
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FBXW11-SPIDR (FusionGDB2 ID:29889) |
Fusion Gene Summary for FBXW11-SPIDR |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FBXW11-SPIDR | Fusion gene ID: 29889 | Hgene | Tgene | Gene symbol | FBXW11 | SPIDR | Gene ID | 23291 | 23514 |
| Gene name | F-box and WD repeat domain containing 11 | scaffold protein involved in DNA repair | |
| Synonyms | BTRC2|BTRCP2|FBW1B|FBXW1B|Fbw11|Hos | KIAA0146 | |
| Cytomap | 5q35.1 | 8q11.21 | |
| Type of gene | protein-coding | protein-coding | |
| Description | F-box/WD repeat-containing protein 11F-box and WD repeats protein beta-TrCP2F-box and WD-40 domain protein 11F-box and WD-40 domain protein 1BF-box protein Fbw1bF-box/WD repeat-containing protein 1Bbeta-transducin repeat-containing protein 2homolog | DNA repair-scaffolding proteinscaffolding protein involved in DNA repair | |
| Modification date | 20200329 | 20200320 | |
| UniProtAcc | Q9UKB1 | Q14159 | |
| Ensembl transtripts involved in fusion gene | ENST00000265094, ENST00000296933, ENST00000393802, ENST00000425623, ENST00000522891, | ENST00000521214, ENST00000517693, ENST00000518060, ENST00000297423, ENST00000518074, ENST00000541342, | |
| Fusion gene scores | * DoF score | 13 X 6 X 9=702 | 19 X 18 X 7=2394 |
| # samples | 13 | 22 | |
| ** MAII score | log2(13/702*10)=-2.43295940727611 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(22/2394*10)=-3.44384772341884 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FBXW11 [Title/Abstract] AND SPIDR [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FBXW11(171337576)-SPIDR(48352885), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | FBXW11-SPIDR seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. FBXW11-SPIDR seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | FBXW11 | GO:0000209 | protein polyubiquitination | 20347421 |
| Hgene | FBXW11 | GO:0016567 | protein ubiquitination | 16885022 |
| Hgene | FBXW11 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 20347421 |
| Hgene | FBXW11 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 20347421 |
| Tgene | SPIDR | GO:0006974 | cellular response to DNA damage stimulus | 23509288 |
| Tgene | SPIDR | GO:0010569 | regulation of double-strand break repair via homologous recombination | 23754376 |
| Tgene | SPIDR | GO:0031334 | positive regulation of protein complex assembly | 23509288 |
| Tgene | SPIDR | GO:0070202 | regulation of establishment of protein localization to chromosome | 23509288 |
| Tgene | SPIDR | GO:0071479 | cellular response to ionizing radiation | 23509288|23754376 |
| Tgene | SPIDR | GO:0072711 | cellular response to hydroxyurea | 23509288 |
| Tgene | SPIDR | GO:0072757 | cellular response to camptothecin | 23509288 |
 Fusion gene breakpoints across FBXW11 (5'-gene) Fusion gene breakpoints across FBXW11 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
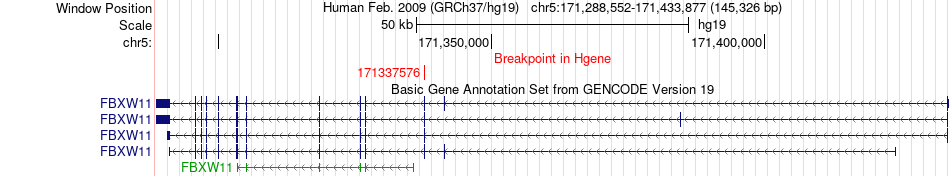 |
 Fusion gene breakpoints across SPIDR (3'-gene) Fusion gene breakpoints across SPIDR (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
