
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FDPS-KHSRP (FusionGDB2 ID:30059) |
Fusion Gene Summary for FDPS-KHSRP |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FDPS-KHSRP | Fusion gene ID: 30059 | Hgene | Tgene | Gene symbol | FDPS | KHSRP | Gene ID | 2224 | 8570 |
| Gene name | farnesyl diphosphate synthase | KH-type splicing regulatory protein | |
| Synonyms | FPPS|FPS|POROK9 | FBP2|FUBP2|KSRP|p75 | |
| Cytomap | 1q22 | 19p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | farnesyl pyrophosphate synthase(2E,6E)-farnesyl diphosphate synthaseFPP synthaseFPP synthetasefarnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase | far upstream element-binding protein 2FUSE-binding protein 2epididymis secretory sperm binding protein | |
| Modification date | 20200329 | 20200329 | |
| UniProtAcc | P14324 | Q92945 | |
| Ensembl transtripts involved in fusion gene | ENST00000487002, ENST00000356657, ENST00000368356, ENST00000447866, | ENST00000398148, | |
| Fusion gene scores | * DoF score | 6 X 2 X 6=72 | 4 X 4 X 3=48 |
| # samples | 7 | 4 | |
| ** MAII score | log2(7/72*10)=-0.0406419844973459 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/48*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FDPS [Title/Abstract] AND KHSRP [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FDPS(155282186)-KHSRP(6422447), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | KHSRP | GO:0043488 | regulation of mRNA stability | 16126846 |
| Tgene | KHSRP | GO:0045019 | negative regulation of nitric oxide biosynthetic process | 16126846 |
| Tgene | KHSRP | GO:0061014 | positive regulation of mRNA catabolic process | 16126846 |
| Tgene | KHSRP | GO:0061158 | 3'-UTR-mediated mRNA destabilization | 16126846 |
 Fusion gene breakpoints across FDPS (5'-gene) Fusion gene breakpoints across FDPS (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
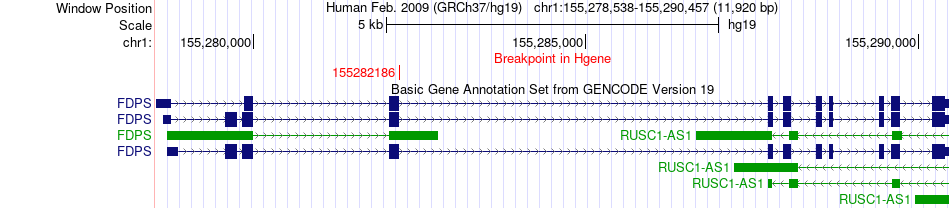 |
 Fusion gene breakpoints across KHSRP (3'-gene) Fusion gene breakpoints across KHSRP (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
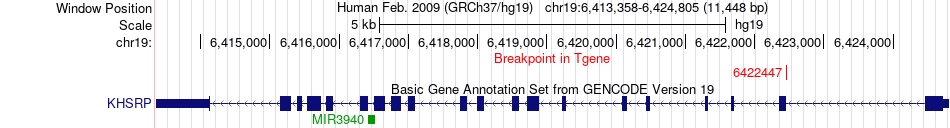 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-A4IZ | FDPS | chr1 | 155282186 | + | KHSRP | chr19 | 6422447 | - |
Top |
Fusion Gene ORF analysis for FDPS-KHSRP |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000487002 | ENST00000398148 | FDPS | chr1 | 155282186 | + | KHSRP | chr19 | 6422447 | - |
