
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FAM65A-EGFR (FusionGDB2 ID:29135) |
Fusion Gene Summary for FAM65A-EGFR |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FAM65A-EGFR | Fusion gene ID: 29135 | Hgene | Tgene | Gene symbol | FAM65A | EGFR | Gene ID | 79567 | 1956 |
| Gene name | RHO family interacting cell polarization regulator 1 | epidermal growth factor receptor | |
| Synonyms | FAM65A | ERBB|ERBB1|HER1|NISBD2|PIG61|mENA | |
| Cytomap | 16q22.1 | 7p11.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | rho family-interacting cell polarization regulator 1family with sequence similarity 65 member Aprotein FAM65A | epidermal growth factor receptoravian erythroblastic leukemia viral (v-erb-b) oncogene homologcell growth inhibiting protein 40cell proliferation-inducing protein 61epidermal growth factor receptor tyrosine kinase domainerb-b2 receptor tyrosine kinas | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | . | P00533 | |
| Ensembl transtripts involved in fusion gene | ENST00000042381, ENST00000379312, ENST00000422602, ENST00000428437, ENST00000540839, ENST00000566522, | ENST00000275493, ENST00000342916, ENST00000344576, ENST00000420316, ENST00000442591, ENST00000454757, ENST00000463948, ENST00000455089, | |
| Fusion gene scores | * DoF score | 6 X 11 X 3=198 | 17 X 20 X 8=2720 |
| # samples | 15 | 22 | |
| ** MAII score | log2(15/198*10)=-0.400537929583729 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(22/2720*10)=-3.62803122261304 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FAM65A [Title/Abstract] AND EGFR [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FAM65A(67576379)-EGFR(55269015), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | EGFR | GO:0001934 | positive regulation of protein phosphorylation | 20551055 |
| Tgene | EGFR | GO:0007165 | signal transduction | 10572067 |
| Tgene | EGFR | GO:0007166 | cell surface receptor signaling pathway | 7736574 |
| Tgene | EGFR | GO:0007173 | epidermal growth factor receptor signaling pathway | 7736574|12435727 |
| Tgene | EGFR | GO:0008283 | cell proliferation | 17115032 |
| Tgene | EGFR | GO:0008284 | positive regulation of cell proliferation | 7736574 |
| Tgene | EGFR | GO:0010750 | positive regulation of nitric oxide mediated signal transduction | 12828935 |
| Tgene | EGFR | GO:0018108 | peptidyl-tyrosine phosphorylation | 22732145 |
| Tgene | EGFR | GO:0030307 | positive regulation of cell growth | 15467833 |
| Tgene | EGFR | GO:0042177 | negative regulation of protein catabolic process | 17115032 |
| Tgene | EGFR | GO:0042327 | positive regulation of phosphorylation | 15082764 |
| Tgene | EGFR | GO:0043406 | positive regulation of MAP kinase activity | 10572067 |
| Tgene | EGFR | GO:0045739 | positive regulation of DNA repair | 17115032 |
| Tgene | EGFR | GO:0045740 | positive regulation of DNA replication | 17115032 |
| Tgene | EGFR | GO:0045944 | positive regulation of transcription by RNA polymerase II | 20551055 |
| Tgene | EGFR | GO:0050679 | positive regulation of epithelial cell proliferation | 10572067 |
| Tgene | EGFR | GO:0050999 | regulation of nitric-oxide synthase activity | 12828935 |
| Tgene | EGFR | GO:0070141 | response to UV-A | 18483258 |
| Tgene | EGFR | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 20551055 |
| Tgene | EGFR | GO:0071392 | cellular response to estradiol stimulus | 20551055 |
| Tgene | EGFR | GO:1900020 | positive regulation of protein kinase C activity | 22732145 |
| Tgene | EGFR | GO:1903078 | positive regulation of protein localization to plasma membrane | 22732145 |
 Fusion gene breakpoints across FAM65A (5'-gene) Fusion gene breakpoints across FAM65A (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
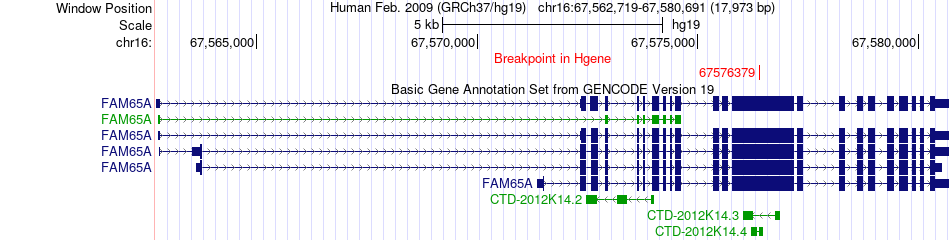 |
 Fusion gene breakpoints across EGFR (3'-gene) Fusion gene breakpoints across EGFR (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
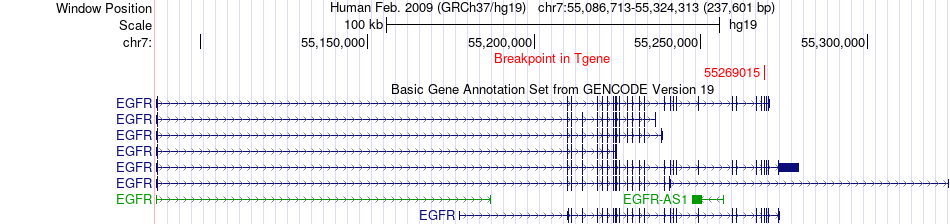 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChiTaRS5.0 | N/A | BF844939 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
Top |