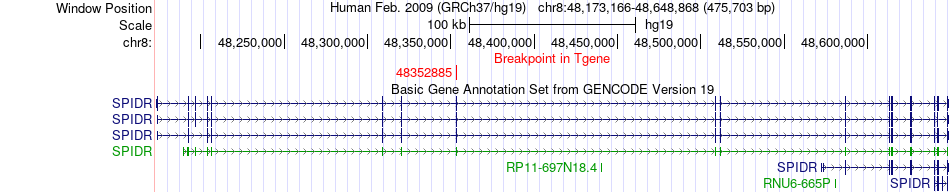 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-8078-01A | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
Top |
Fusion Gene ORF analysis for FBXW11-SPIDR |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000265094 | ENST00000521214 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| 5CDS-3UTR | ENST00000296933 | ENST00000521214 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| 5CDS-3UTR | ENST00000393802 | ENST00000521214 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| 5CDS-3UTR | ENST00000425623 | ENST00000521214 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| 5CDS-intron | ENST00000265094 | ENST00000517693 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| 5CDS-intron | ENST00000265094 | ENST00000518060 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| 5CDS-intron | ENST00000296933 | ENST00000517693 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| 5CDS-intron | ENST00000296933 | ENST00000518060 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| 5CDS-intron | ENST00000393802 | ENST00000517693 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| 5CDS-intron | ENST00000393802 | ENST00000518060 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| 5CDS-intron | ENST00000425623 | ENST00000517693 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| 5CDS-intron | ENST00000425623 | ENST00000518060 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| Frame-shift | ENST00000265094 | ENST00000297423 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| Frame-shift | ENST00000265094 | ENST00000518074 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| Frame-shift | ENST00000265094 | ENST00000541342 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| Frame-shift | ENST00000296933 | ENST00000297423 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| Frame-shift | ENST00000296933 | ENST00000518074 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| Frame-shift | ENST00000296933 | ENST00000541342 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| Frame-shift | ENST00000393802 | ENST00000297423 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| Frame-shift | ENST00000393802 | ENST00000518074 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| Frame-shift | ENST00000393802 | ENST00000541342 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| In-frame | ENST00000425623 | ENST00000297423 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| In-frame | ENST00000425623 | ENST00000518074 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| In-frame | ENST00000425623 | ENST00000541342 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| intron-3CDS | ENST00000522891 | ENST00000297423 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| intron-3CDS | ENST00000522891 | ENST00000518074 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| intron-3CDS | ENST00000522891 | ENST00000541342 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| intron-3UTR | ENST00000522891 | ENST00000521214 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| intron-intron | ENST00000522891 | ENST00000517693 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