| In-frame | ENST00000356657 | ENST00000398148 | FDPS | chr1 | 155282186 | + | KHSRP | chr19 | 6422447 | - |
| In-frame | ENST00000368356 | ENST00000398148 | FDPS | chr1 | 155282186 | + | KHSRP | chr19 | 6422447 | - |
| In-frame | ENST00000447866 | ENST00000398148 | FDPS | chr1 | 155282186 | + | KHSRP | chr19 | 6422447 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000447866 | FDPS | chr1 | 155282186 | + | ENST00000398148 | KHSRP | chr19 | 6422447 | - | 3152 | 501 | 219 | 2387 | 722 |
| ENST00000368356 | FDPS | chr1 | 155282186 | + | ENST00000398148 | KHSRP | chr19 | 6422447 | - | 3246 | 595 | 115 | 2481 | 788 |
| ENST00000356657 | FDPS | chr1 | 155282186 | + | ENST00000398148 | KHSRP | chr19 | 6422447 | - | 3293 | 642 | 162 | 2528 | 788 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000447866 | ENST00000398148 | FDPS | chr1 | 155282186 | + | KHSRP | chr19 | 6422447 | - | 0.005805729 | 0.9941943 |
| ENST00000368356 | ENST00000398148 | FDPS | chr1 | 155282186 | + | KHSRP | chr19 | 6422447 | - | 0.002864284 | 0.99713576 |
| ENST00000356657 | ENST00000398148 | FDPS | chr1 | 155282186 | + | KHSRP | chr19 | 6422447 | - | 0.002336527 | 0.9976635 |
Top |
Fusion Genomic Features for FDPS-KHSRP |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
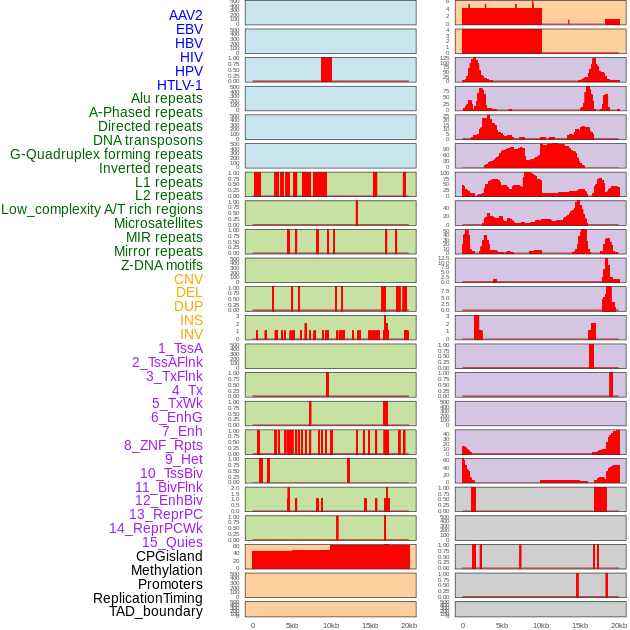 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for FDPS-KHSRP |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:155282186/chr19:6422447) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FDPS | KHSRP |
| FUNCTION: Key enzyme in isoprenoid biosynthesis which catalyzes the formation of farnesyl diphosphate (FPP), a precursor for several classes of essential metabolites including sterols, dolichols, carotenoids, and ubiquinones. FPP also serves as substrate for protein farnesylation and geranylgeranylation. Catalyzes the sequential condensation of isopentenyl pyrophosphate with the allylic pyrophosphates, dimethylallyl pyrophosphate, and then with the resultant geranylpyrophosphate to the ultimate product farnesyl pyrophosphate. | FUNCTION: Binds to the dendritic targeting element and may play a role in mRNA trafficking (By similarity). Part of a ternary complex that binds to the downstream control sequence (DCS) of the pre-mRNA. Mediates exon inclusion in transcripts that are subject to tissue-specific alternative splicing. May interact with single-stranded DNA from the far-upstream element (FUSE). May activate gene expression. Also involved in degradation of inherently unstable mRNAs that contain AU-rich elements (AREs) in their 3'-UTR, possibly by recruiting degradation machinery to ARE-containing mRNAs. {ECO:0000250, ECO:0000269|PubMed:11003644, ECO:0000269|PubMed:8940189, ECO:0000269|PubMed:9136930}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | KHSRP | chr1:155282186 | chr19:6422447 | ENST00000398148 | 0 | 20 | 498_612 | 83 | 712.0 | Compositional bias | Note=Ala/Gly/Pro-rich | |
| Tgene | KHSRP | chr1:155282186 | chr19:6422447 | ENST00000398148 | 0 | 20 | 144_208 | 83 | 712.0 | Domain | KH 1 | |
| Tgene | KHSRP | chr1:155282186 | chr19:6422447 | ENST00000398148 | 0 | 20 | 233_299 | 83 | 712.0 | Domain | KH 2 | |
| Tgene | KHSRP | chr1:155282186 | chr19:6422447 | ENST00000398148 | 0 | 20 | 322_386 | 83 | 712.0 | Domain | KH 3 | |
| Tgene | KHSRP | chr1:155282186 | chr19:6422447 | ENST00000398148 | 0 | 20 | 424_491 | 83 | 712.0 | Domain | KH 4 | |
| Tgene | KHSRP | chr1:155282186 | chr19:6422447 | ENST00000398148 | 0 | 20 | 571_684 | 83 | 712.0 | Region | Note=4 X 12 AA imperfect repeats | |
| Tgene | KHSRP | chr1:155282186 | chr19:6422447 | ENST00000398148 | 0 | 20 | 571_582 | 83 | 712.0 | Repeat | Note=1 | |
| Tgene | KHSRP | chr1:155282186 | chr19:6422447 | ENST00000398148 | 0 | 20 | 617_628 | 83 | 712.0 | Repeat | Note=2 | |
| Tgene | KHSRP | chr1:155282186 | chr19:6422447 | ENST00000398148 | 0 | 20 | 643_654 | 83 | 712.0 | Repeat | Note=3 | |
| Tgene | KHSRP | chr1:155282186 | chr19:6422447 | ENST00000398148 | 0 | 20 | 673_684 | 83 | 712.0 | Repeat | Note=4 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | KHSRP | chr1:155282186 | chr19:6422447 | ENST00000398148 | 0 | 20 | 68_496 | 83 | 712.0 | Compositional bias | Note=Gly-rich | |
| Tgene | KHSRP | chr1:155282186 | chr19:6422447 | ENST00000398148 | 0 | 20 | 7_67 | 83 | 712.0 | Compositional bias | Note=Gly/Pro-rich |
Top |
