
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FARP1-SLC15A1 (FusionGDB2 ID:29384) |
Fusion Gene Summary for FARP1-SLC15A1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FARP1-SLC15A1 | Fusion gene ID: 29384 | Hgene | Tgene | Gene symbol | FARP1 | SLC15A1 | Gene ID | 10160 | 6564 |
| Gene name | FERM, ARH/RhoGEF and pleckstrin domain protein 1 | solute carrier family 15 member 1 | |
| Synonyms | CDEP|FARP1-IT1|PLEKHC2|PPP1R75 | HPECT1|HPEPT1|PEPT1 | |
| Cytomap | 13q32.2 | 13q32.2-q32.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | FERM, ARHGEF and pleckstrin domain-containing protein 1FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)FERM, RhoGEF and pleckstrin domain-containing protein 1PH domain-containing family C member 2chondrocyte-derived ezrin-li | solute carrier family 15 member 1Caco-2 oligopeptide transporterintestinal H+/peptide cotransportermacrophage oligopeptide transporter PEPT1oligopeptide transporter, small intestine isoformpeptide transporter 1solute carrier family 15 (oligopeptide | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | Q9Y4F1 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000319562, ENST00000376586, ENST00000595437, ENST00000376581, ENST00000593990, | ENST00000376503, | |
| Fusion gene scores | * DoF score | 19 X 13 X 10=2470 | 5 X 4 X 3=60 |
| # samples | 22 | 5 | |
| ** MAII score | log2(22/2470*10)=-3.48893561294738 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/60*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FARP1 [Title/Abstract] AND SLC15A1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FARP1(99064288)-SLC15A1(99340831), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | FARP1 | GO:0010923 | negative regulation of phosphatase activity | 19389623 |
 Fusion gene breakpoints across FARP1 (5'-gene) Fusion gene breakpoints across FARP1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
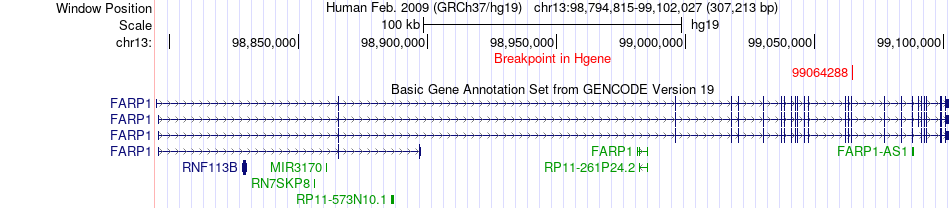 |
 Fusion gene breakpoints across SLC15A1 (3'-gene) Fusion gene breakpoints across SLC15A1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
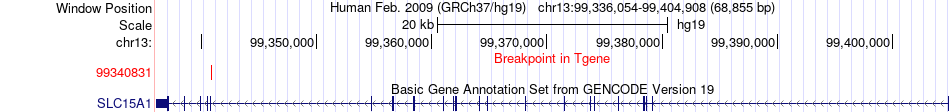 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-13-1411-01A | FARP1 | chr13 | 99064288 | + | SLC15A1 | chr13 | 99340831 | - |
Top |
Fusion Gene ORF analysis for FARP1-SLC15A1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000319562 | ENST00000376503 | FARP1 | chr13 | 99064288 | + | SLC15A1 | chr13 | 99340831 | - |
| In-frame | ENST00000376586 | ENST00000376503 | FARP1 | chr13 | 99064288 | + | SLC15A1 | chr13 | 99340831 | - |
| In-frame | ENST00000595437 | ENST00000376503 | FARP1 | chr13 | 99064288 | + | SLC15A1 | chr13 | 99340831 | - |
| intron-3CDS | ENST00000376581 | ENST00000376503 | FARP1 | chr13 | 99064288 | + | SLC15A1 | chr13 | 99340831 | - |
