 | - | ENST00000222249 | KCNN1 | chr19 | 18099224 | + | 3584 | 1337 | 386 | 1909 | 507 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000337318 | ENST00000222249 | FAM53B | chr10 | 126370176 | - | KCNN1 | chr19 | 18099224 | + | 0.14733042 | 0.8526696 |
| ENST00000280780 | ENST00000222249 | FAM53B | chr10 | 126370176 | - | KCNN1 | chr19 | 18099224 | + | 0.13644525 | 0.8635548 |
Top |
Fusion Genomic Features for FAM53B-KCNN1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| FAM53B | chr10 | 126370175 | - | KCNN1 | chr19 | 18099223 | + | 0.001672572 | 0.99832743 |
| FAM53B | chr10 | 126370175 | - | KCNN1 | chr19 | 18099223 | + | 0.001672572 | 0.99832743 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
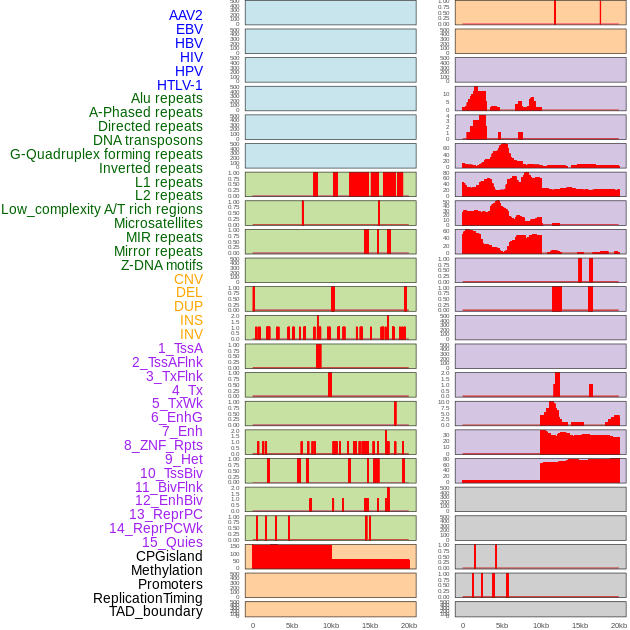 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
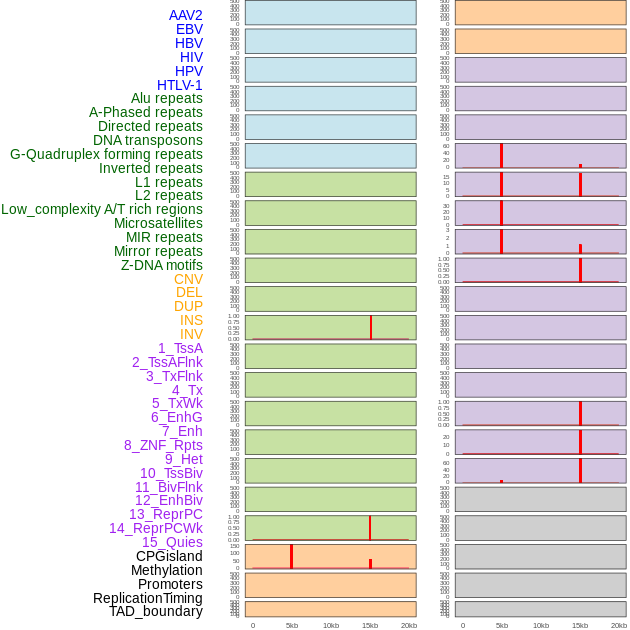 |
Top |
Fusion Protein Features for FAM53B-KCNN1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:126370176/chr19:18099224) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FAM53B | KCNN1 |
| FUNCTION: Acts as a regulator of Wnt signaling pathway by regulating beta-catenin (CTNNB1) nuclear localization. {ECO:0000269|PubMed:25183871}. | FUNCTION: Forms a voltage-independent potassium channel activated by intracellular calcium. Activation is followed by membrane hyperpolarization. Thought to regulate neuronal excitability by contributing to the slow component of synaptic afterhyperpolarization. The channel is blocked by apamin (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FAM53B | chr10:126370176 | chr19:18099224 | ENST00000280780 | - | 4 | 5 | 281_284 | 302 | 306.0 | Motif | Nuclear localization signal |
| Hgene | FAM53B | chr10:126370176 | chr19:18099224 | ENST00000337318 | - | 4 | 5 | 281_284 | 302 | 423.0 | Motif | Nuclear localization signal |
| Hgene | FAM53B | chr10:126370176 | chr19:18099224 | ENST00000392754 | - | 4 | 5 | 281_284 | 302 | 423.0 | Motif | Nuclear localization signal |