| intron-intron | ENST00000522891 | ENST00000518060 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000425623 | FBXW11 | chr5 | 171337576 | - | ENST00000297423 | SPIDR | chr8 | 48352885 | + | 3114 | 387 | 110 | 2257 | 715 |
| ENST00000425623 | FBXW11 | chr5 | 171337576 | - | ENST00000518074 | SPIDR | chr8 | 48352885 | + | 2338 | 387 | 110 | 2320 | 736 |
| ENST00000425623 | FBXW11 | chr5 | 171337576 | - | ENST00000541342 | SPIDR | chr8 | 48352885 | + | 2319 | 387 | 110 | 2257 | 715 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000425623 | ENST00000297423 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + | 0.000804142 | 0.9991959 |
| ENST00000425623 | ENST00000518074 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + | 0.002421513 | 0.99757856 |
| ENST00000425623 | ENST00000541342 | FBXW11 | chr5 | 171337576 | - | SPIDR | chr8 | 48352885 | + | 0.001495602 | 0.9985044 |
Top |
Fusion Genomic Features for FBXW11-SPIDR |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| FBXW11 | chr5 | 171337575 | - | SPIDR | chr8 | 48352884 | + | 0.000571163 | 0.9994288 |
| FBXW11 | chr5 | 171337575 | - | SPIDR | chr8 | 48352884 | + | 0.000571163 | 0.9994288 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
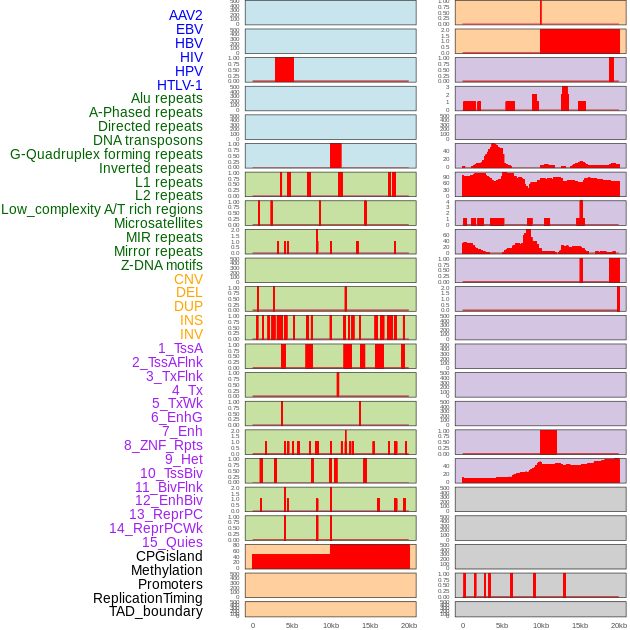 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
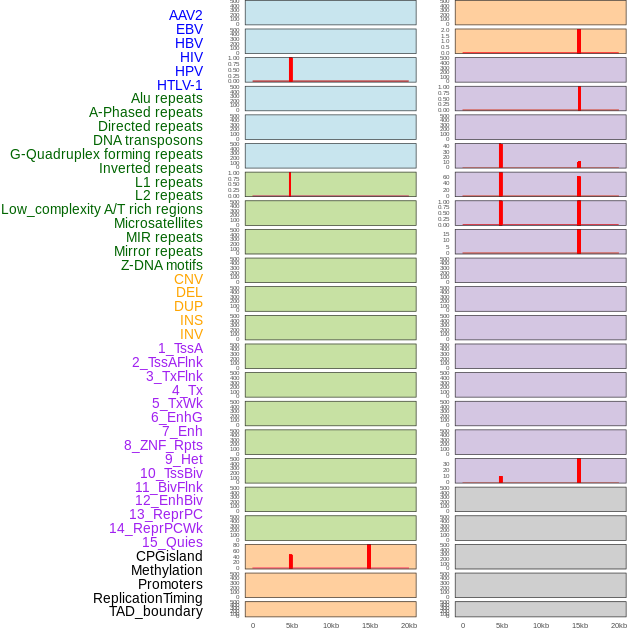 |
Top |
Fusion Protein Features for FBXW11-SPIDR |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:171337576/chr8:48352885) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FBXW11 | SPIDR |
| FUNCTION: Substrate recognition component of a SCF (SKP1-CUL1-F-box protein) E3 ubiquitin-protein ligase complex which mediates the ubiquitination and subsequent proteasomal degradation of target proteins. Probably recognizes and binds to phosphorylated target proteins. SCF(FBXW11) mediates the ubiquitination of phosphorylated CTNNB1 and participates in Wnt signaling. SCF(FBXW11) mediates the ubiquitination of phosphorylated NFKBIA, which degradation frees the associated NFKB1 to translocate into the nucleus and to activate transcription. SCF(FBXW11) mediates the ubiquitination of IFNAR1. SCF(FBXW11) mediates the ubiquitination of CEP68; this is required for centriole separation during mitosis (PubMed:25503564). Involved in the oxidative stress-induced a ubiquitin-mediated decrease in RCAN1. Mediates the degradation of CDC25A induced by ionizing radiation in cells progressing through S phase and thus may function in the intra-S-phase checkpoint. Has an essential role in the control of the clock-dependent transcription via degradation of phosphorylated PER1 and phosphorylated PER2. SCF(FBXW11) mediates the ubiquitination of CYTH1, and probably CYTH2 (PubMed:29420262). {ECO:0000269|PubMed:10321728, ECO:0000269|PubMed:10437795, ECO:0000269|PubMed:10644755, ECO:0000269|PubMed:10648623, ECO:0000269|PubMed:14532120, ECO:0000269|PubMed:14603323, ECO:0000269|PubMed:15917222, ECO:0000269|PubMed:18575781, ECO:0000269|PubMed:19966869, ECO:0000269|PubMed:20347421, ECO:0000269|PubMed:25503564, ECO:0000269|PubMed:29420262}.; FUNCTION: (Microbial infection) Target of human immunodeficiency virus type 1 (HIV-1) protein VPU to polyubiquitinate and deplete BST2 from cells and antagonize its antiviral action. {ECO:0000269|PubMed:19730691}. | FUNCTION: Plays a role in DNA double-strand break (DBS) repair via homologous recombination (HR). Serves as a scaffolding protein that helps to promote the recruitment of DNA-processing enzymes like the helicase BLM and recombinase RAD51 to site of DNA damage, and hence contributes to maintain genomic integrity. {ECO:0000269|PubMed:23509288, ECO:0000269|PubMed:23754376}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000265094 | - | 3 | 13 | 67_116 | 124 | 1360.3333333333333 | Region | Homodimerization domain D |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000265094 | - | 3 | 13 | 129_167 | 124 | 1360.3333333333333 | Domain | F-box |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000296933 | - | 3 | 13 | 129_167 | 111 | 1283.6666666666667 | Domain | F-box |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000393802 | - | 2 | 12 | 129_167 | 90 | 592.0 | Domain | F-box |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000296933 | - | 3 | 13 | 67_116 | 111 | 1283.6666666666667 | Region | Homodimerization domain D |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000393802 | - | 2 | 12 | 67_116 | 90 | 592.0 | Region | Homodimerization domain D |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000265094 | - | 3 | 13 | 238_275 | 124 | 1360.3333333333333 | Repeat | Note=WD 1 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000265094 | - | 3 | 13 | 278_315 | 124 | 1360.3333333333333 | Repeat | Note=WD 2 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000265094 | - | 3 | 13 | 318_355 | 124 | 1360.3333333333333 | Repeat | Note=WD 3 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000265094 | - | 3 | 13 | 361_398 | 124 | 1360.3333333333333 | Repeat | Note=WD 4 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000265094 | - | 3 | 13 | 401_440 | 124 | 1360.3333333333333 | Repeat | Note=WD 5 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000265094 | - | 3 | 13 | 442_478 | 124 | 1360.3333333333333 | Repeat | Note=WD 6 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000265094 | - | 3 | 13 | 490_527 | 124 | 1360.3333333333333 | Repeat | Note=WD 7 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000296933 | - | 3 | 13 | 238_275 | 111 | 1283.6666666666667 | Repeat | Note=WD 1 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000296933 | - | 3 | 13 | 278_315 | 111 | 1283.6666666666667 | Repeat | Note=WD 2 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000296933 | - | 3 | 13 | 318_355 | 111 | 1283.6666666666667 | Repeat | Note=WD 3 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000296933 | - | 3 | 13 | 361_398 | 