Fusion Gene ORF analysis for FAM65A-EGFR |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000042381 | ENST00000275493 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000042381 | ENST00000342916 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000042381 | ENST00000344576 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000042381 | ENST00000420316 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000042381 | ENST00000442591 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000042381 | ENST00000454757 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000042381 | ENST00000463948 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000379312 | ENST00000275493 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000379312 | ENST00000342916 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000379312 | ENST00000344576 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000379312 | ENST00000420316 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000379312 | ENST00000442591 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000379312 | ENST00000454757 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000379312 | ENST00000463948 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000422602 | ENST00000275493 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000422602 | ENST00000342916 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000422602 | ENST00000344576 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000422602 | ENST00000420316 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000422602 | ENST00000442591 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000422602 | ENST00000454757 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000422602 | ENST00000463948 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000428437 | ENST00000275493 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000428437 | ENST00000342916 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000428437 | ENST00000344576 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000428437 | ENST00000420316 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000428437 | ENST00000442591 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000428437 | ENST00000454757 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000428437 | ENST00000463948 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000540839 | ENST00000275493 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000540839 | ENST00000342916 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000540839 | ENST00000344576 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000540839 | ENST00000420316 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000540839 | ENST00000442591 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000540839 | ENST00000454757 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| 5CDS-intron | ENST00000540839 | ENST00000463948 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| In-frame | ENST00000042381 | ENST00000455089 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| In-frame | ENST00000379312 | ENST00000455089 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| In-frame | ENST00000422602 | ENST00000455089 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| In-frame | ENST00000428437 | ENST00000455089 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| In-frame | ENST00000540839 | ENST00000455089 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| intron-3CDS | ENST00000566522 | ENST00000455089 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| intron-intron | ENST00000566522 | ENST00000275493 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| intron-intron | ENST00000566522 | ENST00000342916 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| intron-intron | ENST00000566522 | ENST00000344576 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| intron-intron | ENST00000566522 | ENST00000420316 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| intron-intron | ENST00000566522 | ENST00000442591 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| intron-intron | ENST00000566522 | ENST00000454757 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
| intron-intron | ENST00000566522 | ENST00000463948 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000379312 | FAM65A | chr16 | 67576379 | - | ENST00000455089 | EGFR | chr7 | 55269015 | - | 2642 | 2001 | 121 | 1257 | 378 |
| ENST00000042381 | FAM65A | chr16 | 67576379 | - | ENST00000455089 | EGFR | chr7 | 55269015 | - | 2577 | 1936 | 68 | 1192 | 374 |
| ENST00000540839 | FAM65A | chr16 | 67576379 | - | ENST00000455089 | EGFR | chr7 | 55269015 | - | 2789 | 2148 | 142 | 1404 | 420 |
| ENST00000422602 | FAM65A | chr16 | 67576379 | - | ENST00000455089 | EGFR | chr7 | 55269015 | - | 2665 | 2024 | 18 | 1280 | 420 |