| Tgene | KCNN1 | chr10:126370176 | chr19:18099224 | ENST00000222249 | 5 | 11 | 509_518 | 353 | 544.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | KCNN1 | chr10:126370176 | chr19:18099224 | ENST00000222249 | 5 | 11 | 384_463 | 353 | 544.0 | Region | Calmodulin-binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | KCNN1 | chr10:126370176 | chr19:18099224 | ENST00000222249 | 5 | 11 | 317_337 | 353 | 544.0 | Intramembrane | Pore-forming%3B Name%3DSegment H5 | |
| Tgene | KCNN1 | chr10:126370176 | chr19:18099224 | ENST00000222249 | 5 | 11 | 111_131 | 353 | 544.0 | Transmembrane | Helical%3B Name%3DSegment S1 | |
| Tgene | KCNN1 | chr10:126370176 | chr19:18099224 | ENST00000222249 | 5 | 11 | 140_160 | 353 | 544.0 | Transmembrane | Helical%3B Name%3DSegment S2 | |
| Tgene | KCNN1 | chr10:126370176 | chr19:18099224 | ENST00000222249 | 5 | 11 | 179_199 | 353 | 544.0 | Transmembrane | Helical%3B Name%3DSegment S3 | |
| Tgene | KCNN1 | chr10:126370176 | chr19:18099224 | ENST00000222249 | 5 | 11 | 228_248 | 353 | 544.0 | Transmembrane | Helical%3B Name%3DSegment S4 | |
| Tgene | KCNN1 | chr10:126370176 | chr19:18099224 | ENST00000222249 | 5 | 11 | 277_297 | 353 | 544.0 | Transmembrane | Helical%3B Name%3DSegment S5 | |
| Tgene | KCNN1 | chr10:126370176 | chr19:18099224 | ENST00000222249 | 5 | 11 | 346_366 | 353 | 544.0 | Transmembrane | Helical%3B Name%3DSegment S6 |
Top |
Fusion Gene Sequence for FAM53B-KCNN1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >29097_29097_1_FAM53B-KCNN1_FAM53B_chr10_126370176_ENST00000280780_KCNN1_chr19_18099224_ENST00000222249_length(transcript)=3584nt_BP=1337nt CACGCAGCCGCGGTCTGGCGGCTCCGCGGCGGCGGCGGGTGCGGGCGGCCTGGCCGGTGCCGGTTAAAGGGACGAGTTGCAAACACTTCA GGAAGTGACAAGTCGATTTCCTCCTCCCCGGGAGTCGCTCGTACAAAGCGCTCGGCGCCGGCAGGCGAGCGTGCGCGCGGCGGACGCGCG GCGGGCACCCCGGACGACTTGGCGAGCGCTGGCGGTGACGGCGCGGGGTCCGCGCCCGGAGCGCCCCGCCGCGCACAGGAGTTGACCACA TTTGGCCATTTCCCAGAAGGGCCCCACCCCAAGGGTGAGTGGCCAATGGGGAGCTGTTTCTGCTGACATCAATTCCCCAGGAGGTACTCA CCCCAAGTCTGCCCAAGTGAAGATGGCTGATACCCACCCTGGGATGGAGCCCAGCGCCTGAGGCCCTTATCATGGTGATGGTCCTAAGTG