| intron-3CDS | ENST00000593990 | ENST00000376503 | FARP1 | chr13 | 99064288 | + | SLC15A1 | chr13 | 99340831 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000595437 | FARP1 | chr13 | 99064288 | + | ENST00000376503 | SLC15A1 | chr13 | 99340831 | - | 3649 | 2065 | 239 | 2065 | 608 |
| ENST00000376586 | FARP1 | chr13 | 99064288 | + | ENST00000376503 | SLC15A1 | chr13 | 99340831 | - | 3746 | 2162 | 336 | 2162 | 608 |
| ENST00000319562 | FARP1 | chr13 | 99064288 | + | ENST00000376503 | SLC15A1 | chr13 | 99340831 | - | 3675 | 2091 | 265 | 2091 | 608 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000595437 | ENST00000376503 | FARP1 | chr13 | 99064288 | + | SLC15A1 | chr13 | 99340831 | - | 0.005504732 | 0.9944952 |
| ENST00000376586 | ENST00000376503 | FARP1 | chr13 | 99064288 | + | SLC15A1 | chr13 | 99340831 | - | 0.005479111 | 0.9945209 |
| ENST00000319562 | ENST00000376503 | FARP1 | chr13 | 99064288 | + | SLC15A1 | chr13 | 99340831 | - | 0.005178219 | 0.9948218 |
Top |
Fusion Genomic Features for FARP1-SLC15A1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
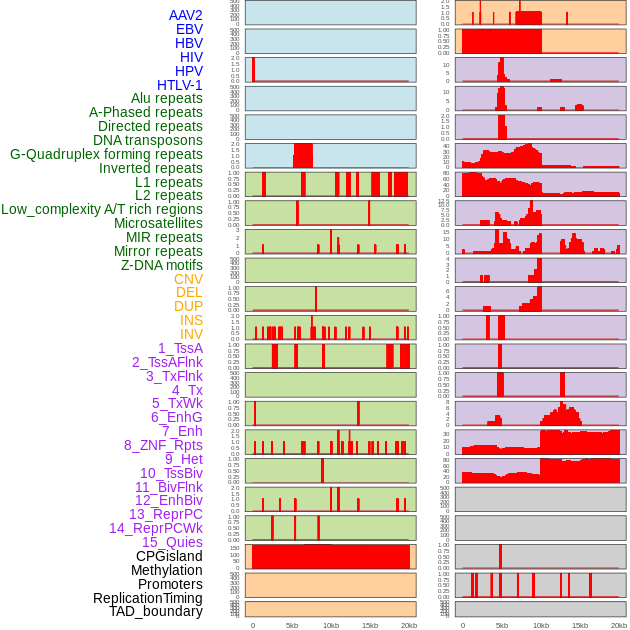 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for FARP1-SLC15A1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr13:99064288/chr13:99340831) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FARP1 | . |
| FUNCTION: Functions as guanine nucleotide exchange factor for RAC1. May play a role in semaphorin signaling. Plays a role in the assembly and disassembly of dendritic filopodia, the formation of dendritic spines, regulation of dendrite length and ultimately the formation of synapses (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FARP1 | chr13:99064288 | chr13:99340831 | ENST00000319562 | + | 16 | 27 | 449_453 | 608 | 1046.0 | Compositional bias | Note=Poly-Glu |
| Hgene | FARP1 | chr13:99064288 | chr13:99340831 | ENST00000319562 | + | 16 | 27 | 40_320 | 608 | 1046.0 | Domain | FERM |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 606_619 | 488 | 709.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 641_645 | 488 | 709.0 | Topological domain | Extracellular | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 667_708 | 488 | 709.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 585_605 | 488 | 709.0 | Transmembrane | Helical | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 620_640 | 488 | 709.0 | Transmembrane | Helical | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 646_666 | 488 | 709.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FARP1 | chr13:99064288 | chr13:99340831 | ENST00000376581 | + | 1 | 3 | 449_453 | 0 | 130.0 | Compositional bias | Note=Poly-Glu |