| ENST00000428437 | FAM65A | chr16 | 67576379 | - | ENST00000455089 | EGFR | chr7 | 55269015 | - | 2676 | 2035 | 26 | 1291 | 421 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000379312 | ENST00000455089 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - | 0.006980599 | 0.9930194 |
| ENST00000042381 | ENST00000455089 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - | 0.003836799 | 0.99616325 |
| ENST00000540839 | ENST00000455089 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - | 0.019462103 | 0.98053795 |
| ENST00000422602 | ENST00000455089 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - | 0.018601604 | 0.9813984 |
| ENST00000428437 | ENST00000455089 | FAM65A | chr16 | 67576379 | - | EGFR | chr7 | 55269015 | - | 0.014910805 | 0.98508924 |
Top |
Fusion Genomic Features for FAM65A-EGFR |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
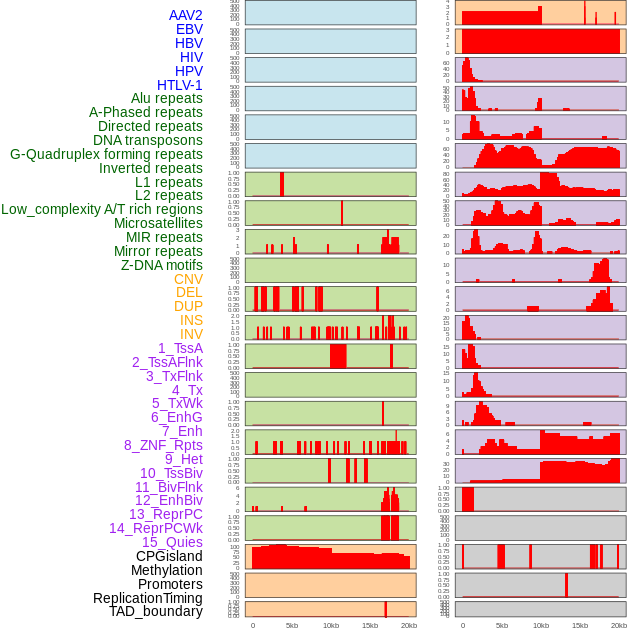 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for FAM65A-EGFR |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:67576379/chr7:55269015) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | EGFR |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Receptor tyrosine kinase binding ligands of the EGF family and activating several signaling cascades to convert extracellular cues into appropriate cellular responses (PubMed:2790960, PubMed:10805725, PubMed:27153536). Known ligands include EGF, TGFA/TGF-alpha, AREG, epigen/EPGN, BTC/betacellulin, epiregulin/EREG and HBEGF/heparin-binding EGF (PubMed:2790960, PubMed:7679104, PubMed:8144591, PubMed:9419975, PubMed:15611079, PubMed:12297049, PubMed:27153536, PubMed:20837704, PubMed:17909029). Ligand binding triggers receptor homo- and/or heterodimerization and autophosphorylation on key cytoplasmic residues. The phosphorylated receptor recruits adapter proteins like GRB2 which in turn activates complex downstream signaling cascades. Activates at least 4 major downstream signaling cascades including the RAS-RAF-MEK-ERK, PI3 kinase-AKT, PLCgamma-PKC and STATs modules (PubMed:27153536). May also activate the NF-kappa-B signaling cascade (PubMed:11116146). Also directly phosphorylates other proteins like RGS16, activating its GTPase activity and probably coupling the EGF receptor signaling to the G protein-coupled receptor signaling (PubMed:11602604). Also phosphorylates MUC1 and increases its interaction with SRC and CTNNB1/beta-catenin (PubMed:11483589). Positively regulates cell migration via interaction with CCDC88A/GIV which retains EGFR at the cell membrane following ligand stimulation, promoting EGFR signaling which triggers cell migration (PubMed:20462955). Plays a role in enhancing learning and memory performance (By similarity). {ECO:0000250|UniProtKB:Q01279, ECO:0000269|PubMed:10805725, ECO:0000269|PubMed:11116146, ECO:0000269|PubMed:11483589, ECO:0000269|PubMed:11602604, ECO:0000269|PubMed:12297049, ECO:0000269|PubMed:12297050, ECO:0000269|PubMed:12620237, ECO:0000269|PubMed:12873986, ECO:0000269|PubMed:15374980, ECO:0000269|PubMed:15590694, ECO:0000269|PubMed:15611079, ECO:0000269|PubMed:17115032, ECO:0000269|PubMed:17909029, ECO:0000269|PubMed:19560417, ECO:0000269|PubMed:20462955, ECO:0000269|PubMed:20837704, ECO:0000269|PubMed:21258366, ECO:0000269|PubMed:27153536, ECO:0000269|PubMed:2790960, ECO:0000269|PubMed:7679104, ECO:0000269|PubMed:8144591, ECO:0000269|PubMed:9419975}.; FUNCTION: Isoform 2 may act as an antagonist of EGF action.; FUNCTION: (Microbial infection) Acts as a receptor for hepatitis C virus (HCV) in hepatocytes and facilitates its cell entry. Mediates HCV entry by promoting the formation of the CD81-CLDN1 receptor complexes that are essential for HCV entry and by enhancing membrane fusion of cells expressing HCV envelope glycoproteins. {ECO:0000269|PubMed:21516087}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000275493 | 0 | 28 | 712_979 | 0 | 1211.0 | Domain | Protein kinase | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000342916 | 0 | 16 | 712_979 | 0 | 629.0 | Domain | Protein kinase | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000344576 | 0 | 16 | 712_979 | 0 | 706.0 | Domain | Protein kinase | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000420316 | 0 | 10 | 712_979 | 0 | 406.0 | Domain | Protein kinase | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000275493 | 0 | 28 | 718_726 | 0 | 1211.0 | Nucleotide