AAAGCCTCAGCACCCGGGGAGCTGACTCCATTGCATGTGGGACCTTCAGCCGTGAACTGCACACGCCAAAGAAGATGAGTCAAGGACCTA CACTTTTCTCTTGTGGAATTATGGAAAATGACAGATGGCGAGACCTGGACAGGAAATGCCCTCTTCAGATTGACCAACCGAGCACCAGCA TCTGGGAATGCCTGCCTGAAAAGGACAGCTCACTATGGCACCGGGAGGCAGTGACCGCCTGCGCTGTGACCAGTCTGATCAAAGACCTCA GCATCAGCGACCACAACGGGAACCCCTCAGCACCCCCTAGCAAGCGCCAGTGCCGCTCACTGTCCTTCTCCGATGAGATGTCCAGTTGCC GGACATCATGGAGGCCCTTGGGCTCCAAAGTCTGGACTCCCGTGGAAAAGAGACGCTGCTACAGCGGGGGCAGCGTCCAGCGCTATTCCA ACGGCTTCAGCACCATGCAGAGGAGTTCCAGCTTCAGCCTCCCTTCCCGGGCCAACGTGCTCTCCTCACCCTGCGACCAGGCAGGACTCC ACCACCGATTTGGAGGGCAGCCCTGCCAAGGGGTGCCAGGCTCAGCCCCGTGTGGACAGGCAGGTGACACCTGGAGCCCTGACCTGCACC CCGTGGGAGGAGGCCGGCTGGACCTGCAGCGGTCCCTCTCTTGCTCACATGAGCAGTTTTCCTTTGTGGAATACTGTCCTCCCTCAGCCA ACAGCACACCTGCCTCAACACCAGAGCTGGCGAGACGCTCCAGCGGCCTTTCCCGCAGCCGCTCCCAGCCGTGTGTCCTTAACGACAAGA AGGTCGGTGTTAAAAGGCGGCGCCCTGAAGAAGTGCAAGAGCAGAGGCCTTCTCTAGACCTTGCCAAGATGGCACAGGGAGCTGGCTGTA CCGCGCTCGTGGTGGCTGTGGTGGCTCGGAAGCTGGAGCTCACCAAGGCTGAGAAGCACGTGCACAACTTCATGATGGACACTCAGCTCA CCAAGCGGGTAAAAAACGCCGCTGCTAACGTTCTCAGGGAGACGTGGCTCATCTACAAACATACCAGGCTGGTGAAGAAGCCAGACCAAG CCCGGGTTCGGAAACACCAGCGTAAGTTCCTCCAAGCCATCCATCAGGCTCAGAAGCTCCGGAGTGTGAAGATCGAGCAAGGGAAGCTGA ACGACCAGGCTAACACGCTTACCGACCTAGCCAAGACCCAGACCGTCATGTACGACCTTGTATCGGAGCTGCACGCTCAGCACGAGGAGC TGGAGGCCCGCCTGGCCACCCTGGAAAGCCGCTTGGATGCGCTGGGTGCCTCTCTACAGGCCCTGCCTGGCCTCATCGCCCAAGCCATAC GCCCACCCCCGCCTCCCCTGCCTCCCAGGCCCGGCCCCGGCCCCCAAGACCAGGCAGCCCGGAGCTCCCCCTGCCGGTGGACGCCCGTGG CCCCCTCGGACTGCGGGTGACGGCCCTGCCCGCCACCAGACCCCTAAATCTTGGCCATCGTGTGGCCGCCACCTCCGGGAAGCCTTGTAC AGTGGCGCCTCTTGGAGTTCAAGAAGCCAACGCTGAGTCAGGCTGAGTGGACTGAGGCCTGCCCCGCCCAGACTGCCCAGGCAGAGGGCA GGGCTGGACCATGGGTGAGGGCAGGGGAGCCCGGAGCTTCCTCTGGTCACCTGGTCCCCCGACTCTCCCCAGGCCCCCGGTGGGCATGGA GCAGCCCGGGGAGGGGTCCGTGCTGGTTCTGAATAAAGCAGGACCCGCCTAGTGGCTGCCTGTGTGCATGGCTGGAAGGCACTGGTGATG TCCCAGGAGGTAGACCTCCAGCCCTGGGTACCAAGATGAATGTGGGAATCAGAAAAACCTGTTCCCATCACCGGCCTAGCCTAGAATCCT AGCCTAGAAGCCCTCTCTCCCTCTGGGCTGGAGCTCAGTGAGGGACAACTCTCTAGGGACACCTGTACCAGCCCCACCTGGCGCTGAGAT