| Hgene | FARP1 | chr13:99064288 | chr13:99340831 | ENST00000319562 | + | 16 | 27 | 540_730 | 608 | 1046.0 | Domain | DH |
| Hgene | FARP1 | chr13:99064288 | chr13:99340831 | ENST00000319562 | + | 16 | 27 | 759_856 | 608 | 1046.0 | Domain | PH 1 |
| Hgene | FARP1 | chr13:99064288 | chr13:99340831 | ENST00000319562 | + | 16 | 27 | 932_1029 | 608 | 1046.0 | Domain | PH 2 |
| Hgene | FARP1 | chr13:99064288 | chr13:99340831 | ENST00000376581 | + | 1 | 3 | 40_320 | 0 | 130.0 | Domain | FERM |
| Hgene | FARP1 | chr13:99064288 | chr13:99340831 | ENST00000376581 | + | 1 | 3 | 540_730 | 0 | 130.0 | Domain | DH |
| Hgene | FARP1 | chr13:99064288 | chr13:99340831 | ENST00000376581 | + | 1 | 3 | 759_856 | 0 | 130.0 | Domain | PH 1 |
| Hgene | FARP1 | chr13:99064288 | chr13:99340831 | ENST00000376581 | + | 1 | 3 | 932_1029 | 0 | 130.0 | Domain | PH 2 |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 383_584 | 488 | 709.0 | Region | Extracellular domain (ECD) | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 104_118 | 488 | 709.0 | Topological domain | Extracellular | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 140_161 | 488 | 709.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 183_198 | 488 | 709.0 | Topological domain | Extracellular | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 220_276 | 488 | 709.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 22_53 | 488 | 709.0 | Topological domain | Extracellular | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 298_327 | 488 | 709.0 | Topological domain | Extracellular | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 349_361 | 488 | 709.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 75_82 | 488 | 709.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 119_139 | 488 | 709.0 | Transmembrane | Helical | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 162_182 | 488 | 709.0 | Transmembrane | Helical | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 199_219 | 488 | 709.0 | Transmembrane | Helical | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 1_21 | 488 | 709.0 | Transmembrane | Helical | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 277_297 | 488 | 709.0 | Transmembrane | Helical | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 328_348 | 488 | 709.0 | Transmembrane | Helical | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 362_382 | 488 | 709.0 | Transmembrane | Helical | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 54_74 | 488 | 709.0 | Transmembrane | Helical | |