Fusion Gene Sequence for FDPS-KHSRP |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >30059_30059_1_FDPS-KHSRP_FDPS_chr1_155282186_ENST00000356657_KHSRP_chr19_6422447_ENST00000398148_length(transcript)=3293nt_BP=642nt GAGTACCCGCCAACAAGCGGGACCGAGCAGGAATCCGTATCTGGGAACAGGTGAGAGAGGATGTGTGCTGGGCCTTGGAGGAAGGGGGCC GAGACCGGGCCTTACTTCTGTAACGATACTGTGAGGCATCGGAAGGCCAGCCTGTTGTGTCCGTTTTGAAGGATGCCCCTGTCCCGCTGG TTGAGATCTGTGGGGGTCTTCCTGCTGCCAGCCCCCTACTGGGCACCCCGGGAGAGGTGGCTGGGTTCCCTACGGCGGCCCTCCCTGGTG CACGGGTACCCAGTCCTGGCCTGGCACAGTGCCCGCTGCTGGTGCCAAGCGTGGACAGAGGAACCTCGAGCCCTTTGCTCCTCCCTCAGA ATGAACGGAGACCAGAATTCAGATGTTTATGCCCAAGAAAAGCAGGATTTCGTTCAGCACTTCTCCCAGATCGTTAGGGTGCTGACTGAG GATGAGATGGGGCACCCAGAGATAGGAGATGCTATTGCCCGGCTCAAGGAGGTCCTGGAGTACAATGCCATTGGAGGCAAGTATAACCGG GGTTTGACGGTGGTAGTAGCATTCCGGGAGCTGGTGGAGCCAAGGAAACAGGATGCTGATAGTCTCCAGCGGGCCTGGACTGTGGGCTGG TGTGTGGAACTGATTGCAGCCAAAATTGGAGGCGATGCTGCCACGACAGTGAATAACAGCACTCCTGATTTTGGTTTTGGGGGCCAAAAG AGACAGTTGGAAGATGGAGATCAACCGGAGAGCAAGAAGCTGGCTTCCCAGGGAGACTCAATCAGTTCTCAACTTGGACCCATCCATCCT CCCCCAAGGACTTCAATGACAGAAGAGTACAGGGTCCCAGACGGCATGGTGGGCCTGATCATTGGCAGAGGAGGTGAACAAATTAACAAA ATCCAACAGGATTCAGGCTGCAAAGTACAGATTTCTCCAGACAGCGGTGGCCTACCCGAGCGCAGTGTGTCCTTGACAGGAGCCCCAGAA TCTGTCCAGAAAGCCAAGATGATGCTGGATGACATTGTGTCTCGGGGTCGTGGGGGCCCCCCAGGACAGTTCCACGACAACGCCAACGGG GGCCAGAACGGCACCGTGCAGGAGATCATGATCCCCGCGGGCAAGGCCGGCCTGGTCATTGGCAAGGGCGGGGAGACCATTAAGCAGCTG CAGGAACGCGCTGGAGTGAAGATGATCTTAATTCAGGACGGATCTCAGAATACGAATGTGGACAAACCTCTCCGCATCATTGGGGATCCT TACAAAGTGCAGCAAGCCTGTGAGATGGTGATGGACATCCTCCGGGAACGTGACCAAGGCGGCTTTGGGGACCGGAATGAGTACGGATCT CGGATTGGCGGAGGCATCGATGTGCCAGTGCCCAGGCATTCTGTTGGCGTGGTCATTGGCCGGAGTGGAGAGATGATCAAGAAGATCCAG AATGATGCTGGCGTGCGGATACAGTTCAAGCAAGATGACGGGACAGGGCCCGAGAAGATTGCTCATATAATGGGGCCCCCAGACAGGTGC GAGCACGCAGCCCGGATCATCAACGACCTCCTCCAGAGCCTCAGGAGTGGTCCCCCAGGTCCTCCAGGGGGTCCAGGCATGCCCCCGGGG GGCCGAGGCCGAGGAAGAGGCCAAGGCAATTGGGGTCCCCCTGGCGGGGAGATGACCTTCTCCATCCCCACTCACAAGTGTGGGCTGGTC ATCGGCCGAGGTGGCGAGAATGTGAAAGCCATAAACCAGCAGACGGGAGCCTTCGTAGAGATCTCCCGGCAGCTGCCACCCAACGGGGAC CCCAACTTCAAGTTGTTCATCATCCGGGGTTCACCCCAGCAGATTGACCACGCCAAGCAGCTTATCGAGGAAAAGATCGAGGGTCCTCTC TGCCCAGTTGGACCAGGCCCAGGTGGCCCAGGCCCTGCTGGCCCAATGGGGCCCTTCAATCCTGGGCCCTTCAACCAGGGGCCACCCGGG GCTCCCCCACATGCCGGGGGGCCCCCTCCTCACCAGTACCCACCCCAGGGCTGGGGCAATACCTACCCCCAGTGGCAGCCGCCTGCTCCT CATGACCCAAGCAAAGCAGCTGCAGCGGCCGCGGACCCCAACGCCGCGTGGGCCGCCTACTACTCACACTACTACCAGCAGCCCCCGGGC CCCGTCCCCGGCCCCGCACCGGCCCCTGCGGCCCCACCGGCTCAGGGTGAGCCCCCTCAGCCCCCACCCACCGGCCAGTCGGACTACACT