binding | ATP | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000275493 | 0 | 28 | 790_791 | 0 | 1211.0 | Nucleotide binding | ATP | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000342916 | 0 | 16 | 718_726 | 0 | 629.0 | Nucleotide binding | ATP | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000342916 | 0 | 16 | 790_791 | 0 | 629.0 | Nucleotide binding | ATP | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000344576 | 0 | 16 | 718_726 | 0 | 706.0 | Nucleotide binding | ATP | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000344576 | 0 | 16 | 790_791 | 0 | 706.0 | Nucleotide binding | ATP | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000420316 | 0 | 10 | 718_726 | 0 | 406.0 | Nucleotide binding | ATP | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000420316 | 0 | 10 | 790_791 | 0 | 406.0 | Nucleotide binding | ATP | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000275493 | 0 | 28 | 688_704 | 0 | 1211.0 | Region | Note=Important for dimerization%2C phosphorylation and activation | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000342916 | 0 | 16 | 688_704 | 0 | 629.0 | Region | Note=Important for dimerization%2C phosphorylation and activation | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000344576 | 0 | 16 | 688_704 | 0 | 706.0 | Region | Note=Important for dimerization%2C phosphorylation and activation | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000420316 | 0 | 10 | 688_704 | 0 | 406.0 | Region | Note=Important for dimerization%2C phosphorylation and activation | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000275493 | 0 | 28 | 390_600 | 0 | 1211.0 | Repeat | Note=Approximate | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000275493 | 0 | 28 | 75_300 | 0 | 1211.0 | Repeat | Note=Approximate | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000342916 | 0 | 16 | 390_600 | 0 | 629.0 | Repeat | Note=Approximate | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000342916 | 0 | 16 | 75_300 | 0 | 629.0 | Repeat | Note=Approximate | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000344576 | 0 | 16 | 390_600 | 0 | 706.0 | Repeat | Note=Approximate | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000344576 | 0 | 16 | 75_300 | 0 | 706.0 | Repeat | Note=Approximate | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000420316 | 0 | 10 | 390_600 | 0 | 406.0 | Repeat | Note=Approximate | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000420316 | 0 | 10 | 75_300 | 0 | 406.0 | Repeat | Note=Approximate | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000275493 | 0 | 28 | 25_645 | 0 | 1211.0 | Topological domain | Extracellular | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000275493 | 0 | 28 | 669_1210 | 0 | 1211.0 | Topological domain | Cytoplasmic | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000342916 | 0 | 16 | 25_645 | 0 | 629.0 | Topological domain | Extracellular | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000342916 | 0 | 16 | 669_1210 | 0 | 629.0 | Topological domain | Cytoplasmic | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000344576 | 0 | 16 | 25_645 | 0 | 706.0 | Topological domain | Extracellular | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000344576 | 0 | 16 | 669_1210 | 0 | 706.0 | Topological domain | Cytoplasmic | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000420316 | 0 | 10 | 25_645 | 0 | 406.0 | Topological domain | Extracellular | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000420316 | 0 | 10 | 669_1210 | 0 | 406.0 | Topological domain | Cytoplasmic | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000275493 | 0 | 28 | 646_668 | 0 | 1211.0 | Transmembrane | Helical | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000342916 | 0 | 16 | 646_668 | 0 | 629.0 | Transmembrane | Helical | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000344576 | 0 | 16 | 646_668 | 0 | 706.0 | Transmembrane | Helical | |
| Tgene | EGFR | chr16:67576379 | chr7:55269015 | ENST00000420316 | 0 | 10 | 646_668 | 0 | 406.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FAM65A | chr16:67576379 | chr7:55269015 | ENST00000042381 | - | 1 | 22 | 89_114 | 0 | 1220.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | FAM65A | chr16:67576379 | chr7:55269015 | ENST00000379312 | - | 1 | 22 | 89_114 | 0 | 1224.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | FAM65A | chr16:67576379 | chr7:55269015 | ENST00000422602 | - | 1 | 22 | 89_114 | 0 | 1240.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | FAM65A | chr16:67576379 | chr7:55269015 | ENST00000428437 | - | 1 | 22 | 89_114 | 0 | 1234.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | FAM65A | chr16:67576379 | chr7:55269015 | ENST00000042381 | - | 1 | 22 | 481_767 | 0 | 1220.0 | Compositional bias | Note=Pro-rich |