| Tgene | SLC15A1 | chr13:99064288 | chr13:99340831 | ENST00000376503 | 17 | 23 | 83_103 | 488 | 709.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for FARP1-SLC15A1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >29384_29384_1_FARP1-SLC15A1_FARP1_chr13_99064288_ENST00000319562_SLC15A1_chr13_99340831_ENST00000376503_length(transcript)=3675nt_BP=2091nt GCGACCCGGAGCCCGCTCCCCACCCACCCCGCCTGCTCCGCCCTCCCCTCCGCCCCGCGCCACCTTTGATGGCTCGGACCTCAGCCGGCC ACCGCCAGCCCTGCTCGCGCGCCCGCGCCGCCGCCGCCCGCGGGTATTAATAGCCGGCGCCGCCGCGCCCTCGGCCGCCGGGGGCTTGGG AGCCGCCGATCCCGGAGCCCGAGCCGGGAGAGGGAGCCGCCGCAGCCGCCGGCGCTGTGGAGATATTCTCTAAGCCGCTTTCATCATGGG AGAAATAGAGCAGAGGCCGACCCCAGGATCACGACTGGGGGCCCCGGAAAATTCGGGGATCAGTACCTTGGAACGTGGACAGAAGCCGCC CCCAACACCTTCAGGAAAACTCGTGTCCATCAAAATCCAGATGCTGGATGACACCCAGGAGGCATTTGAAGTTCCACAAAGAGCTCCTGG GAAGGTGCTGCTGGATGCAGTTTGCAACCACCTCAACCTCGTGGAAGGTGACTATTTTGGCCTCGAGTTTCCTGATCACAAAAAGATCAC GGTGTGGCTGGATCTCCTAAAACCCATTGTGAAACAGATTAGAAGGCCAAAGCACGTTGTTGTTAAGTTTGTGGTGAAATTCTTTCCGCC TGACCACACACAACTCCAAGAAGAACTCACAAGGTACCTGTTCGCGCTGCAGGTGAAGCAGGACTTGGCTCAAGGCAGGTTGACGTGTAA TGACACCAGCGCAGCTCTCTTGATTTCACACATTGTGCAATCTGAGATTGGGGATTTTGATGAAGCCTTGGACAGAGAGCACTTAGCAAA AAATAAATACATACCTCAGCAAGACGCACTAGAGGACAAAATCGTGGAATTTCACCATAACCACATTGGACAAACACCAGCAGAATCAGA TTTCCAGCTCCTAGAGATTGCCCGTCGGCTAGAGATGTATGGAATCCGGTTGCACCCGGCCAAGGACAGGGAAGGCACGAAGATCAATCT GGCCGTTGCCAACACGGGAATTCTAGTGTTTCAGGGTTTCACTAAGATCAATGCCTTCAACTGGGCCAAGGTGCGGAAGCTGAGCTTCAA GAGGAAGCGCTTTCTCATCAAGCTCCGGCCAGATGCCAATAGTGCGTACCAGGATACCTTGGAATTCCTGATGGCCAGTCGGGATTTCTG CAAGTCCTTCTGGAAAATCTGTGTTGAACATCATGCCTTCTTTAGACTTTTTGAAGAGCCCAAACCAAAGCCCAAGCCCGTCCTCTTTAG CCGGGGGTCATCATTTCGGTTCAGTGGTCGGACTCAGAAGCAGGTTCTCGACTATGTTAAAGAAGGAGGACATAAGAAGGTGCAGTTTGA AAGGAAGCACAGCAAGATTCATTCTATCCGGAGCCTTGCTTCACAGCCTACAGAACTGAATTCGGAAGTGCTGGAGCAGTCTCAGCAGAG CACCAGCCTTACATTTGGAGAAGGTGCCGAATCTCCAGGGGGCCAGAGCTGCCGGCGAGGAAAGGAACCGAAGGTTTCCGCCGGGGAGCC GGGGTCGCACCCGAGCCCTGCGCCGAGGAGAAGCCCCGCGGGTAACAAGCAGGCGGACGGAGCCGCCTCGGCGCCCACGGAGGAAGAGGA GGAGGTCGTTAAGGATAGGACCCAGCAGAGTAAACCTCAGCCCCCGCAGCCAAGCACAGGCTCCCTGACTGGCAGTCCTCACCTTTCCGA GCTGTCTGTGAACTCGCAGGGGGGAGTGGCCCCTGCCAACGTGACCTTGTCTCCCAACCTGAGCCCCGACACCAAGCAGGCCTCTCCCTT GATCAGCCCGCTGCTGAATGACCAGGCCTGCCCCCGGACGGACGATGAGGATGAGGGCCGGAGGAAGAGATTCCCAACTGATAAAGCGTA CTTCATAGCTAAGGAAGTGTCTACCACCGAGCGAACATATCTGAAGGATCTCGAAGTTATCACTTCGTGGTTTCAGAGCACAGTGAGCAA AGAGGACGCCATGCCGGAAGCACTGAAAAGTCTCATATTCCCGAATTTTGAACCTTTGCACAAATTTCATACTAATTTTCTCAAGGAAAT TGAGCAACGACTTGCCCTGTGATTTGTAAATACTTTTAACGAGCTCATCACCATCACAATGAGTGGGAAAGTTTATGCAAACATCAGCAG CTACAATGCCAGCACATACCAGTTTTTTCCTTCTGGCATAAAAGGCTTCACAATAAGCTCAACAGAGATTCCGCCACAATGTCAACCTAA TTTCAATACTTTCTACCTTGAATTTGGTAGTGCTTATACCTATATAGTCCAAAGGAAGAATGACAGCTGCCCTGAAGTGAAGGTGTTTGA AGATATTTCAGCCAACACAGTTAACATGGCTCTGCAAATCCCGCAGTATTTTCTTCTCACCTGTGGCGAAGTGGTCTTCTCTGTCACGGG ATTGGAATTCTCATATTCTCAGGCTCCTTCCAACATGAAGTCGGTGCTTCAGGCAGGATGGCTGCTGACCGTGGCTGTTGGCAACATCAT TGTGCTCATCGTGGCAGGGGCAGGCCAGTTCAGCAAACAGTGGGCCGAGTACATTCTATTTGCCGCGTTGCTTCTGGTCGTCTGTGTAAT