AAGGCCTGGGAAGAGTATTACAAAAAGATCGGCCAGCAGCCCCAGCAGCCCGGAGCACCCCCACAGCAGGACTACACGAAGGCTTGGGAG GAGTACTACAAGAAGCAAGCGCAAGTGGCCACCGGAGGGGGTCCAGGAGCTCCCCCAGGCTCCCAGCCAGACTACAGTGCCGCCTGGGCG GAATATTACAGACAGCAGGCCGCTTACTACGGACAGACCCCAGGTCCTGGCGGCCCCCAGCCGCCGCCCACGCAGCAGGGACAGCAGCAG GCTCAATGAATCGAATGAATGTGAACTTCTTCATCTGTGAAAAATCTTTTTTTTTTCCATTTTGTTCTGTTTGGGGGCTTCTGTTTTGTT TGGCGAGAGAGCGATGGCTGCCGTGGGGAGTACTGGGGAGCCCTCGCGGCAAGCAGGGTGGGGGGGACTTGGGGGCATGCCGGGCCCTCA CTCTCTCGCCTGTTCTGTGTCTCACATGCTTTTTCTTTCAAAATTGGGATCCTTCCATGTTGAGCCAGCCAGAGAAGATAGCGAGATCTA AATCTCTGCCAAAAAAAAAAAAAAACTTAAAAATTAAAAACACAAAGAGCAAAGCAGAACTTATAAAATTATATATATATATATTAAAAA GTCTCTATTCTTCACCCCCCAGCCTTCCTGAACCTGCCTCTCTGAGGATAAAGCAATTCATTTTCTCCCACCCTCGGCCCTCTTGTTTTT AAAATAAACTTTTAAAAAGGAAAAAAAAAAGTCACTCTTGCTATTTCTTTTTTTTAGTTAGAGGTGGAACATTCCTTGGACCAGGTGTTG TATTGCAGGACCCCTTCCCCCAGCAGCCAAGCCCCCTCTTCTCTCCCTCCCGCCCTGGCTCAGCTCCCGCGGCCCCGCCCGTCCCCCCTC CCAGGACTGGTCTGTTGTCTTTTCATCTGTTCAAGAGGAGATTGAAACTGAAAACAAAATGAGAACAACAAAAAAAATTGTATGGCAGTT >30059_30059_1_FDPS-KHSRP_FDPS_chr1_155282186_ENST00000356657_KHSRP_chr19_6422447_ENST00000398148_length(amino acids)=788AA_BP=160 MPLSRWLRSVGVFLLPAPYWAPRERWLGSLRRPSLVHGYPVLAWHSARCWCQAWTEEPRALCSSLRMNGDQNSDVYAQEKQDFVQHFSQI VRVLTEDEMGHPEIGDAIARLKEVLEYNAIGGKYNRGLTVVVAFRELVEPRKQDADSLQRAWTVGWCVELIAAKIGGDAATTVNNSTPDF GFGGQKRQLEDGDQPESKKLASQGDSISSQLGPIHPPPRTSMTEEYRVPDGMVGLIIGRGGEQINKIQQDSGCKVQISPDSGGLPERSVS LTGAPESVQKAKMMLDDIVSRGRGGPPGQFHDNANGGQNGTVQEIMIPAGKAGLVIGKGGETIKQLQERAGVKMILIQDGSQNTNVDKPL RIIGDPYKVQQACEMVMDILRERDQGGFGDRNEYGSRIGGGIDVPVPRHSVGVVIGRSGEMIKKIQNDAGVRIQFKQDDGTGPEKIAHIM GPPDRCEHAARIINDLLQSLRSGPPGPPGGPGMPPGGRGRGRGQGNWGPPGGEMTFSIPTHKCGLVIGRGGENVKAINQQTGAFVEISRQ LPPNGDPNFKLFIIRGSPQQIDHAKQLIEEKIEGPLCPVGPGPGGPGPAGPMGPFNPGPFNQGPPGAPPHAGGPPPHQYPPQGWGNTYPQ WQPPAPHDPSKAAAAAADPNAAWAAYYSHYYQQPPGPVPGPAPAPAAPPAQGEPPQPPPTGQSDYTKAWEEYYKKIGQQPQQPGAPPQQD -------------------------------------------------------------- >30059_30059_2_FDPS-KHSRP_FDPS_chr1_155282186_ENST00000368356_KHSRP_chr19_6422447_ENST00000398148_length(transcript)=3246nt_BP=595nt AGTGTGGGGAGAGCGGGAACTACTCGACCCACAGAGCCGATCGCGGAGCGGATTCTGCTTTTAGGAGTACCCGCCAACAAGCGGGACCGA GCAGGAATCCGTATCTGGGAACAGGATGCCCCTGTCCCGCTGGTTGAGATCTGTGGGGGTCTTCCTGCTGCCAGCCCCCTACTGGGCACC CCGGGAGAGGTGGCTGGGTTCCCTACGGCGGCCCTCCCTGGTGCACGGGTACCCAGTCCTGGCCTGGCACAGTGCCCGCTGCTGGTGCCA