111 | 1283.6666666666667 | Repeat | Note=WD 4 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000296933 | - | 3 | 13 | 401_440 | 111 | 1283.6666666666667 | Repeat | Note=WD 5 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000296933 | - | 3 | 13 | 442_478 | 111 | 1283.6666666666667 | Repeat | Note=WD 6 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000296933 | - | 3 | 13 | 490_527 | 111 | 1283.6666666666667 | Repeat | Note=WD 7 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000393802 | - | 2 | 12 | 238_275 | 90 | 592.0 | Repeat | Note=WD 1 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000393802 | - | 2 | 12 | 278_315 | 90 | 592.0 | Repeat | Note=WD 2 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000393802 | - | 2 | 12 | 318_355 | 90 | 592.0 | Repeat | Note=WD 3 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000393802 | - | 2 | 12 | 361_398 | 90 | 592.0 | Repeat | Note=WD 4 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000393802 | - | 2 | 12 | 401_440 | 90 | 592.0 | Repeat | Note=WD 5 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000393802 | - | 2 | 12 | 442_478 | 90 | 592.0 | Repeat | Note=WD 6 |
| Hgene | FBXW11 | chr5:171337576 | chr8:48352885 | ENST00000393802 | - | 2 | 12 | 490_527 | 90 | 592.0 | Repeat | Note=WD 7 |
Top |
Fusion Gene Sequence for FBXW11-SPIDR |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >29889_29889_1_FBXW11-SPIDR_FBXW11_chr5_171337576_ENST00000425623_SPIDR_chr8_48352885_ENST00000297423_length(transcript)=3114nt_BP=387nt GGAAGATGCTTGACTGCTGTCTTTTCTCAGACTAGTCTGAAAGGGAAAACCCCCATTCATTCTGGATTGGTGGATCTGAAGTACAAAATA TTTTCCAGAACACTTCAGTTATGGAAGATCAAAATGAAGATGAGTCCCCAAAGAAAAATACTCTTTGGCAGATAAGTAATGGAACATCAT CTGTGATCGTCTCCAGAAAGAGGCCATCAGAAGGAAACTATCAAAAAGAAAAAGACTTGTGTATTAAATATTTTGACCAGTGGTCTGAAT CAGATCAAGTGGAATTTGTGGAACATCTTATTTCACGAATGTGTCATTATCAGCATGGACATATTAACTCTTACCTGAAGCCCATGTTGC AGCGGGACTTTATTACCGCTTTACCAGGTAGAAAATCTGGTGTATTAACTGTGAAAATTTTAGAGCTGCATGAGGAATGTGCCATGCAAG TTGCCATGTGTGAGCAGTTATTGGGGTCACCAGCCACCAGCTCCTCCCAAAGTGTGGCTCCCAGGCCTGGAGCTGGCCTGAAAGTTCTCT TCACCAAGGAGACTGCAGGCTACCTCAGGGGCCGTCCCCAGGACACTGTCCGGATCTTCCCTCCCTGGCAAAAACTGATTATTCCAAGTG GAAGTTGCCCTGTTATTCTGAATACTTACTTTTGTGAGAAAGTTGTTGCCAAAGAAGATTCAGAAAAAACTTGTGAAGTGTACTGTCCGG ACATACCCCTTCCAAGAAGAAGCATCTCTTTGGCCCAGATGTTTGTAATTAAGGGTCTAACAAATAATTCACCTGAAATCCAGGTTGTGT GTAGTGGTGTAGCCACTACAGGGACAGCCTGGACCCATGGGCACAAAGAAGCAAAACAGCGCATCCCAACCAGCACTCCCCTGAGGGATT CTCTCCTGGATGTGGTGGAAAGCCAGGGAGCTGCCTCGTGGCCAGGAGCTGGAGTCCGAGTGGTGGTGCAAAGAGTGTATTCTCTTCCCA GCAGAGACAGCACCAGGGGTCAGCAGGGGGCCAGCTCAGGACACACAGACCCAGCTGGAACTCGAGCCTGCCTTCTGGTACAAGATGCCT GTGGAATGTTCGGTGAAGTGCACTTGGAGTTCACCATGTCGAAGGCAAGACAGTTGGAAGGGAAGTCTTGCAGCCTGGTGGGAATGAAGG TTCTACAGAAAGTCACCAGAGGAAGGACAGCGGGGATTTTCAGTTTGATTGACACCCTGTGGCCCCCAGCGATACCTCTGAAAACACCTG GCCGCGACCAGCCCTGTGAAGAGATAAAAACTCATCTGCCTCCTCCAGCCTTGTGTTACATCCTCACAGCTCATCCAAATCTGGGACAAA TTGATATAATTGACGAAGACCCCATTTATAAGCTTTACCAGCCTCCAGTTACCCGCTGCTTAAGAGACATTCTCCAGATGAATGATCTTG GTACCCGTTGCAGTTTCTATGCCACGGTGATTTACCAAAAACCACAGCTGAAGAGTCTGCTGCTTCTGGAGCAAAGGGAGATCTGGCTGC