TTTTGCCATCATGGCTCGGTTCTATACTTACATCAACCCAGCGGAGATCGAAGCTCAATTTGATGAGGATGAAAAGAAAAACAGACTGGA AAAGAGTAACCCATATTTCATGTCAGGGGCCAATTCACAGAAACAGATGTGAAGGTCAGGAGGCAAGTGGAGGATGGACTGGGCCCGCAG ATGCCCTGACCTCTGCCCCCAGGTAGCAGGACACTCCATTGGATGGCCCCTGATGAGGAAGACTTCAGAATTGGGAACTAAACCATGAAT GCTATTTTCTTTTTTCTTTTTCTTTTCTTTTTTTTTTTTTTTTTTTTTTTGAGACAGAGTTTTGCTCTTGTTGTCCAGGCTGGAGTGCAA TGGCACGATCTCAGCTCACTGCAACCTCCGCCTCCCAGGTTCAAGTAATTCTCCTGCCTCAGCCTCCCGAGTGGCTGGGATTAGCGGCAT GCACCACCACGCCCAGCTATTTTTGTATTTTTAGTAGAGATGGGGTTTCACCATGTTGGCCAGGATGGTCTCGATCTCTTGACCTGGTGA TCTGCCCACCTCGGCCTGCCAAAGTGCTGGGATTACAGGCTTGAGCTACCGCGCCCGGCCGTGAACGCTATTTTCTAAGCAGCCAGCAGT GAATCTAAAACTCTGGAAGAAGTCTTCTGTTTGAAAGGCTTATTTAAGCCACACGTACACACACTGTCTTAGAGTACTGTGAGCCCACCC CACATTGGTCATCTTCCCTATCACACAAATGATGTTATTTTGGACTAGCTTAATTTTGAAATGGTAACAAAGTTTCCTATTCCATACTGT TCATTTCTAATACTCTTACGAAAACTATTCTAAAGGAGGCAGGAGCCAAGGCCAAAAGTGAACGTACAGGTTTGAAATGGCTGTGATAAG GACCAGCTGGTATTAACTGATAACTTTACCTTTGGGTTTTTGTTATTTTGTTTTTCTAGTCCCTACCTGTGTTTAAATTATGGATAACTC >29384_29384_1_FARP1-SLC15A1_FARP1_chr13_99064288_ENST00000319562_SLC15A1_chr13_99340831_ENST00000376503_length(amino acids)=608AA_BP= MGEIEQRPTPGSRLGAPENSGISTLERGQKPPPTPSGKLVSIKIQMLDDTQEAFEVPQRAPGKVLLDAVCNHLNLVEGDYFGLEFPDHKK ITVWLDLLKPIVKQIRRPKHVVVKFVVKFFPPDHTQLQEELTRYLFALQVKQDLAQGRLTCNDTSAALLISHIVQSEIGDFDEALDREHL AKNKYIPQQDALEDKIVEFHHNHIGQTPAESDFQLLEIARRLEMYGIRLHPAKDREGTKINLAVANTGILVFQGFTKINAFNWAKVRKLS FKRKRFLIKLRPDANSAYQDTLEFLMASRDFCKSFWKICVEHHAFFRLFEEPKPKPKPVLFSRGSSFRFSGRTQKQVLDYVKEGGHKKVQ FERKHSKIHSIRSLASQPTELNSEVLEQSQQSTSLTFGEGAESPGGQSCRRGKEPKVSAGEPGSHPSPAPRRSPAGNKQADGAASAPTEE EEEVVKDRTQQSKPQPPQPSTGSLTGSPHLSELSVNSQGGVAPANVTLSPNLSPDTKQASPLISPLLNDQACPRTDDEDEGRRKRFPTDK -------------------------------------------------------------- >29384_29384_2_FARP1-SLC15A1_FARP1_chr13_99064288_ENST00000376586_SLC15A1_chr13_99340831_ENST00000376503_length(transcript)=3746nt_BP=2162nt GCGGCCGCTGCCCGCTTTGCGCCGCTCCTCCCTGCGCGAGTAGCGCTGGCCCCGGCGTCGAGGCGGCCATGGCGACCCGGAGCCCGCTCC CCACCCACCCCGCCTGCTCCGCCCTCCCCTCCGCCCCGCGCCACCTTTGATGGCTCGGACCTCAGCCGGCCACCGCCAGCCCTGCTCGCG CGCCCGCGCCGCCGCCGCCCGCGGGTATTAATAGCCGGCGCCGCCGCGCCCTCGGCCGCCGGGGGCTTGGGAGCCGCCGATCCCGGAGCC CGAGCCGGGAGAGGGAGCCGCCGCAGCCGCCGGCGCTGTGGAGATATTCTCTAAGCCGCTTTCATCATGGGAGAAATAGAGCAGAGGCCG ACCCCAGGATCACGACTGGGGGCCCCGGAAAATTCGGGGATCAGTACCTTGGAACGTGGACAGAAGCCGCCCCCAACACCTTCAGGAAAA CTCGTGTCCATCAAAATCCAGATGCTGGATGACACCCAGGAGGCATTTGAAGTTCCACAAAGAGCTCCTGGGAAGGTGCTGCTGGATGCA GTTTGCAACCACCTCAACCTCGTGGAAGGTGACTATTTTGGCCTCGAGTTTCCTGATCACAAAAAGATCACGGTGTGGCTGGATCTCCTA AAACCCATTGTGAAACAGATTAGAAGGCCAAAGCACGTTGTTGTTAAGTTTGTGGTGAAATTCTTTCCGCCTGACCACACACAACTCCAA GAAGAACTCACAAGGTACCTGTTCGCGCTGCAGGTGAAGCAGGACTTGGCTCAAGGCAGGTTGACGTGTAATGACACCAGCGCAGCTCTC TTGATTTCACACATTGTGCAATCTGAGATTGGGGATTTTGATGAAGCCTTGGACAGAGAGCACTTAGCAAAAAATAAATACATACCTCAG