| Hgene | FAM65A | chr16:67576379 | chr7:55269015 | ENST00000379312 | - | 1 | 22 | 481_767 | 0 | 1224.0 | Compositional bias | Note=Pro-rich |
| Hgene | FAM65A | chr16:67576379 | chr7:55269015 | ENST00000422602 | - | 1 | 22 | 481_767 | 0 | 1240.0 | Compositional bias | Note=Pro-rich |
| Hgene | FAM65A | chr16:67576379 | chr7:55269015 | ENST00000428437 | - | 1 | 22 | 481_767 | 0 | 1234.0 | Compositional bias | Note=Pro-rich |
Top |
Fusion Gene Sequence for FAM65A-EGFR |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >29135_29135_1_FAM65A-EGFR_FAM65A_chr16_67576379_ENST00000042381_EGFR_chr7_55269015_ENST00000455089_length(transcript)=2577nt_BP=1936nt CGAGCGCAAGGACAGCGCGAGGTCCGCGCAGCCCAGCGCAGCCATGGAGCCCCGCGCGGACTCACTCTATGATGTCCCTGTCGGTGCGGC CGCAGCGCCGTCTGCTCAGCGCCCGGGTCAATAGGAGCCAGTCCTTCGCAGGCGTCCTCGGCAGCCACGAGCGGGGGCCCAGGAGTTTCC CGGTCTTCAGCCCGCCGGGGCCCCCACGGAAGCCCCCCGCGCTCTCCCGAGTGTCCAGGATGTTTTCCGTGGCTCACCCAGCCGCCAAGG TGCCGCAGCCCGAGCGGCTGGACCTGGTGTACACGGCGCTGAAGCGGGGCCTGACGGCCTACTTGGAAGTGCACCAGCAGGAGCAAGAGA AACTCCAGGGGCAGATAAGGGAGTCCAAGAGGAATTCCCGCTTGGGCTTCCTGTATGATCTGGACAAGCAAGTCAAGTCCATTGAACGCT TCCTGCGACGACTGGAGTTCCATGCCAGCAAGATCGATGAGCTGTATGAGGCATACTGTGTCCAGCGGCGTCTCCGGGATGGTGCCTACA ACATGGTCCGTGCCTACACCACTGGGTCCCCGGGAAGCCGAGAGGCCCGGGACAGCCTGGCAGAGGCCACTCGGGGGCATCGCGAGTACA CGGAGAGCATGTGTCTGCTGGAGAGCGAGCTGGAGGCACAGCTGGGCGAGTTTCATCTCCGAATGAAAGGGCTGGCTGGCTTCGCCAGGC TGTGTGTAGGCGATCAGTATGAGATCTGCATGAAATATGGGCGTCAGCGCTGGAAACTACGGGGCCGAATTGAGGGTAGTGGAAAGCAGG TGTGGGACAGTGAAGAAACCATCTTTCTCCCTCTACTCACGGAATTTCTGTCTATTAAGGTGACAGAACTGAAGGGCCTGGCCAACCATG TGGTTGTGGGCAGTGTCTCCTGTGAGACCAAGGACCTGTTTGCCGCCCTGCCCCAGGTTGTGGCTGTGGATATCAATGACCTTGGTACCA TCAAGCTCAGCCTGGAAGTCACATGGAGCCCCTTCGACAAGGATGACCAGCCCTCAGCTGCTTCTTCTGTCAACAAGGCCTCCACAGTCA CCAAGCGCTTCTCCACCTATAGCCAGAGCCCACCGGACACACCCTCACTTCGGGAACAGGCTTTCTATGCCCCCTCACTCACACTACTAC AGGCTCCACCCACAAGCCCATAATCTCTACCCTTACTACTACAGGCCCTACCCTCAATATCATAGGCCCAGTCCAGACTACCACAAGCCC CACCCACACTATGCCAAGCCCTACCCATACCACAGCAAGCCCCACTCATACTTCCACAAGCCCCACCCATACCCCCACAAGTCCCACCCA CAAAACCAGTATGTCACCTCCCACCACTACAAGTCCTACCCCCAGTGGTATGGGCCTAGTCCAGACTGCCACAAGTCCCACCCATCCTAC CACAAGCCCCACCCATCCCACCACAAGCCCCATCCTTATAAATGTAAGCCCTTCCACTTCTCTAGAACTTGCTACCCTCTCCAGCCCCTC CAAACACTCAGACCCCACCCTCCCAGGCACTGACTCCCTTCCCTGTAGTCCCCCAGTCTCCAATTCCTACACTCAGGCAGACCCTATGGC CCCCAGAACTCCCCACCCAAGTCCTGCCCATTCCAGTAGGAAACCCCTCACAAGCCCTGCCCCAGATCCCTCAGAGTCTACGGTTCAGAG TCTAAGCCCCACTCCCTCACCCCCAACCCCTGCACCCCAGCATTCAGACCTTTGCCTGGCCATGGCTGTCCAGACCCCAGTCCCAACGGC AGCCGGAGGGTCTGGGGACAGGAGCCTGGAGGAGGCACTGGGGGCCCTAATGGCTGCCCTGGATGACTACCGTGGCCAGTTTCCTGAGCT GCAGGGCCTGGAGCAGGAGGTGACCCGCCTAGAAAGTCTGCTCATGTCCACGTCACGGACTCCCCTCCTGAGCTCTCTGAGTGCAACCAG CAACAATTCCACCGTGGCTTGCATTGATAGAAATGGGCTGCAAAGCTGTCCCATCAAGGAAGACAGCTTCTTGCAGCGATACAGCTCAGA CCCCACAGGCGCCTTGACTGAGGACAGCATAGACGACACCTTCCTCCCAGTGCCTGGTGAGTGGCTTGTCTGGAAACAGTCCTGCTCCTC AACCTCCTCGACCCACTCAGCAGCAGCCAGTCTCCAGTGTCCAAGCCAGGTGCTCCCTCCAGCATCTCCAGAGGGGGAAACAGTGGCAGA TTTGCAGACACAGTGAAGGGCGTAAGGAGCAGATAAACACATGACCGAGCCTGCACAAGCTCTTTGTTGTGTCTGGTTGTTTGCTGTACC TCTGTTGTAAGAATGAATCTGCAAAATTTCTAGCTTATGAAGCAAATCACGGACATACACATCTGTGTGTGTGAGTGTTCATGATGTGTG TACATCTGTGTATGTGTGTGTGTGTATGTGTGTGTTTGTGACAGATTTGATCCCTGTTCTCTCTGCTGGCTCTATCTTGACCTGTGAAAC >29135_29135_1_FAM65A-EGFR_FAM65A_chr16_67576379_ENST00000042381_EGFR_chr7_55269015_ENST00000455089_length(amino acids)=374AA_BP= MMSLSVRPQRRLLSARVNRSQSFAGVLGSHERGPRSFPVFSPPGPPRKPPALSRVSRMFSVAHPAAKVPQPERLDLVYTALKRGLTAYLE VHQQEQEKLQGQIRESKRNSRLGFLYDLDKQVKSIERFLRRLEFHASKIDELYEAYCVQRRLRDGAYNMVRAYTTGSPGSREARDSLAEA TRGHREYTESMCLLESELEAQLGEFHLRMKGLAGFARLCVGDQYEICMKYGRQRWKLRGRIEGSGKQVWDSEETIFLPLLTEFLSIKVTE LKGLANHVVVGSVSCETKDLFAALPQVVAVDINDLGTIKLSLEVTWSPFDKDDQPSAASSVNKASTVTKRFSTYSQSPPDTPSLREQAFY -------------------------------------------------------------- >29135_29135_2_FAM65A-EGFR_FAM65A_chr16_67576379_ENST00000379312_EGFR_chr7_55269015_ENST00000455089_length(transcript)=2642nt_BP=2001nt GGCGCCTGCGGCGGCGACAGCGGCAGCTGCGGCGCGACCAGGCCGGGCACCTCCGAGCGCAAGGACAGCGCGAGGTCCGCGCAGCCCAGC GCAGCCATGGAGCCCCGCGCGGACTCACTCTATGATGTCCCTGTCGGTGCGGCCGCAGCGCCGTCTGCTCAGCGCCCGGGTCAATAGGAG