AGCGTGGACAGAGGAACCTCGAGCCCTTTGCTCCTCCCTCAGAATGAACGGAGACCAGAATTCAGATGTTTATGCCCAAGAAAAGCAGGA TTTCGTTCAGCACTTCTCCCAGATCGTTAGGGTGCTGACTGAGGATGAGATGGGGCACCCAGAGATAGGAGATGCTATTGCCCGGCTCAA GGAGGTCCTGGAGTACAATGCCATTGGAGGCAAGTATAACCGGGGTTTGACGGTGGTAGTAGCATTCCGGGAGCTGGTGGAGCCAAGGAA ACAGGATGCTGATAGTCTCCAGCGGGCCTGGACTGTGGGCTGGTGTGTGGAACTGATTGCAGCCAAAATTGGAGGCGATGCTGCCACGAC AGTGAATAACAGCACTCCTGATTTTGGTTTTGGGGGCCAAAAGAGACAGTTGGAAGATGGAGATCAACCGGAGAGCAAGAAGCTGGCTTC CCAGGGAGACTCAATCAGTTCTCAACTTGGACCCATCCATCCTCCCCCAAGGACTTCAATGACAGAAGAGTACAGGGTCCCAGACGGCAT GGTGGGCCTGATCATTGGCAGAGGAGGTGAACAAATTAACAAAATCCAACAGGATTCAGGCTGCAAAGTACAGATTTCTCCAGACAGCGG TGGCCTACCCGAGCGCAGTGTGTCCTTGACAGGAGCCCCAGAATCTGTCCAGAAAGCCAAGATGATGCTGGATGACATTGTGTCTCGGGG TCGTGGGGGCCCCCCAGGACAGTTCCACGACAACGCCAACGGGGGCCAGAACGGCACCGTGCAGGAGATCATGATCCCCGCGGGCAAGGC CGGCCTGGTCATTGGCAAGGGCGGGGAGACCATTAAGCAGCTGCAGGAACGCGCTGGAGTGAAGATGATCTTAATTCAGGACGGATCTCA GAATACGAATGTGGACAAACCTCTCCGCATCATTGGGGATCCTTACAAAGTGCAGCAAGCCTGTGAGATGGTGATGGACATCCTCCGGGA ACGTGACCAAGGCGGCTTTGGGGACCGGAATGAGTACGGATCTCGGATTGGCGGAGGCATCGATGTGCCAGTGCCCAGGCATTCTGTTGG CGTGGTCATTGGCCGGAGTGGAGAGATGATCAAGAAGATCCAGAATGATGCTGGCGTGCGGATACAGTTCAAGCAAGATGACGGGACAGG GCCCGAGAAGATTGCTCATATAATGGGGCCCCCAGACAGGTGCGAGCACGCAGCCCGGATCATCAACGACCTCCTCCAGAGCCTCAGGAG TGGTCCCCCAGGTCCTCCAGGGGGTCCAGGCATGCCCCCGGGGGGCCGAGGCCGAGGAAGAGGCCAAGGCAATTGGGGTCCCCCTGGCGG GGAGATGACCTTCTCCATCCCCACTCACAAGTGTGGGCTGGTCATCGGCCGAGGTGGCGAGAATGTGAAAGCCATAAACCAGCAGACGGG AGCCTTCGTAGAGATCTCCCGGCAGCTGCCACCCAACGGGGACCCCAACTTCAAGTTGTTCATCATCCGGGGTTCACCCCAGCAGATTGA CCACGCCAAGCAGCTTATCGAGGAAAAGATCGAGGGTCCTCTCTGCCCAGTTGGACCAGGCCCAGGTGGCCCAGGCCCTGCTGGCCCAAT GGGGCCCTTCAATCCTGGGCCCTTCAACCAGGGGCCACCCGGGGCTCCCCCACATGCCGGGGGGCCCCCTCCTCACCAGTACCCACCCCA GGGCTGGGGCAATACCTACCCCCAGTGGCAGCCGCCTGCTCCTCATGACCCAAGCAAAGCAGCTGCAGCGGCCGCGGACCCCAACGCCGC GTGGGCCGCCTACTACTCACACTACTACCAGCAGCCCCCGGGCCCCGTCCCCGGCCCCGCACCGGCCCCTGCGGCCCCACCGGCTCAGGG TGAGCCCCCTCAGCCCCCACCCACCGGCCAGTCGGACTACACTAAGGCCTGGGAAGAGTATTACAAAAAGATCGGCCAGCAGCCCCAGCA GCCCGGAGCACCCCCACAGCAGGACTACACGAAGGCTTGGGAGGAGTACTACAAGAAGCAAGCGCAAGTGGCCACCGGAGGGGGTCCAGG AGCTCCCCCAGGCTCCCAGCCAGACTACAGTGCCGCCTGGGCGGAATATTACAGACAGCAGGCCGCTTACTACGGACAGACCCCAGGTCC TGGCGGCCCCCAGCCGCCGCCCACGCAGCAGGGACAGCAGCAGGCTCAATGAATCGAATGAATGTGAACTTCTTCATCTGTGAAAAATCT TTTTTTTTTCCATTTTGTTCTGTTTGGGGGCTTCTGTTTTGTTTGGCGAGAGAGCGATGGCTGCCGTGGGGAGTACTGGGGAGCCCTCGC