CCCTCAGACAGCATGGCCCAGCCCTGGCCAGAAGCATCGCTCCCCTTCAACCAACCGCGTTGATGGACACCCACTGTGTGCCAGGCCCCA GCGGGGCCCATGGGGGAGGTGACCTGGGTGAGGAAGGTCATTTGGGTTTTTGTGAGATTTGTATTGAGCACCTGCTGTCTACAGAGAATG TGATGATGTCAATTACCACACTGATTCCTCATCAGAACTTGACCATGGTGGGATGGGGGGGAAACTGAGGCCCAGAGAGGTGAATCTACC TACCTGGGACCACACTGCGAGGAAGGATGGTGCATTCAAACTGGAAATCCCCTAGGCCAAAGTAAAGAACCGGACTGGCTGGGCACAGTG GCTCACACCTGTAATCCCAGCACTTTGGGAGGGTAAAGCGGGCGGATCACCTGAGGTCAGGAGTTCAAGACCAGCCTGGGCAACATGGTG AAACCCTGTCTCTACTAAAAATATAAAAATTAGCTGGGTGTGGTGGTGCACGCCTGTAATCCCAGCTACTTGGGAGGCTGAGGCAGGATA ATCGCTTGAACCCAGGAGGTGGAGGTTGCAGTGAGAAAAAACCTGGACTTGGTGGAAGGGACTCCATGGAGGAGCCATGGGGACGTCAGG AGGAGGGGTTATTTGGTGACATCCAGGGGTCAGAGAGACCCTTCAGGGGTGGGCTACAGGGGAGGGTTCCAGACACAAGTAGAATCAGAA AGGTCCCTGGAGGCCATGGGCTTGGGTGCTGGGAGCAACAGGCATTTACTTGGCAGATGCTGAGCCCTGGGTGGGGCGAGGAGGCCTGGC TCCTGGGGAGACGACGCTGTGCATAGCCGAGGAGCCCCAGGTCCTGGCAGCCTCTGATAAGGCCATCCATCGCCTGGCTCAACGAGATAA CTAGGCCGTGGCTGGTGCATGAATGACCAGCTTGGGCCCACGCACGAGAAGGGTATGAGGAGGTGCTGGTCAGACCCGGGTTCGAGTCCT GACTCTGCCACTGCTGGGTGACTCTGAGCAGTGGCTGCCCCTCTGGGGCCTTGGTTGTTTCATCTGTAAAATGGGGTGACAGTCTATTTA >29097_29097_1_FAM53B-KCNN1_FAM53B_chr10_126370176_ENST00000280780_KCNN1_chr19_18099224_ENST00000222249_length(amino acids)=507AA_BP=316 MIPTLGWSPAPEALIMVMVLSESLSTRGADSIACGTFSRELHTPKKMSQGPTLFSCGIMENDRWRDLDRKCPLQIDQPSTSIWECLPEKD SSLWHREAVTACAVTSLIKDLSISDHNGNPSAPPSKRQCRSLSFSDEMSSCRTSWRPLGSKVWTPVEKRRCYSGGSVQRYSNGFSTMQRS SSFSLPSRANVLSSPCDQAGLHHRFGGQPCQGVPGSAPCGQAGDTWSPDLHPVGGGRLDLQRSLSCSHEQFSFVEYCPPSANSTPASTPE LARRSSGLSRSRSQPCVLNDKKVGVKRRRPEEVQEQRPSLDLAKMAQGAGCTALVVAVVARKLELTKAEKHVHNFMMDTQLTKRVKNAAA NVLRETWLIYKHTRLVKKPDQARVRKHQRKFLQAIHQAQKLRSVKIEQGKLNDQANTLTDLAKTQTVMYDLVSELHAQHEELEARLATLE -------------------------------------------------------------- >29097_29097_2_FAM53B-KCNN1_FAM53B_chr10_126370176_ENST00000337318_KCNN1_chr19_18099224_ENST00000222249_length(transcript)=3365nt_BP=1118nt GGCGCGGGGTCCGCGCCCGGAGCGCCCCGCCGCGCACAGGAGTTGACCACATTTGGCCATTTCCCAGAAGGGCCCCACCCCAAGGGTGAG TGGCCAATGGGGAGCTGTTTCTGCTGACATCAATTCCCCAGGAGGTACTCACCCCAAGTCTGCCCAAGTGAAGATGGCTGATACCCACCC