CCAGTCCTTCGCAGGCGTCCTCGGCAGCCACGAGCGGGGGCCCAGCCTCTCCTTCAGGAGTTTCCCGGTCTTCAGCCCGCCGGGGCCCCC ACGGAAGCCCCCCGCGCTCTCCCGAGTGTCCAGGATGTTTTCCGTGGCTCACCCAGCCGCCAAGGTGCCGCAGCCCGAGCGGCTGGACCT GGTGTACACGGCGCTGAAGCGGGGCCTGACGGCCTACTTGGAAGTGCACCAGCAGGAGCAAGAGAAACTCCAGGGGCAGATAAGGGAGTC CAAGAGGAATTCCCGCTTGGGCTTCCTGTATGATCTGGACAAGCAAGTCAAGTCCATTGAACGCTTCCTGCGACGACTGGAGTTCCATGC CAGCAAGATCGATGAGCTGTATGAGGCATACTGTGTCCAGCGGCGTCTCCGGGATGGTGCCTACAACATGGTCCGTGCCTACACCACTGG GTCCCCGGGAAGCCGAGAGGCCCGGGACAGCCTGGCAGAGGCCACTCGGGGGCATCGCGAGTACACGGAGAGCATGTGTCTGCTGGAGAG CGAGCTGGAGGCACAGCTGGGCGAGTTTCATCTCCGAATGAAAGGGCTGGCTGGCTTCGCCAGGCTGTGTGTAGGCGATCAGTATGAGAT CTGCATGAAATATGGGCGTCAGCGCTGGAAACTACGGGGCCGAATTGAGGGTAGTGGAAAGCAGGTGTGGGACAGTGAAGAAACCATCTT TCTCCCTCTACTCACGGAATTTCTGTCTATTAAGGTGACAGAACTGAAGGGCCTGGCCAACCATGTGGTTGTGGGCAGTGTCTCCTGTGA GACCAAGGACCTGTTTGCCGCCCTGCCCCAGGTTGTGGCTGTGGATATCAATGACCTTGGTACCATCAAGCTCAGCCTGGAAGTCACATG GAGCCCCTTCGACAAGGATGACCAGCCCTCAGCTGCTTCTTCTGTCAACAAGGCCTCCACAGTCACCAAGCGCTTCTCCACCTATAGCCA GAGCCCACCGGACACACCCTCACTTCGGGAACAGGCTTTCTATGCCCCCTCACTCACACTACTACAGGCTCCACCCACAAGCCCATAATC TCTACCCTTACTACTACAGGCCCTACCCTCAATATCATAGGCCCAGTCCAGACTACCACAAGCCCCACCCACACTATGCCAAGCCCTACC CATACCACAGCAAGCCCCACTCATACTTCCACAAGCCCCACCCATACCCCCACAAGTCCCACCCACAAAACCAGTATGTCACCTCCCACC ACTACAAGTCCTACCCCCAGTGGTATGGGCCTAGTCCAGACTGCCACAAGTCCCACCCATCCTACCACAAGCCCCACCCATCCCACCACA AGCCCCATCCTTATAAATGTAAGCCCTTCCACTTCTCTAGAACTTGCTACCCTCTCCAGCCCCTCCAAACACTCAGACCCCACCCTCCCA GGCACTGACTCCCTTCCCTGTAGTCCCCCAGTCTCCAATTCCTACACTCAGGCAGACCCTATGGCCCCCAGAACTCCCCACCCAAGTCCT GCCCATTCCAGTAGGAAACCCCTCACAAGCCCTGCCCCAGATCCCTCAGAGTCTACGGTTCAGAGTCTAAGCCCCACTCCCTCACCCCCA ACCCCTGCACCCCAGCATTCAGACCTTTGCCTGGCCATGGCTGTCCAGACCCCAGTCCCAACGGCAGCCGGAGGGTCTGGGGACAGGAGC CTGGAGGAGGCACTGGGGGCCCTAATGGCTGCCCTGGATGACTACCGTGGCCAGTTTCCTGAGCTGCAGGGCCTGGAGCAGGAGGTGACC CGCCTAGAAAGTCTGCTCATGTCCACGTCACGGACTCCCCTCCTGAGCTCTCTGAGTGCAACCAGCAACAATTCCACCGTGGCTTGCATT GATAGAAATGGGCTGCAAAGCTGTCCCATCAAGGAAGACAGCTTCTTGCAGCGATACAGCTCAGACCCCACAGGCGCCTTGACTGAGGAC AGCATAGACGACACCTTCCTCCCAGTGCCTGGTGAGTGGCTTGTCTGGAAACAGTCCTGCTCCTCAACCTCCTCGACCCACTCAGCAGCA GCCAGTCTCCAGTGTCCAAGCCAGGTGCTCCCTCCAGCATCTCCAGAGGGGGAAACAGTGGCAGATTTGCAGACACAGTGAAGGGCGTAA GGAGCAGATAAACACATGACCGAGCCTGCACAAGCTCTTTGTTGTGTCTGGTTGTTTGCTGTACCTCTGTTGTAAGAATGAATCTGCAAA ATTTCTAGCTTATGAAGCAAATCACGGACATACACATCTGTGTGTGTGAGTGTTCATGATGTGTGTACATCTGTGTATGTGTGTGTGTGT ATGTGTGTGTTTGTGACAGATTTGATCCCTGTTCTCTCTGCTGGCTCTATCTTGACCTGTGAAACGTATATTTAACTAATTAAATATTAG >29135_29135_2_FAM65A-EGFR_FAM65A_chr16_67576379_ENST00000379312_EGFR_chr7_55269015_ENST00000455089_length(amino acids)=378AA_BP= MMSLSVRPQRRLLSARVNRSQSFAGVLGSHERGPSLSFRSFPVFSPPGPPRKPPALSRVSRMFSVAHPAAKVPQPERLDLVYTALKRGLT AYLEVHQQEQEKLQGQIRESKRNSRLGFLYDLDKQVKSIERFLRRLEFHASKIDELYEAYCVQRRLRDGAYNMVRAYTTGSPGSREARDS LAEATRGHREYTESMCLLESELEAQLGEFHLRMKGLAGFARLCVGDQYEICMKYGRQRWKLRGRIEGSGKQVWDSEETIFLPLLTEFLSI KVTELKGLANHVVVGSVSCETKDLFAALPQVVAVDINDLGTIKLSLEVTWSPFDKDDQPSAASSVNKASTVTKRFSTYSQSPPDTPSLRE -------------------------------------------------------------- >29135_29135_3_FAM65A-EGFR_FAM65A_chr16_67576379_ENST00000422602_EGFR_chr7_55269015_ENST00000455089_length(transcript)=2665nt_BP=2024nt AGCCTCGTGTAACAATACTTGTGCCCTGGCTGCAGTCTGCGGGGCCGCGCCCTGGGCCTGCCGCATTCGGCCAACGCACAGCATCTGAGG AGGGTTATGACCATCTGGCAGATGCAGAAACAGGCCCAGAGAGGGAGCCCCGCGCGGACTCACTCTATGATGTCCCTGTCGGTGCGGCCG CAGCGCCGTCTGCTCAGCGCCCGGGTCAATAGGAGCCAGTCCTTCGCAGGCGTCCTCGGCAGCCACGAGCGGGGGCCCAGGAGTTTCCCG GTCTTCAGCCCGCCGGGGCCCCCACGGAAGCCCCCCGCGCTCTCCCGAGTGTCCAGGATGTTTTCCGTGGCTCACCCAGCCGCCAAGGTG CCGCAGCCCGAGCGGCTGGACCTGGTGTACACGGCGCTGAAGCGGGGCCTGACGGCCTACTTGGAAGTGCACCAGCAGGAGCAAGAGAAA CTCCAGGGGCAGATAAGGGAGTCCAAGAGGAATTCCCGCTTGGGCTTCCTGTATGATCTGGACAAGCAAGTCAAGTCCATTGAACGCTTC CTGCGACGACTGGAGTTCCATGCCAGCAAGATCGATGAGCTGTATGAGGCATACTGTGTCCAGCGGCGTCTCCGGGATGGTGCCTACAAC ATGGTCCGTGCCTACACCACTGGGTCCCCGGGAAGCCGAGAGGCCCGGGACAGCCTGGCAGAGGCCACTCGGGGGCATCGCGAGTACACG GAGAGCATGTGTCTGCTGGAGAGCGAGCTGGAGGCACAGCTGGGCGAGTTTCATCTCCGAATGAAAGGGCTGGCTGGCTTCGCCAGGCTG TGTGTAGGCGATCAGTATGAGATCTGCATGAAATATGGGCGTCAGCGCTGGAAACTACGGGGCCGAATTGAGGGTAGTGGAAAGCAGGTG TGGGACAGTGAAGAAACCATCTTTCTCCCTCTACTCACGGAATTTCTGTCTATTAAGGTGACAGAACTGAAGGGCCTGGCCAACCATGTG GTTGTGGGCAGTGTCTCCTGTGAGACCAAGGACCTGTTTGCCGCCCTGCCCCAGGTTGTGGCTGTGGATATCAATGACCTTGGTACCATC AAGCTCAGCCTGGAAGTCACATGGAGCCCCTTCGACAAGGATGACCAGCCCTCAGCTGCTTCTTCTGTCAACAAGGCCTCCACAGTCACC AAGCGCTTCTCCACCTATAGCCAGAGCCCACCGGACACACCCTCACTTCGGGAACAGGCTTTCTATGCCCCCTCACTCACACTACTACAG