GGCAAGCAGGGTGGGGGGGACTTGGGGGCATGCCGGGCCCTCACTCTCTCGCCTGTTCTGTGTCTCACATGCTTTTTCTTTCAAAATTGG GATCCTTCCATGTTGAGCCAGCCAGAGAAGATAGCGAGATCTAAATCTCTGCCAAAAAAAAAAAAAAACTTAAAAATTAAAAACACAAAG AGCAAAGCAGAACTTATAAAATTATATATATATATATTAAAAAGTCTCTATTCTTCACCCCCCAGCCTTCCTGAACCTGCCTCTCTGAGG ATAAAGCAATTCATTTTCTCCCACCCTCGGCCCTCTTGTTTTTAAAATAAACTTTTAAAAAGGAAAAAAAAAAGTCACTCTTGCTATTTC TTTTTTTTAGTTAGAGGTGGAACATTCCTTGGACCAGGTGTTGTATTGCAGGACCCCTTCCCCCAGCAGCCAAGCCCCCTCTTCTCTCCC TCCCGCCCTGGCTCAGCTCCCGCGGCCCCGCCCGTCCCCCCTCCCAGGACTGGTCTGTTGTCTTTTCATCTGTTCAAGAGGAGATTGAAA CTGAAAACAAAATGAGAACAACAAAAAAAATTGTATGGCAGTTTTTACTTTTTATCGCTCGTTTTTAACTTCACAAATAAATGATAACAA >30059_30059_2_FDPS-KHSRP_FDPS_chr1_155282186_ENST00000368356_KHSRP_chr19_6422447_ENST00000398148_length(amino acids)=788AA_BP=160 MPLSRWLRSVGVFLLPAPYWAPRERWLGSLRRPSLVHGYPVLAWHSARCWCQAWTEEPRALCSSLRMNGDQNSDVYAQEKQDFVQHFSQI VRVLTEDEMGHPEIGDAIARLKEVLEYNAIGGKYNRGLTVVVAFRELVEPRKQDADSLQRAWTVGWCVELIAAKIGGDAATTVNNSTPDF GFGGQKRQLEDGDQPESKKLASQGDSISSQLGPIHPPPRTSMTEEYRVPDGMVGLIIGRGGEQINKIQQDSGCKVQISPDSGGLPERSVS LTGAPESVQKAKMMLDDIVSRGRGGPPGQFHDNANGGQNGTVQEIMIPAGKAGLVIGKGGETIKQLQERAGVKMILIQDGSQNTNVDKPL RIIGDPYKVQQACEMVMDILRERDQGGFGDRNEYGSRIGGGIDVPVPRHSVGVVIGRSGEMIKKIQNDAGVRIQFKQDDGTGPEKIAHIM GPPDRCEHAARIINDLLQSLRSGPPGPPGGPGMPPGGRGRGRGQGNWGPPGGEMTFSIPTHKCGLVIGRGGENVKAINQQTGAFVEISRQ LPPNGDPNFKLFIIRGSPQQIDHAKQLIEEKIEGPLCPVGPGPGGPGPAGPMGPFNPGPFNQGPPGAPPHAGGPPPHQYPPQGWGNTYPQ WQPPAPHDPSKAAAAAADPNAAWAAYYSHYYQQPPGPVPGPAPAPAAPPAQGEPPQPPPTGQSDYTKAWEEYYKKIGQQPQQPGAPPQQD -------------------------------------------------------------- >30059_30059_3_FDPS-KHSRP_FDPS_chr1_155282186_ENST00000447866_KHSRP_chr19_6422447_ENST00000398148_length(transcript)=3152nt_BP=501nt TCAGCGCGGCTCATCTTCGCCCAGCCAGGACACGCAGGATCGCGTTATCTGTCCCTCCCGCGGGCACATTGGGAGTTGTAGTCTAATTAT ATATTTCCGCCCTTAGTGTGGGGAGAGCGGGAACTACTCGACCCACAGAGCCGATCGCGGAGCGGATTCTGCTTTTAGGAGTACCCGCCA ACAAGCGGGACCGAGCAGGAATCCGTATCTGGGAACAGAATGAACGGAGACCAGAATTCAGATGTTTATGCCCAAGAAAAGCAGGATTTC GTTCAGCACTTCTCCCAGATCGTTAGGGTGCTGACTGAGGATGAGATGGGGCACCCAGAGATAGGAGATGCTATTGCCCGGCTCAAGGAG GTCCTGGAGTACAATGCCATTGGAGGCAAGTATAACCGGGGTTTGACGGTGGTAGTAGCATTCCGGGAGCTGGTGGAGCCAAGGAAACAG GATGCTGATAGTCTCCAGCGGGCCTGGACTGTGGGCTGGTGTGTGGAACTGATTGCAGCCAAAATTGGAGGCGATGCTGCCACGACAGTG AATAACAGCACTCCTGATTTTGGTTTTGGGGGCCAAAAGAGACAGTTGGAAGATGGAGATCAACCGGAGAGCAAGAAGCTGGCTTCCCAG