TAGTGACCGATGTCACTCTGCAAACGAAGGAGGAGAGAGACCCCAGGCTCCCCAAAACCCTGCTGGTCTATGTGGCCCCCTTGTGTGTGC TGGGCTCTGAAGTCCTGGAGGCACTCGCTGGGGCTGCCCCTCACAGCCTCTTCTTCAAGGACGCTCTCCGTGACCAGGGTCGGATTGTTT GTGCTGAACGAACTGTCCTCTTGCTTCAGAAGCCCCTTTTGAGTGTGGTCTCTGGTGCAAGTTCCTGTGAGCTGCCTGGCCCGGTGATGC TCGACAGCCTGGACTCTGCAACACCTGTCAACTCCATCTGCAGTGTTCAAGGCACTGTGGTTGGCGTGGACGAGAGCACTGCTTTCTCAT GGCCTGTGTGTGACATGTGTGGCAACGGGAGATTGGAACAGAGGCCGGAAGACAGAGGCGCCTTTTCCTGTGGGGACTGCTCCCGGGTGG TCACATCTCCTGTTCTCAAGAGGCACCTGCAGGTCTTCCTGGACTGCCGCTCAAGACCGCAGTGCAGAGTGAAGGTCAAGCTGTTGCAGC GCAGCATTTCCTCCCTGCTGAGGTTTGCCGCCGGTGAAGATGGGAGCTACGAAGTGAAGAGTGTCCTCGGAAAGGAAGTGGGGTTGTTAA ATTGTTTTGTCCAGTCCGTAACCGCCCACCCGACCAGCTGCATTGGATTGGAGGAAATCGAGCTTCTGAGTGCAGGAGGGGCCTCTGCAG AACACTAGCGGTTGCCGCAGGATCTGTGAACTTTGCAATGTGGCTGCAAGGGTGGTGGTGGTGGTGGTGATTTGGGGTAGTTATTTGTTA ACTATGGACACAGTGAACGTAGTTTACGATCTTGAAATGAAACTTAGATTTTTCTGGGGAAATGTTCAGATACAGTTTTGTGAACTGTAA ATCAAAATACCTTTTTCTACAGTTTATCTTTTATTTTCTGCAAATTTAGGAACATATTTACTCGTTTTCACATTGAATCTTAAGTTTAAG CTCTTCATTTGGTATTTAGGCAATATATGAGAAAAAAATTTTTTTTGTTCATTTGTAATTTTAACAAGTTGAACATTTTACCATGATTGA ACATGTTTTTATTACAGTATTTAACATTCCCCCAAAGAATACCCTGCAAAGTGTAAACCTTTGTCCCATACTGTGATATTACTGTTCTGC TACAATAAATGTCAAACCTAAGCACTTTGCAGTTCACTACTTTTGGGAAAATGTTCTAGGGAACTGTATCACAGGTGAAACTGTTACCCA TAAAGTGTAGCTCTCTGAACTGGTGTGATCGTGTCTCTGCTGAATGGTGGTGGGGATAGCTGGCTCCACCACACACACCTGTTTTAATTA GGCTGGGTAATGTGCACGGAGAGCAAAGATCACAGACTAAGGAGCAGTCAGTCGGATATTTGCTTTGCTGGCAAAGATTTAAACCAAGAA TTAATTATCTTTGGACACTATAAAGATTTTGTTCCCTTCCTTATAGCAATATAATCATGACAGGCCATGGTTAACTACATCAGATACACG >29889_29889_1_FBXW11-SPIDR_FBXW11_chr5_171337576_ENST00000425623_SPIDR_chr8_48352885_ENST00000297423_length(amino acids)=715AA_BP=90 MEDQNEDESPKKNTLWQISNGTSSVIVSRKRPSEGNYQKEKDLCIKYFDQWSESDQVEFVEHLISRMCHYQHGHINSYLKPMLQRDFITA LPGRKSGVLTVKILELHEECAMQVAMCEQLLGSPATSSSQSVAPRPGAGLKVLFTKETAGYLRGRPQDTVRIFPPWQKLIIPSGSCPVIL NTYFCEKVVAKEDSEKTCEVYCPDIPLPRRSISLAQMFVIKGLTNNSPEIQVVCSGVATTGTAWTHGHKEAKQRIPTSTPLRDSLLDVVE SQGAASWPGAGVRVVVQRVYSLPSRDSTRGQQGASSGHTDPAGTRACLLVQDACGMFGEVHLEFTMSKARQLEGKSCSLVGMKVLQKVTR GRTAGIFSLIDTLWPPAIPLKTPGRDQPCEEIKTHLPPPALCYILTAHPNLGQIDIIDEDPIYKLYQPPVTRCLRDILQMNDLGTRCSFY ATVIYQKPQLKSLLLLEQREIWLLVTDVTLQTKEERDPRLPKTLLVYVAPLCVLGSEVLEALAGAAPHSLFFKDALRDQGRIVCAERTVL LLQKPLLSVVSGASSCELPGPVMLDSLDSATPVNSICSVQGTVVGVDESTAFSWPVCDMCGNGRLEQRPEDRGAFSCGDCSRVVTSPVLK -------------------------------------------------------------- >29889_29889_2_FBXW11-SPIDR_FBXW11_chr5_171337576_ENST00000425623_SPIDR_chr8_48352885_ENST00000518074_length(transcript)=2338nt_BP=387nt GGAAGATGCTTGACTGCTGTCTTTTCTCAGACTAGTCTGAAAGGGAAAACCCCCATTCATTCTGGATTGGTGGATCTGAAGTACAAAATA TTTTCCAGAACACTTCAGTTATGGAAGATCAAAATGAAGATGAGTCCCCAAAGAAAAATACTCTTTGGCAGATAAGTAATGGAACATCAT CTGTGATCGTCTCCAGAAAGAGGCCATCAGAAGGAAACTATCAAAAAGAAAAAGACTTGTGTATTAAATATTTTGACCAGTGGTCTGAAT CAGATCAAGTGGAATTTGTGGAACATCTTATTTCACGAATGTGTCATTATCAGCATGGACATATTAACTCTTACCTGAAGCCCATGTTGC AGCGGGACTTTATTACCGCTTTACCAGGTAGAAAATCTGGTGTATTAACTGTGAAAATTTTAGAGCTGCATGAGGAATGTGCCATGCAAG TTGCCATGTGTGAGCAGTTATTGGGGTCACCAGCCACCAGCTCCTCCCAAAGTGTGGCTCCCAGGCCTGGAGCTGGCCTGAAAGTTCTCT