CAAGACGCACTAGAGGACAAAATCGTGGAATTTCACCATAACCACATTGGACAAACACCAGCAGAATCAGATTTCCAGCTCCTAGAGATT GCCCGTCGGCTAGAGATGTATGGAATCCGGTTGCACCCGGCCAAGGACAGGGAAGGCACGAAGATCAATCTGGCCGTTGCCAACACGGGA ATTCTAGTGTTTCAGGGTTTCACTAAGATCAATGCCTTCAACTGGGCCAAGGTGCGGAAGCTGAGCTTCAAGAGGAAGCGCTTTCTCATC AAGCTCCGGCCAGATGCCAATAGTGCGTACCAGGATACCTTGGAATTCCTGATGGCCAGTCGGGATTTCTGCAAGTCCTTCTGGAAAATC TGTGTTGAACATCATGCCTTCTTTAGACTTTTTGAAGAGCCCAAACCAAAGCCCAAGCCCGTCCTCTTTAGCCGGGGGTCATCATTTCGG TTCAGTGGTCGGACTCAGAAGCAGGTTCTCGACTATGTTAAAGAAGGAGGACATAAGAAGGTGCAGTTTGAAAGGAAGCACAGCAAGATT CATTCTATCCGGAGCCTTGCTTCACAGCCTACAGAACTGAATTCGGAAGTGCTGGAGCAGTCTCAGCAGAGCACCAGCCTTACATTTGGA GAAGGTGCCGAATCTCCAGGGGGCCAGAGCTGCCGGCGAGGAAAGGAACCGAAGGTTTCCGCCGGGGAGCCGGGGTCGCACCCGAGCCCT GCGCCGAGGAGAAGCCCCGCGGGTAACAAGCAGGCGGACGGAGCCGCCTCGGCGCCCACGGAGGAAGAGGAGGAGGTCGTTAAGGATAGG ACCCAGCAGAGTAAACCTCAGCCCCCGCAGCCAAGCACAGGCTCCCTGACTGGCAGTCCTCACCTTTCCGAGCTGTCTGTGAACTCGCAG GGGGGAGTGGCCCCTGCCAACGTGACCTTGTCTCCCAACCTGAGCCCCGACACCAAGCAGGCCTCTCCCTTGATCAGCCCGCTGCTGAAT GACCAGGCCTGCCCCCGGACGGACGATGAGGATGAGGGCCGGAGGAAGAGATTCCCAACTGATAAAGCGTACTTCATAGCTAAGGAAGTG TCTACCACCGAGCGAACATATCTGAAGGATCTCGAAGTTATCACTTCGTGGTTTCAGAGCACAGTGAGCAAAGAGGACGCCATGCCGGAA GCACTGAAAAGTCTCATATTCCCGAATTTTGAACCTTTGCACAAATTTCATACTAATTTTCTCAAGGAAATTGAGCAACGACTTGCCCTG TGATTTGTAAATACTTTTAACGAGCTCATCACCATCACAATGAGTGGGAAAGTTTATGCAAACATCAGCAGCTACAATGCCAGCACATAC CAGTTTTTTCCTTCTGGCATAAAAGGCTTCACAATAAGCTCAACAGAGATTCCGCCACAATGTCAACCTAATTTCAATACTTTCTACCTT GAATTTGGTAGTGCTTATACCTATATAGTCCAAAGGAAGAATGACAGCTGCCCTGAAGTGAAGGTGTTTGAAGATATTTCAGCCAACACA GTTAACATGGCTCTGCAAATCCCGCAGTATTTTCTTCTCACCTGTGGCGAAGTGGTCTTCTCTGTCACGGGATTGGAATTCTCATATTCT CAGGCTCCTTCCAACATGAAGTCGGTGCTTCAGGCAGGATGGCTGCTGACCGTGGCTGTTGGCAACATCATTGTGCTCATCGTGGCAGGG GCAGGCCAGTTCAGCAAACAGTGGGCCGAGTACATTCTATTTGCCGCGTTGCTTCTGGTCGTCTGTGTAATTTTTGCCATCATGGCTCGG TTCTATACTTACATCAACCCAGCGGAGATCGAAGCTCAATTTGATGAGGATGAAAAGAAAAACAGACTGGAAAAGAGTAACCCATATTTC ATGTCAGGGGCCAATTCACAGAAACAGATGTGAAGGTCAGGAGGCAAGTGGAGGATGGACTGGGCCCGCAGATGCCCTGACCTCTGCCCC CAGGTAGCAGGACACTCCATTGGATGGCCCCTGATGAGGAAGACTTCAGAATTGGGAACTAAACCATGAATGCTATTTTCTTTTTTCTTT TTCTTTTCTTTTTTTTTTTTTTTTTTTTTTTGAGACAGAGTTTTGCTCTTGTTGTCCAGGCTGGAGTGCAATGGCACGATCTCAGCTCAC TGCAACCTCCGCCTCCCAGGTTCAAGTAATTCTCCTGCCTCAGCCTCCCGAGTGGCTGGGATTAGCGGCATGCACCACCACGCCCAGCTA TTTTTGTATTTTTAGTAGAGATGGGGTTTCACCATGTTGGCCAGGATGGTCTCGATCTCTTGACCTGGTGATCTGCCCACCTCGGCCTGC CAAAGTGCTGGGATTACAGGCTTGAGCTACCGCGCCCGGCCGTGAACGCTATTTTCTAAGCAGCCAGCAGTGAATCTAAAACTCTGGAAG AAGTCTTCTGTTTGAAAGGCTTATTTAAGCCACACGTACACACACTGTCTTAGAGTACTGTGAGCCCACCCCACATTGGTCATCTTCCCT ATCACACAAATGATGTTATTTTGGACTAGCTTAATTTTGAAATGGTAACAAAGTTTCCTATTCCATACTGTTCATTTCTAATACTCTTAC GAAAACTATTCTAAAGGAGGCAGGAGCCAAGGCCAAAAGTGAACGTACAGGTTTGAAATGGCTGTGATAAGGACCAGCTGGTATTAACTG