TGGGATGGAGCCCAGCGCCTGAGGCCCTTATCATGGTGATGGTCCTAAGTGAAAGCCTCAGCACCCGGGGAGCTGACTCCATTGCATGTG GGACCTTCAGCCGTGAACTGCACACGCCAAAGAAGATGAGTCAAGGACCTACACTTTTCTCTTGTGGAATTATGGAAAATGACAGATGGC GAGACCTGGACAGGAAATGCCCTCTTCAGATTGACCAACCGAGCACCAGCATCTGGGAATGCCTGCCTGAAAAGGACAGCTCACTATGGC ACCGGGAGGCAGTGACCGCCTGCGCTGTGACCAGTCTGATCAAAGACCTCAGCATCAGCGACCACAACGGGAACCCCTCAGCACCCCCTA GCAAGCGCCAGTGCCGCTCACTGTCCTTCTCCGATGAGATGTCCAGTTGCCGGACATCATGGAGGCCCTTGGGCTCCAAAGTCTGGACTC CCGTGGAAAAGAGACGCTGCTACAGCGGGGGCAGCGTCCAGCGCTATTCCAACGGCTTCAGCACCATGCAGAGGAGTTCCAGCTTCAGCC TCCCTTCCCGGGCCAACGTGCTCTCCTCACCCTGCGACCAGGCAGGACTCCACCACCGATTTGGAGGGCAGCCCTGCCAAGGGGTGCCAG GCTCAGCCCCGTGTGGACAGGCAGGTGACACCTGGAGCCCTGACCTGCACCCCGTGGGAGGAGGCCGGCTGGACCTGCAGCGGTCCCTCT CTTGCTCACATGAGCAGTTTTCCTTTGTGGAATACTGTCCTCCCTCAGCCAACAGCACACCTGCCTCAACACCAGAGCTGGCGAGACGCT CCAGCGGCCTTTCCCGCAGCCGCTCCCAGCCGTGTGTCCTTAACGACAAGAAGGTCGGTGTTAAAAGGCGGCGCCCTGAAGAAGTGCAAG AGCAGAGGCCTTCTCTAGACCTTGCCAAGATGGCACAGGGAGCTGGCTGTACCGCGCTCGTGGTGGCTGTGGTGGCTCGGAAGCTGGAGC TCACCAAGGCTGAGAAGCACGTGCACAACTTCATGATGGACACTCAGCTCACCAAGCGGGTAAAAAACGCCGCTGCTAACGTTCTCAGGG AGACGTGGCTCATCTACAAACATACCAGGCTGGTGAAGAAGCCAGACCAAGCCCGGGTTCGGAAACACCAGCGTAAGTTCCTCCAAGCCA TCCATCAGGCTCAGAAGCTCCGGAGTGTGAAGATCGAGCAAGGGAAGCTGAACGACCAGGCTAACACGCTTACCGACCTAGCCAAGACCC AGACCGTCATGTACGACCTTGTATCGGAGCTGCACGCTCAGCACGAGGAGCTGGAGGCCCGCCTGGCCACCCTGGAAAGCCGCTTGGATG CGCTGGGTGCCTCTCTACAGGCCCTGCCTGGCCTCATCGCCCAAGCCATACGCCCACCCCCGCCTCCCCTGCCTCCCAGGCCCGGCCCCG GCCCCCAAGACCAGGCAGCCCGGAGCTCCCCCTGCCGGTGGACGCCCGTGGCCCCCTCGGACTGCGGGTGACGGCCCTGCCCGCCACCAG ACCCCTAAATCTTGGCCATCGTGTGGCCGCCACCTCCGGGAAGCCTTGTACAGTGGCGCCTCTTGGAGTTCAAGAAGCCAACGCTGAGTC AGGCTGAGTGGACTGAGGCCTGCCCCGCCCAGACTGCCCAGGCAGAGGGCAGGGCTGGACCATGGGTGAGGGCAGGGGAGCCCGGAGCTT CCTCTGGTCACCTGGTCCCCCGACTCTCCCCAGGCCCCCGGTGGGCATGGAGCAGCCCGGGGAGGGGTCCGTGCTGGTTCTGAATAAAGC AGGACCCGCCTAGTGGCTGCCTGTGTGCATGGCTGGAAGGCACTGGTGATGTCCCAGGAGGTAGACCTCCAGCCCTGGGTACCAAGATGA ATGTGGGAATCAGAAAAACCTGTTCCCATCACCGGCCTAGCCTAGAATCCTAGCCTAGAAGCCCTCTCTCCCTCTGGGCTGGAGCTCAGT