GCTCCACCCACAAGCCCATAATCTCTACCCTTACTACTACAGGCCCTACCCTCAATATCATAGGCCCAGTCCAGACTACCACAAGCCCCA CCCACACTATGCCAAGCCCTACCCATACCACAGCAAGCCCCACTCATACTTCCACAAGCCCCACCCATACCCCCACAAGTCCCACCCACA AAACCAGTATGTCACCTCCCACCACTACAAGTCCTACCCCCAGTGGTATGGGCCTAGTCCAGACTGCCACAAGTCCCACCCATCCTACCA CAAGCCCCACCCATCCCACCACAAGCCCCATCCTTATAAATGTAAGCCCTTCCACTTCTCTAGAACTTGCTACCCTCTCCAGCCCCTCCA AACACTCAGACCCCACCCTCCCAGGCACTGACTCCCTTCCCTGTAGTCCCCCAGTCTCCAATTCCTACACTCAGGCAGACCCTATGGCCC CCAGAACTCCCCACCCAAGTCCTGCCCATTCCAGTAGGAAACCCCTCACAAGCCCTGCCCCAGATCCCTCAGAGTCTACGGTTCAGAGTC TAAGCCCCACTCCCTCACCCCCAACCCCTGCACCCCAGCATTCAGACCTTTGCCTGGCCATGGCTGTCCAGACCCCAGTCCCAACGGCAG CCGGAGGGTCTGGGGACAGGAGCCTGGAGGAGGCACTGGGGGCCCTAATGGCTGCCCTGGATGACTACCGTGGCCAGTTTCCTGAGCTGC AGGGCCTGGAGCAGGAGGTGACCCGCCTAGAAAGTCTGCTCATGTCCACGTCACGGACTCCCCTCCTGAGCTCTCTGAGTGCAACCAGCA ACAATTCCACCGTGGCTTGCATTGATAGAAATGGGCTGCAAAGCTGTCCCATCAAGGAAGACAGCTTCTTGCAGCGATACAGCTCAGACC CCACAGGCGCCTTGACTGAGGACAGCATAGACGACACCTTCCTCCCAGTGCCTGGTGAGTGGCTTGTCTGGAAACAGTCCTGCTCCTCAA CCTCCTCGACCCACTCAGCAGCAGCCAGTCTCCAGTGTCCAAGCCAGGTGCTCCCTCCAGCATCTCCAGAGGGGGAAACAGTGGCAGATT TGCAGACACAGTGAAGGGCGTAAGGAGCAGATAAACACATGACCGAGCCTGCACAAGCTCTTTGTTGTGTCTGGTTGTTTGCTGTACCTC TGTTGTAAGAATGAATCTGCAAAATTTCTAGCTTATGAAGCAAATCACGGACATACACATCTGTGTGTGTGAGTGTTCATGATGTGTGTA CATCTGTGTATGTGTGTGTGTGTATGTGTGTGTTTGTGACAGATTTGATCCCTGTTCTCTCTGCTGGCTCTATCTTGACCTGTGAAACGT >29135_29135_3_FAM65A-EGFR_FAM65A_chr16_67576379_ENST00000422602_EGFR_chr7_55269015_ENST00000455089_length(amino acids)=420AA_BP= MCPGCSLRGRALGLPHSANAQHLRRVMTIWQMQKQAQRGSPARTHSMMSLSVRPQRRLLSARVNRSQSFAGVLGSHERGPRSFPVFSPPG PPRKPPALSRVSRMFSVAHPAAKVPQPERLDLVYTALKRGLTAYLEVHQQEQEKLQGQIRESKRNSRLGFLYDLDKQVKSIERFLRRLEF HASKIDELYEAYCVQRRLRDGAYNMVRAYTTGSPGSREARDSLAEATRGHREYTESMCLLESELEAQLGEFHLRMKGLAGFARLCVGDQY EICMKYGRQRWKLRGRIEGSGKQVWDSEETIFLPLLTEFLSIKVTELKGLANHVVVGSVSCETKDLFAALPQVVAVDINDLGTIKLSLEV -------------------------------------------------------------- >29135_29135_4_FAM65A-EGFR_FAM65A_chr16_67576379_ENST00000428437_EGFR_chr7_55269015_ENST00000455089_length(transcript)=2676nt_BP=2035nt GAGGAGCCGAAGCCGAGCCAGAGCCGCTGGGAGCGAGCCCGGAGCCCAGCCGGGCGGCTCGAAGTGGCCAGGGCCGGAAGGTCCGCGGGG GGCGAGCGCGGGTCGGGGGCCGTCCCGGACCCACCATGAACACCAAGAAGAGAGGGAGCCCCGCGCGGACTCACTCTATGATGTCCCTGT CGGTGCGGCCGCAGCGCCGTCTGCTCAGCGCCCGGGTCAATAGGAGCCAGTCCTTCGCAGGCGTCCTCGGCAGCCACGAGCGGGGGCCCA GGAGTTTCCCGGTCTTCAGCCCGCCGGGGCCCCCACGGAAGCCCCCCGCGCTCTCCCGAGTGTCCAGGATGTTTTCCGTGGCTCACCCAG CCGCCAAGGTGCCGCAGCCCGAGCGGCTGGACCTGGTGTACACGGCGCTGAAGCGGGGCCTGACGGCCTACTTGGAAGTGCACCAGCAGG AGCAAGAGAAACTCCAGGGGCAGATAAGGGAGTCCAAGAGGAATTCCCGCTTGGGCTTCCTGTATGATCTGGACAAGCAAGTCAAGTCCA TTGAACGCTTCCTGCGACGACTGGAGTTCCATGCCAGCAAGATCGATGAGCTGTATGAGGCATACTGTGTCCAGCGGCGTCTCCGGGATG GTGCCTACAACATGGTCCGTGCCTACACCACTGGGTCCCCGGGAAGCCGAGAGGCCCGGGACAGCCTGGCAGAGGCCACTCGGGGGCATC GCGAGTACACGGAGAGCATGTGTCTGCTGGAGAGCGAGCTGGAGGCACAGCTGGGCGAGTTTCATCTCCGAATGAAAGGGCTGGCTGGCT TCGCCAGGCTGTGTGTAGGCGATCAGTATGAGATCTGCATGAAATATGGGCGTCAGCGCTGGAAACTACGGGGCCGAATTGAGGGTAGTG GAAAGCAGGTGTGGGACAGTGAAGAAACCATCTTTCTCCCTCTACTCACGGAATTTCTGTCTATTAAGGTGACAGAACTGAAGGGCCTGG CCAACCATGTGGTTGTGGGCAGTGTCTCCTGTGAGACCAAGGACCTGTTTGCCGCCCTGCCCCAGGTTGTGGCTGTGGATATCAATGACC TTGGTACCATCAAGCTCAGCCTGGAAGTCACATGGAGCCCCTTCGACAAGGATGACCAGCCCTCAGCTGCTTCTTCTGTCAACAAGGCCT CCACAGTCACCAAGCGCTTCTCCACCTATAGCCAGAGCCCACCGGACACACCCTCACTTCGGGAACAGGCTTTCTATGCCCCCTCACTCA CACTACTACAGGCTCCACCCACAAGCCCATAATCTCTACCCTTACTACTACAGGCCCTACCCTCAATATCATAGGCCCAGTCCAGACTAC CACAAGCCCCACCCACACTATGCCAAGCCCTACCCATACCACAGCAAGCCCCACTCATACTTCCACAAGCCCCACCCATACCCCCACAAG TCCCACCCACAAAACCAGTATGTCACCTCCCACCACTACAAGTCCTACCCCCAGTGGTATGGGCCTAGTCCAGACTGCCACAAGTCCCAC CCATCCTACCACAAGCCCCACCCATCCCACCACAAGCCCCATCCTTATAAATGTAAGCCCTTCCACTTCTCTAGAACTTGCTACCCTCTC CAGCCCCTCCAAACACTCAGACCCCACCCTCCCAGGCACTGACTCCCTTCCCTGTAGTCCCCCAGTCTCCAATTCCTACACTCAGGCAGA CCCTATGGCCCCCAGAACTCCCCACCCAAGTCCTGCCCATTCCAGTAGGAAACCCCTCACAAGCCCTGCCCCAGATCCCTCAGAGTCTAC GGTTCAGAGTCTAAGCCCCACTCCCTCACCCCCAACCCCTGCACCCCAGCATTCAGACCTTTGCCTGGCCATGGCTGTCCAGACCCCAGT CCCAACGGCAGCCGGAGGGTCTGGGGACAGGAGCCTGGAGGAGGCACTGGGGGCCCTAATGGCTGCCCTGGATGACTACCGTGGCCAGTT TCCTGAGCTGCAGGGCCTGGAGCAGGAGGTGACCCGCCTAGAAAGTCTGCTCATGTCCACGTCACGGACTCCCCTCCTGAGCTCTCTGAG TGCAACCAGCAACAATTCCACCGTGGCTTGCATTGATAGAAATGGGCTGCAAAGCTGTCCCATCAAGGAAGACAGCTTCTTGCAGCGATA CAGCTCAGACCCCACAGGCGCCTTGACTGAGGACAGCATAGACGACACCTTCCTCCCAGTGCCTGGTGAGTGGCTTGTCTGGAAACAGTC CTGCTCCTCAACCTCCTCGACCCACTCAGCAGCAGCCAGTCTCCAGTGTCCAAGCCAGGTGCTCCCTCCAGCATCTCCAGAGGGGGAAAC