ATAACTTTACCTTTGGGTTTTTGTTATTTTGTTTTTCTAGTCCCTACCTGTGTTTAAATTATGGATAACTCGAAAGACAGCTCAGGTGAA >29384_29384_2_FARP1-SLC15A1_FARP1_chr13_99064288_ENST00000376586_SLC15A1_chr13_99340831_ENST00000376503_length(amino acids)=608AA_BP= MGEIEQRPTPGSRLGAPENSGISTLERGQKPPPTPSGKLVSIKIQMLDDTQEAFEVPQRAPGKVLLDAVCNHLNLVEGDYFGLEFPDHKK ITVWLDLLKPIVKQIRRPKHVVVKFVVKFFPPDHTQLQEELTRYLFALQVKQDLAQGRLTCNDTSAALLISHIVQSEIGDFDEALDREHL AKNKYIPQQDALEDKIVEFHHNHIGQTPAESDFQLLEIARRLEMYGIRLHPAKDREGTKINLAVANTGILVFQGFTKINAFNWAKVRKLS FKRKRFLIKLRPDANSAYQDTLEFLMASRDFCKSFWKICVEHHAFFRLFEEPKPKPKPVLFSRGSSFRFSGRTQKQVLDYVKEGGHKKVQ FERKHSKIHSIRSLASQPTELNSEVLEQSQQSTSLTFGEGAESPGGQSCRRGKEPKVSAGEPGSHPSPAPRRSPAGNKQADGAASAPTEE EEEVVKDRTQQSKPQPPQPSTGSLTGSPHLSELSVNSQGGVAPANVTLSPNLSPDTKQASPLISPLLNDQACPRTDDEDEGRRKRFPTDK -------------------------------------------------------------- >29384_29384_3_FARP1-SLC15A1_FARP1_chr13_99064288_ENST00000595437_SLC15A1_chr13_99340831_ENST00000376503_length(transcript)=3649nt_BP=2065nt AAGTCCCGCGCAGCCCGGGGTGGTGCCGGAAGCCCGTGCCTGTGGCTCCGGGGGAGGGGAGGGCTCGGTGGGTCCCTGAAGACTGTCCTC CCCGCGCGACGTCGCAGGCGAAGCAGCCCCAGGCAGGACCTCAAAAAGCGACCTGCAGAGTCCGAGCCCCTGACCGCCCGCCTGCCGCTC CCGGCTTCGCCGTGCACACGTCTCCCGCTCTCCCCGATATTCTCTAAGCCGCTTTCATCATGGGAGAAATAGAGCAGAGGCCGACCCCAG GATCACGACTGGGGGCCCCGGAAAATTCGGGGATCAGTACCTTGGAACGTGGACAGAAGCCGCCCCCAACACCTTCAGGAAAACTCGTGT CCATCAAAATCCAGATGCTGGATGACACCCAGGAGGCATTTGAAGTTCCACAAAGAGCTCCTGGGAAGGTGCTGCTGGATGCAGTTTGCA ACCACCTCAACCTCGTGGAAGGTGACTATTTTGGCCTCGAGTTTCCTGATCACAAAAAGATCACGGTGTGGCTGGATCTCCTAAAACCCA TTGTGAAACAGATTAGAAGGCCAAAGCACGTTGTTGTTAAGTTTGTGGTGAAATTCTTTCCGCCTGACCACACACAACTCCAAGAAGAAC TCACAAGGTACCTGTTCGCGCTGCAGGTGAAGCAGGACTTGGCTCAAGGCAGGTTGACGTGTAATGACACCAGCGCAGCTCTCTTGATTT CACACATTGTGCAATCTGAGATTGGGGATTTTGATGAAGCCTTGGACAGAGAGCACTTAGCAAAAAATAAATACATACCTCAGCAAGACG CACTAGAGGACAAAATCGTGGAATTTCACCATAACCACATTGGACAAACACCAGCAGAATCAGATTTCCAGCTCCTAGAGATTGCCCGTC GGCTAGAGATGTATGGAATCCGGTTGCACCCGGCCAAGGACAGGGAAGGCACGAAGATCAATCTGGCCGTTGCCAACACGGGAATTCTAG TGTTTCAGGGTTTCACTAAGATCAATGCCTTCAACTGGGCCAAGGTGCGGAAGCTGAGCTTCAAGAGGAAGCGCTTTCTCATCAAGCTCC GGCCAGATGCCAATAGTGCGTACCAGGATACCTTGGAATTCCTGATGGCCAGTCGGGATTTCTGCAAGTCCTTCTGGAAAATCTGTGTTG AACATCATGCCTTCTTTAGACTTTTTGAAGAGCCCAAACCAAAGCCCAAGCCCGTCCTCTTTAGCCGGGGGTCATCATTTCGGTTCAGTG GTCGGACTCAGAAGCAGGTTCTCGACTATGTTAAAGAAGGAGGACATAAGAAGGTGCAGTTTGAAAGGAAGCACAGCAAGATTCATTCTA TCCGGAGCCTTGCTTCACAGCCTACAGAACTGAATTCGGAAGTGCTGGAGCAGTCTCAGCAGAGCACCAGCCTTACATTTGGAGAAGGTG CCGAATCTCCAGGGGGCCAGAGCTGCCGGCGAGGAAAGGAACCGAAGGTTTCCGCCGGGGAGCCGGGGTCGCACCCGAGCCCTGCGCCGA GGAGAAGCCCCGCGGGTAACAAGCAGGCGGACGGAGCCGCCTCGGCGCCCACGGAGGAAGAGGAGGAGGTCGTTAAGGATAGGACCCAGC AGAGTAAACCTCAGCCCCCGCAGCCAAGCACAGGCTCCCTGACTGGCAGTCCTCACCTTTCCGAGCTGTCTGTGAACTCGCAGGGGGGAG TGGCCCCTGCCAACGTGACCTTGTCTCCCAACCTGAGCCCCGACACCAAGCAGGCCTCTCCCTTGATCAGCCCGCTGCTGAATGACCAGG