TCACCAAGGAGACTGCAGGCTACCTCAGGGGCCGTCCCCAGGACACTGTCCGGATCTTCCCTCCCTGGCAAAAACTGATTATTCCAAGTG GAAGTTGCCCTGTTATTCTGAATACTTACTTTTGTGAGAAAGTTGTTGCCAAAGAAGATTCAGAAAAAACTTGTGAAGTGTACTGTCCGG ACATACCCCTTCCAAGAAGAAGCATCTCTTTGGCCCAGATGTTTGTAATTAAGGGTCTAACAAATAATTCACCTGAAATCCAGGTTGTGT GTAGTGGTGTAGCCACTACAGGGACAGCCTGGACCCATGGGCACAAAGAAGCAAAACAGCGCATCCCAACCAGCACTCCCCTGAGGGATT CTCTCCTGGATGTGGTGGAAAGCCAGGGAGCTGCCTCGTGGCCAGGAGCTGGAGTCCGAGTGGTGGTGCAAAGAGTGTATTCTCTTCCCA GCAGAGACAGCACCAGGGGTCAGCAGGGGGCCAGCTCAGGACACACAGACCCAGCTGGAACTCGAGCCTGCCTTCTGGTACAAGATGCCT GTGGAATGTTCGGTGAAGTGCACTTGGAGTTCACCATGTCGAAGGCAAGACAGTTGGAAGGGAAGTCTTGCAGCCTGGTGGGAATGAAGG TTCTACAGAAAGTCACCAGAGGAAGGACAGCGGGGATTTTCAGTTTGATTGACACCCTGTGGCCCCCAGCGATACCTCTGAAAACACCTG GCCGCGACCAGCCCTGTGAAGAGATAAAAACTCATCTGCCTCCTCCAGCCTTGTGTTACATCCTCACAGCTCATCCAAATCTGGGACAAA TTGATATAATTGACGAAGACCCCATTTATAAGCTTTACCAGCCTCCAGTTACCCGCTGCTTAAGAGACATTCTCCAGATGAATGATCTTG GTACCCGTTGCAGTTTCTATGCCACGGTGATTTACCAAAAACCACAGCTGAAGAGTCTGCTGCTTCTGGAGCAAAGGGAGATCTGGCTGC TAGTGACCGATGTCACTCTGCAAACGAAGGAGGAGAGAGACCCCAGGCTCCCCAAAACCCTGCTGGTCTATGTGGCCCCCTTGTGTGTGC TGGGCTCTGAAGTCCTGGAGGCACTCGCTGGGGCTGCCCCTCACAGCCTCTTCTTCAAGGACGCTCTCCGTGACCAGGGTCGGATTGTTT GTGCTGAACGAACTGTCCTCTTGCTTCAGAAGCCCCTTTTGAGTGTGGTCTCTGGTGCAAGTTCCTGTGAGCTGCCTGGCCCGGTGATGC TCGACAGCCTGGACTCTGCAACACCTGTCAACTCCATCTGCAGTGTTCAAGGCACTGTGGTTGGCGTGGACGAGAGCACTGCTTTCTCAT GGCCTGTGTGTGACATGTGTGGCAACGGGAGATTGGAACAGAGGCCGGAAGACAGAGGCGCCTTTTCCTGTGGGGACTGCTCCCGGGTGG TCACATCTCCTGTTCTCAAGAGGCACCTGCAGGTCTTCCTGGACTGCCGCTCAAGACCGCAGTGCAGAGTGAAGGTCAAGGTAGGAGCCA GGCCAGAGCACGCACGCACTCCTAGCTCACTCCAACATAGCGAAGAGCTACGAAGTGAAGAGTGTCCTCGGAAAGGAAGTGGGGTTGTTA AATTGTTTTGTCCAGTCCGTAACCGCCCACCCGACCAGCTGCATTGGATTGGAGGAAATCGAGCTTCTGAGTGCAGGAGGGGCCTCTGCA >29889_29889_2_FBXW11-SPIDR_FBXW11_chr5_171337576_ENST00000425623_SPIDR_chr8_48352885_ENST00000518074_length(amino acids)=736AA_BP=90 MEDQNEDESPKKNTLWQISNGTSSVIVSRKRPSEGNYQKEKDLCIKYFDQWSESDQVEFVEHLISRMCHYQHGHINSYLKPMLQRDFITA LPGRKSGVLTVKILELHEECAMQVAMCEQLLGSPATSSSQSVAPRPGAGLKVLFTKETAGYLRGRPQDTVRIFPPWQKLIIPSGSCPVIL NTYFCEKVVAKEDSEKTCEVYCPDIPLPRRSISLAQMFVIKGLTNNSPEIQVVCSGVATTGTAWTHGHKEAKQRIPTSTPLRDSLLDVVE SQGAASWPGAGVRVVVQRVYSLPSRDSTRGQQGASSGHTDPAGTRACLLVQDACGMFGEVHLEFTMSKARQLEGKSCSLVGMKVLQKVTR GRTAGIFSLIDTLWPPAIPLKTPGRDQPCEEIKTHLPPPALCYILTAHPNLGQIDIIDEDPIYKLYQPPVTRCLRDILQMNDLGTRCSFY ATVIYQKPQLKSLLLLEQREIWLLVTDVTLQTKEERDPRLPKTLLVYVAPLCVLGSEVLEALAGAAPHSLFFKDALRDQGRIVCAERTVL LLQKPLLSVVSGASSCELPGPVMLDSLDSATPVNSICSVQGTVVGVDESTAFSWPVCDMCGNGRLEQRPEDRGAFSCGDCSRVVTSPVLK RHLQVFLDCRSRPQCRVKVKVGARPEHARTPSSLQHSEELRSEECPRKGSGVVKLFCPVRNRPPDQLHWIGGNRASECRRGLCRTLAVAA -------------------------------------------------------------- >29889_29889_3_FBXW11-SPIDR_FBXW11_chr5_171337576_ENST00000425623_SPIDR_chr8_48352885_ENST00000541342_length(transcript)=2319nt_BP=387nt GGAAGATGCTTGACTGCTGTCTTTTCTCAGACTAGTCTGAAAGGGAAAACCCCCATTCATTCTGGATTGGTGGATCTGAAGTACAAAATA TTTTCCAGAACACTTCAGTTATGGAAGATCAAAATGAAGATGAGTCCCCAAAGAAAAATACTCTTTGGCAGATAAGTAATGGAACATCAT CTGTGATCGTCTCCAGAAAGAGGCCATCAGAAGGAAACTATCAAAAAGAAAAAGACTTGTGTATTAAATATTTTGACCAGTGGTCTGAAT