AGTGGCAGATTTGCAGACACAGTGAAGGGCGTAAGGAGCAGATAAACACATGACCGAGCCTGCACAAGCTCTTTGTTGTGTCTGGTTGTT TGCTGTACCTCTGTTGTAAGAATGAATCTGCAAAATTTCTAGCTTATGAAGCAAATCACGGACATACACATCTGTGTGTGTGAGTGTTCA TGATGTGTGTACATCTGTGTATGTGTGTGTGTGTATGTGTGTGTTTGTGACAGATTTGATCCCTGTTCTCTCTGCTGGCTCTATCTTGAC >29135_29135_4_FAM65A-EGFR_FAM65A_chr16_67576379_ENST00000428437_EGFR_chr7_55269015_ENST00000455089_length(amino acids)=421AA_BP= MGASPEPSRAARSGQGRKVRGGRARVGGRPGPTMNTKKRGSPARTHSMMSLSVRPQRRLLSARVNRSQSFAGVLGSHERGPRSFPVFSPP GPPRKPPALSRVSRMFSVAHPAAKVPQPERLDLVYTALKRGLTAYLEVHQQEQEKLQGQIRESKRNSRLGFLYDLDKQVKSIERFLRRLE FHASKIDELYEAYCVQRRLRDGAYNMVRAYTTGSPGSREARDSLAEATRGHREYTESMCLLESELEAQLGEFHLRMKGLAGFARLCVGDQ YEICMKYGRQRWKLRGRIEGSGKQVWDSEETIFLPLLTEFLSIKVTELKGLANHVVVGSVSCETKDLFAALPQVVAVDINDLGTIKLSLE -------------------------------------------------------------- >29135_29135_5_FAM65A-EGFR_FAM65A_chr16_67576379_ENST00000540839_EGFR_chr7_55269015_ENST00000455089_length(transcript)=2789nt_BP=2148nt ACAGCGCGAGGTCCGCGCAGCCCAGCGCAGCCATGGTGAGCCCTGAGTCCGGCCTCCCCTACAGACCCTCCCCAGCCAGTTCTAACGTGT GTCCAGGCTGGCCGCCCCAGCACCTACTGTGCGCAGCCTCGTGTAACAATACTTGTGCCCTGGCTGCAGTCTGCGGGGCCGCGCCCTGGG CCTGCCGCATTCGGCCAACGCACAGCATCTGAGGAGGGTTATGACCATCTGGCAGATGCAGAAACAGGCCCAGAGAGGGAGCCCCGCGCG GACTCACTCTATGATGTCCCTGTCGGTGCGGCCGCAGCGCCGTCTGCTCAGCGCCCGGGTCAATAGGAGCCAGTCCTTCGCAGGCGTCCT CGGCAGCCACGAGCGGGGGCCCAGGAGTTTCCCGGTCTTCAGCCCGCCGGGGCCCCCACGGAAGCCCCCCGCGCTCTCCCGAGTGTCCAG GATGTTTTCCGTGGCTCACCCAGCCGCCAAGGTGCCGCAGCCCGAGCGGCTGGACCTGGTGTACACGGCGCTGAAGCGGGGCCTGACGGC CTACTTGGAAGTGCACCAGCAGGAGCAAGAGAAACTCCAGGGGCAGATAAGGGAGTCCAAGAGGAATTCCCGCTTGGGCTTCCTGTATGA TCTGGACAAGCAAGTCAAGTCCATTGAACGCTTCCTGCGACGACTGGAGTTCCATGCCAGCAAGATCGATGAGCTGTATGAGGCATACTG TGTCCAGCGGCGTCTCCGGGATGGTGCCTACAACATGGTCCGTGCCTACACCACTGGGTCCCCGGGAAGCCGAGAGGCCCGGGACAGCCT GGCAGAGGCCACTCGGGGGCATCGCGAGTACACGGAGAGCATGTGTCTGCTGGAGAGCGAGCTGGAGGCACAGCTGGGCGAGTTTCATCT CCGAATGAAAGGGCTGGCTGGCTTCGCCAGGCTGTGTGTAGGCGATCAGTATGAGATCTGCATGAAATATGGGCGTCAGCGCTGGAAACT ACGGGGCCGAATTGAGGGTAGTGGAAAGCAGGTGTGGGACAGTGAAGAAACCATCTTTCTCCCTCTACTCACGGAATTTCTGTCTATTAA GGTGACAGAACTGAAGGGCCTGGCCAACCATGTGGTTGTGGGCAGTGTCTCCTGTGAGACCAAGGACCTGTTTGCCGCCCTGCCCCAGGT TGTGGCTGTGGATATCAATGACCTTGGTACCATCAAGCTCAGCCTGGAAGTCACATGGAGCCCCTTCGACAAGGATGACCAGCCCTCAGC TGCTTCTTCTGTCAACAAGGCCTCCACAGTCACCAAGCGCTTCTCCACCTATAGCCAGAGCCCACCGGACACACCCTCACTTCGGGAACA GGCTTTCTATGCCCCCTCACTCACACTACTACAGGCTCCACCCACAAGCCCATAATCTCTACCCTTACTACTACAGGCCCTACCCTCAAT ATCATAGGCCCAGTCCAGACTACCACAAGCCCCACCCACACTATGCCAAGCCCTACCCATACCACAGCAAGCCCCACTCATACTTCCACA AGCCCCACCCATACCCCCACAAGTCCCACCCACAAAACCAGTATGTCACCTCCCACCACTACAAGTCCTACCCCCAGTGGTATGGGCCTA GTCCAGACTGCCACAAGTCCCACCCATCCTACCACAAGCCCCACCCATCCCACCACAAGCCCCATCCTTATAAATGTAAGCCCTTCCACT TCTCTAGAACTTGCTACCCTCTCCAGCCCCTCCAAACACTCAGACCCCACCCTCCCAGGCACTGACTCCCTTCCCTGTAGTCCCCCAGTC TCCAATTCCTACACTCAGGCAGACCCTATGGCCCCCAGAACTCCCCACCCAAGTCCTGCCCATTCCAGTAGGAAACCCCTCACAAGCCCT GCCCCAGATCCCTCAGAGTCTACGGTTCAGAGTCTAAGCCCCACTCCCTCACCCCCAACCCCTGCACCCCAGCATTCAGACCTTTGCCTG GCCATGGCTGTCCAGACCCCAGTCCCAACGGCAGCCGGAGGGTCTGGGGACAGGAGCCTGGAGGAGGCACTGGGGGCCCTAATGGCTGCC CTGGATGACTACCGTGGCCAGTTTCCTGAGCTGCAGGGCCTGGAGCAGGAGGTGACCCGCCTAGAAAGTCTGCTCATGTCCACGTCACGG ACTCCCCTCCTGAGCTCTCTGAGTGCAACCAGCAACAATTCCACCGTGGCTTGCATTGATAGAAATGGGCTGCAAAGCTGTCCCATCAAG GAAGACAGCTTCTTGCAGCGATACAGCTCAGACCCCACAGGCGCCTTGACTGAGGACAGCATAGACGACACCTTCCTCCCAGTGCCTGGT GAGTGGCTTGTCTGGAAACAGTCCTGCTCCTCAACCTCCTCGACCCACTCAGCAGCAGCCAGTCTCCAGTGTCCAAGCCAGGTGCTCCCT CCAGCATCTCCAGAGGGGGAAACAGTGGCAGATTTGCAGACACAGTGAAGGGCGTAAGGAGCAGATAAACACATGACCGAGCCTGCACAA GCTCTTTGTTGTGTCTGGTTGTTTGCTGTACCTCTGTTGTAAGAATGAATCTGCAAAATTTCTAGCTTATGAAGCAAATCACGGACATAC ACATCTGTGTGTGTGAGTGTTCATGATGTGTGTACATCTGTGTATGTGTGTGTGTGTATGTGTGTGTTTGTGACAGATTTGATCCCTGTT >29135_29135_5_FAM65A-EGFR_FAM65A_chr16_67576379_ENST00000540839_EGFR_chr7_55269015_ENST00000455089_length(amino acids)=420AA_BP= MCPGCSLRGRALGLPHSANAQHLRRVMTIWQMQKQAQRGSPARTHSMMSLSVRPQRRLLSARVNRSQSFAGVLGSHERGPRSFPVFSPPG PPRKPPALSRVSRMFSVAHPAAKVPQPERLDLVYTALKRGLTAYLEVHQQEQEKLQGQIRESKRNSRLGFLYDLDKQVKSIERFLRRLEF HASKIDELYEAYCVQRRLRDGAYNMVRAYTTGSPGSREARDSLAEATRGHREYTESMCLLESELEAQLGEFHLRMKGLAGFARLCVGDQY EICMKYGRQRWKLRGRIEGSGKQVWDSEETIFLPLLTEFLSIKVTELKGLANHVVVGSVSCETKDLFAALPQVVAVDINDLGTIKLSLEV -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FAM65A-EGFR |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FAM65A-EGFR |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FAM65A-EGFR |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