CCTGCCCCCGGACGGACGATGAGGATGAGGGCCGGAGGAAGAGATTCCCAACTGATAAAGCGTACTTCATAGCTAAGGAAGTGTCTACCA CCGAGCGAACATATCTGAAGGATCTCGAAGTTATCACTTCGTGGTTTCAGAGCACAGTGAGCAAAGAGGACGCCATGCCGGAAGCACTGA AAAGTCTCATATTCCCGAATTTTGAACCTTTGCACAAATTTCATACTAATTTTCTCAAGGAAATTGAGCAACGACTTGCCCTGTGATTTG TAAATACTTTTAACGAGCTCATCACCATCACAATGAGTGGGAAAGTTTATGCAAACATCAGCAGCTACAATGCCAGCACATACCAGTTTT TTCCTTCTGGCATAAAAGGCTTCACAATAAGCTCAACAGAGATTCCGCCACAATGTCAACCTAATTTCAATACTTTCTACCTTGAATTTG GTAGTGCTTATACCTATATAGTCCAAAGGAAGAATGACAGCTGCCCTGAAGTGAAGGTGTTTGAAGATATTTCAGCCAACACAGTTAACA TGGCTCTGCAAATCCCGCAGTATTTTCTTCTCACCTGTGGCGAAGTGGTCTTCTCTGTCACGGGATTGGAATTCTCATATTCTCAGGCTC CTTCCAACATGAAGTCGGTGCTTCAGGCAGGATGGCTGCTGACCGTGGCTGTTGGCAACATCATTGTGCTCATCGTGGCAGGGGCAGGCC AGTTCAGCAAACAGTGGGCCGAGTACATTCTATTTGCCGCGTTGCTTCTGGTCGTCTGTGTAATTTTTGCCATCATGGCTCGGTTCTATA CTTACATCAACCCAGCGGAGATCGAAGCTCAATTTGATGAGGATGAAAAGAAAAACAGACTGGAAAAGAGTAACCCATATTTCATGTCAG GGGCCAATTCACAGAAACAGATGTGAAGGTCAGGAGGCAAGTGGAGGATGGACTGGGCCCGCAGATGCCCTGACCTCTGCCCCCAGGTAG CAGGACACTCCATTGGATGGCCCCTGATGAGGAAGACTTCAGAATTGGGAACTAAACCATGAATGCTATTTTCTTTTTTCTTTTTCTTTT CTTTTTTTTTTTTTTTTTTTTTTTGAGACAGAGTTTTGCTCTTGTTGTCCAGGCTGGAGTGCAATGGCACGATCTCAGCTCACTGCAACC TCCGCCTCCCAGGTTCAAGTAATTCTCCTGCCTCAGCCTCCCGAGTGGCTGGGATTAGCGGCATGCACCACCACGCCCAGCTATTTTTGT ATTTTTAGTAGAGATGGGGTTTCACCATGTTGGCCAGGATGGTCTCGATCTCTTGACCTGGTGATCTGCCCACCTCGGCCTGCCAAAGTG CTGGGATTACAGGCTTGAGCTACCGCGCCCGGCCGTGAACGCTATTTTCTAAGCAGCCAGCAGTGAATCTAAAACTCTGGAAGAAGTCTT CTGTTTGAAAGGCTTATTTAAGCCACACGTACACACACTGTCTTAGAGTACTGTGAGCCCACCCCACATTGGTCATCTTCCCTATCACAC AAATGATGTTATTTTGGACTAGCTTAATTTTGAAATGGTAACAAAGTTTCCTATTCCATACTGTTCATTTCTAATACTCTTACGAAAACT ATTCTAAAGGAGGCAGGAGCCAAGGCCAAAAGTGAACGTACAGGTTTGAAATGGCTGTGATAAGGACCAGCTGGTATTAACTGATAACTT TACCTTTGGGTTTTTGTTATTTTGTTTTTCTAGTCCCTACCTGTGTTTAAATTATGGATAACTCGAAAGACAGCTCAGGTGAAGGCCAGT >29384_29384_3_FARP1-SLC15A1_FARP1_chr13_99064288_ENST00000595437_SLC15A1_chr13_99340831_ENST00000376503_length(amino acids)=608AA_BP= MGEIEQRPTPGSRLGAPENSGISTLERGQKPPPTPSGKLVSIKIQMLDDTQEAFEVPQRAPGKVLLDAVCNHLNLVEGDYFGLEFPDHKK ITVWLDLLKPIVKQIRRPKHVVVKFVVKFFPPDHTQLQEELTRYLFALQVKQDLAQGRLTCNDTSAALLISHIVQSEIGDFDEALDREHL AKNKYIPQQDALEDKIVEFHHNHIGQTPAESDFQLLEIARRLEMYGIRLHPAKDREGTKINLAVANTGILVFQGFTKINAFNWAKVRKLS FKRKRFLIKLRPDANSAYQDTLEFLMASRDFCKSFWKICVEHHAFFRLFEEPKPKPKPVLFSRGSSFRFSGRTQKQVLDYVKEGGHKKVQ FERKHSKIHSIRSLASQPTELNSEVLEQSQQSTSLTFGEGAESPGGQSCRRGKEPKVSAGEPGSHPSPAPRRSPAGNKQADGAASAPTEE EEEVVKDRTQQSKPQPPQPSTGSLTGSPHLSELSVNSQGGVAPANVTLSPNLSPDTKQASPLISPLLNDQACPRTDDEDEGRRKRFPTDK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FARP1-SLC15A1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FARP1-SLC15A1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FARP1-SLC15A1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