GAGGGACAACTCTCTAGGGACACCTGTACCAGCCCCACCTGGCGCTGAGATCCCTCAGACAGCATGGCCCAGCCCTGGCCAGAAGCATCG CTCCCCTTCAACCAACCGCGTTGATGGACACCCACTGTGTGCCAGGCCCCAGCGGGGCCCATGGGGGAGGTGACCTGGGTGAGGAAGGTC ATTTGGGTTTTTGTGAGATTTGTATTGAGCACCTGCTGTCTACAGAGAATGTGATGATGTCAATTACCACACTGATTCCTCATCAGAACT TGACCATGGTGGGATGGGGGGGAAACTGAGGCCCAGAGAGGTGAATCTACCTACCTGGGACCACACTGCGAGGAAGGATGGTGCATTCAA ACTGGAAATCCCCTAGGCCAAAGTAAAGAACCGGACTGGCTGGGCACAGTGGCTCACACCTGTAATCCCAGCACTTTGGGAGGGTAAAGC GGGCGGATCACCTGAGGTCAGGAGTTCAAGACCAGCCTGGGCAACATGGTGAAACCCTGTCTCTACTAAAAATATAAAAATTAGCTGGGT GTGGTGGTGCACGCCTGTAATCCCAGCTACTTGGGAGGCTGAGGCAGGATAATCGCTTGAACCCAGGAGGTGGAGGTTGCAGTGAGAAAA AACCTGGACTTGGTGGAAGGGACTCCATGGAGGAGCCATGGGGACGTCAGGAGGAGGGGTTATTTGGTGACATCCAGGGGTCAGAGAGAC CCTTCAGGGGTGGGCTACAGGGGAGGGTTCCAGACACAAGTAGAATCAGAAAGGTCCCTGGAGGCCATGGGCTTGGGTGCTGGGAGCAAC AGGCATTTACTTGGCAGATGCTGAGCCCTGGGTGGGGCGAGGAGGCCTGGCTCCTGGGGAGACGACGCTGTGCATAGCCGAGGAGCCCCA GGTCCTGGCAGCCTCTGATAAGGCCATCCATCGCCTGGCTCAACGAGATAACTAGGCCGTGGCTGGTGCATGAATGACCAGCTTGGGCCC ACGCACGAGAAGGGTATGAGGAGGTGCTGGTCAGACCCGGGTTCGAGTCCTGACTCTGCCACTGCTGGGTGACTCTGAGCAGTGGCTGCC CCTCTGGGGCCTTGGTTGTTTCATCTGTAAAATGGGGTGACAGTCTATTTAACCAGAACTCCCTAAAAAGGGCCAGGCAACTGGTAGGTG >29097_29097_2_FAM53B-KCNN1_FAM53B_chr10_126370176_ENST00000337318_KCNN1_chr19_18099224_ENST00000222249_length(amino acids)=507AA_BP=316 MIPTLGWSPAPEALIMVMVLSESLSTRGADSIACGTFSRELHTPKKMSQGPTLFSCGIMENDRWRDLDRKCPLQIDQPSTSIWECLPEKD SSLWHREAVTACAVTSLIKDLSISDHNGNPSAPPSKRQCRSLSFSDEMSSCRTSWRPLGSKVWTPVEKRRCYSGGSVQRYSNGFSTMQRS SSFSLPSRANVLSSPCDQAGLHHRFGGQPCQGVPGSAPCGQAGDTWSPDLHPVGGGRLDLQRSLSCSHEQFSFVEYCPPSANSTPASTPE LARRSSGLSRSRSQPCVLNDKKVGVKRRRPEEVQEQRPSLDLAKMAQGAGCTALVVAVVARKLELTKAEKHVHNFMMDTQLTKRVKNAAA NVLRETWLIYKHTRLVKKPDQARVRKHQRKFLQAIHQAQKLRSVKIEQGKLNDQANTLTDLAKTQTVMYDLVSELHAQHEELEARLATLE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FAM53B-KCNN1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FAM53B-KCNN1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FAM53B-KCNN1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