CAGATCAAGTGGAATTTGTGGAACATCTTATTTCACGAATGTGTCATTATCAGCATGGACATATTAACTCTTACCTGAAGCCCATGTTGC AGCGGGACTTTATTACCGCTTTACCAGGTAGAAAATCTGGTGTATTAACTGTGAAAATTTTAGAGCTGCATGAGGAATGTGCCATGCAAG TTGCCATGTGTGAGCAGTTATTGGGGTCACCAGCCACCAGCTCCTCCCAAAGTGTGGCTCCCAGGCCTGGAGCTGGCCTGAAAGTTCTCT TCACCAAGGAGACTGCAGGCTACCTCAGGGGCCGTCCCCAGGACACTGTCCGGATCTTCCCTCCCTGGCAAAAACTGATTATTCCAAGTG GAAGTTGCCCTGTTATTCTGAATACTTACTTTTGTGAGAAAGTTGTTGCCAAAGAAGATTCAGAAAAAACTTGTGAAGTGTACTGTCCGG ACATACCCCTTCCAAGAAGAAGCATCTCTTTGGCCCAGATGTTTGTAATTAAGGGTCTAACAAATAATTCACCTGAAATCCAGGTTGTGT GTAGTGGTGTAGCCACTACAGGGACAGCCTGGACCCATGGGCACAAAGAAGCAAAACAGCGCATCCCAACCAGCACTCCCCTGAGGGATT CTCTCCTGGATGTGGTGGAAAGCCAGGGAGCTGCCTCGTGGCCAGGAGCTGGAGTCCGAGTGGTGGTGCAAAGAGTGTATTCTCTTCCCA GCAGAGACAGCACCAGGGGTCAGCAGGGGGCCAGCTCAGGACACACAGACCCAGCTGGAACTCGAGCCTGCCTTCTGGTACAAGATGCCT GTGGAATGTTCGGTGAAGTGCACTTGGAGTTCACCATGTCGAAGGCAAGACAGTTGGAAGGGAAGTCTTGCAGCCTGGTGGGAATGAAGG TTCTACAGAAAGTCACCAGAGGAAGGACAGCGGGGATTTTCAGTTTGATTGACACCCTGTGGCCCCCAGCGATACCTCTGAAAACACCTG GCCGCGACCAGCCCTGTGAAGAGATAAAAACTCATCTGCCTCCTCCAGCCTTGTGTTACATCCTCACAGCTCATCCAAATCTGGGACAAA TTGATATAATTGACGAAGACCCCATTTATAAGCTTTACCAGCCTCCAGTTACCCGCTGCTTAAGAGACATTCTCCAGATGAATGATCTTG GTACCCGTTGCAGTTTCTATGCCACGGTGATTTACCAAAAACCACAGCTGAAGAGTCTGCTGCTTCTGGAGCAAAGGGAGATCTGGCTGC TAGTGACCGATGTCACTCTGCAAACGAAGGAGGAGAGAGACCCCAGGCTCCCCAAAACCCTGCTGGTCTATGTGGCCCCCTTGTGTGTGC TGGGCTCTGAAGTCCTGGAGGCACTCGCTGGGGCTGCCCCTCACAGCCTCTTCTTCAAGGACGCTCTCCGTGACCAGGGTCGGATTGTTT GTGCTGAACGAACTGTCCTCTTGCTTCAGAAGCCCCTTTTGAGTGTGGTCTCTGGTGCAAGTTCCTGTGAGCTGCCTGGCCCGGTGATGC TCGACAGCCTGGACTCTGCAACACCTGTCAACTCCATCTGCAGTGTTCAAGGCACTGTGGTTGGCGTGGACGAGAGCACTGCTTTCTCAT GGCCTGTGTGTGACATGTGTGGCAACGGGAGATTGGAACAGAGGCCGGAAGACAGAGGCGCCTTTTCCTGTGGGGACTGCTCCCGGGTGG TCACATCTCCTGTTCTCAAGAGGCACCTGCAGGTCTTCCTGGACTGCCGCTCAAGACCGCAGTGCAGAGTGAAGGTCAAGCTGTTGCAGC GCAGCATTTCCTCCCTGCTGAGGTTTGCCGCCGGTGAAGATGGGAGCTACGAAGTGAAGAGTGTCCTCGGAAAGGAAGTGGGGTTGTTAA ATTGTTTTGTCCAGTCCGTAACCGCCCACCCGACCAGCTGCATTGGATTGGAGGAAATCGAGCTTCTGAGTGCAGGAGGGGCCTCTGCAG >29889_29889_3_FBXW11-SPIDR_FBXW11_chr5_171337576_ENST00000425623_SPIDR_chr8_48352885_ENST00000541342_length(amino acids)=715AA_BP=90 MEDQNEDESPKKNTLWQISNGTSSVIVSRKRPSEGNYQKEKDLCIKYFDQWSESDQVEFVEHLISRMCHYQHGHINSYLKPMLQRDFITA LPGRKSGVLTVKILELHEECAMQVAMCEQLLGSPATSSSQSVAPRPGAGLKVLFTKETAGYLRGRPQDTVRIFPPWQKLIIPSGSCPVIL NTYFCEKVVAKEDSEKTCEVYCPDIPLPRRSISLAQMFVIKGLTNNSPEIQVVCSGVATTGTAWTHGHKEAKQRIPTSTPLRDSLLDVVE SQGAASWPGAGVRVVVQRVYSLPSRDSTRGQQGASSGHTDPAGTRACLLVQDACGMFGEVHLEFTMSKARQLEGKSCSLVGMKVLQKVTR GRTAGIFSLIDTLWPPAIPLKTPGRDQPCEEIKTHLPPPALCYILTAHPNLGQIDIIDEDPIYKLYQPPVTRCLRDILQMNDLGTRCSFY ATVIYQKPQLKSLLLLEQREIWLLVTDVTLQTKEERDPRLPKTLLVYVAPLCVLGSEVLEALAGAAPHSLFFKDALRDQGRIVCAERTVL LLQKPLLSVVSGASSCELPGPVMLDSLDSATPVNSICSVQGTVVGVDESTAFSWPVCDMCGNGRLEQRPEDRGAFSCGDCSRVVTSPVLK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FBXW11-SPIDR |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FBXW11-SPIDR |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FBXW11-SPIDR |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