GGAGACTCAATCAGTTCTCAACTTGGACCCATCCATCCTCCCCCAAGGACTTCAATGACAGAAGAGTACAGGGTCCCAGACGGCATGGTG GGCCTGATCATTGGCAGAGGAGGTGAACAAATTAACAAAATCCAACAGGATTCAGGCTGCAAAGTACAGATTTCTCCAGACAGCGGTGGC CTACCCGAGCGCAGTGTGTCCTTGACAGGAGCCCCAGAATCTGTCCAGAAAGCCAAGATGATGCTGGATGACATTGTGTCTCGGGGTCGT GGGGGCCCCCCAGGACAGTTCCACGACAACGCCAACGGGGGCCAGAACGGCACCGTGCAGGAGATCATGATCCCCGCGGGCAAGGCCGGC CTGGTCATTGGCAAGGGCGGGGAGACCATTAAGCAGCTGCAGGAACGCGCTGGAGTGAAGATGATCTTAATTCAGGACGGATCTCAGAAT ACGAATGTGGACAAACCTCTCCGCATCATTGGGGATCCTTACAAAGTGCAGCAAGCCTGTGAGATGGTGATGGACATCCTCCGGGAACGT GACCAAGGCGGCTTTGGGGACCGGAATGAGTACGGATCTCGGATTGGCGGAGGCATCGATGTGCCAGTGCCCAGGCATTCTGTTGGCGTG GTCATTGGCCGGAGTGGAGAGATGATCAAGAAGATCCAGAATGATGCTGGCGTGCGGATACAGTTCAAGCAAGATGACGGGACAGGGCCC GAGAAGATTGCTCATATAATGGGGCCCCCAGACAGGTGCGAGCACGCAGCCCGGATCATCAACGACCTCCTCCAGAGCCTCAGGAGTGGT CCCCCAGGTCCTCCAGGGGGTCCAGGCATGCCCCCGGGGGGCCGAGGCCGAGGAAGAGGCCAAGGCAATTGGGGTCCCCCTGGCGGGGAG ATGACCTTCTCCATCCCCACTCACAAGTGTGGGCTGGTCATCGGCCGAGGTGGCGAGAATGTGAAAGCCATAAACCAGCAGACGGGAGCC TTCGTAGAGATCTCCCGGCAGCTGCCACCCAACGGGGACCCCAACTTCAAGTTGTTCATCATCCGGGGTTCACCCCAGCAGATTGACCAC GCCAAGCAGCTTATCGAGGAAAAGATCGAGGGTCCTCTCTGCCCAGTTGGACCAGGCCCAGGTGGCCCAGGCCCTGCTGGCCCAATGGGG CCCTTCAATCCTGGGCCCTTCAACCAGGGGCCACCCGGGGCTCCCCCACATGCCGGGGGGCCCCCTCCTCACCAGTACCCACCCCAGGGC TGGGGCAATACCTACCCCCAGTGGCAGCCGCCTGCTCCTCATGACCCAAGCAAAGCAGCTGCAGCGGCCGCGGACCCCAACGCCGCGTGG GCCGCCTACTACTCACACTACTACCAGCAGCCCCCGGGCCCCGTCCCCGGCCCCGCACCGGCCCCTGCGGCCCCACCGGCTCAGGGTGAG CCCCCTCAGCCCCCACCCACCGGCCAGTCGGACTACACTAAGGCCTGGGAAGAGTATTACAAAAAGATCGGCCAGCAGCCCCAGCAGCCC GGAGCACCCCCACAGCAGGACTACACGAAGGCTTGGGAGGAGTACTACAAGAAGCAAGCGCAAGTGGCCACCGGAGGGGGTCCAGGAGCT CCCCCAGGCTCCCAGCCAGACTACAGTGCCGCCTGGGCGGAATATTACAGACAGCAGGCCGCTTACTACGGACAGACCCCAGGTCCTGGC GGCCCCCAGCCGCCGCCCACGCAGCAGGGACAGCAGCAGGCTCAATGAATCGAATGAATGTGAACTTCTTCATCTGTGAAAAATCTTTTT TTTTTCCATTTTGTTCTGTTTGGGGGCTTCTGTTTTGTTTGGCGAGAGAGCGATGGCTGCCGTGGGGAGTACTGGGGAGCCCTCGCGGCA AGCAGGGTGGGGGGGACTTGGGGGCATGCCGGGCCCTCACTCTCTCGCCTGTTCTGTGTCTCACATGCTTTTTCTTTCAAAATTGGGATC CTTCCATGTTGAGCCAGCCAGAGAAGATAGCGAGATCTAAATCTCTGCCAAAAAAAAAAAAAAACTTAAAAATTAAAAACACAAAGAGCA AAGCAGAACTTATAAAATTATATATATATATATTAAAAAGTCTCTATTCTTCACCCCCCAGCCTTCCTGAACCTGCCTCTCTGAGGATAA AGCAATTCATTTTCTCCCACCCTCGGCCCTCTTGTTTTTAAAATAAACTTTTAAAAAGGAAAAAAAAAAGTCACTCTTGCTATTTCTTTT TTTTAGTTAGAGGTGGAACATTCCTTGGACCAGGTGTTGTATTGCAGGACCCCTTCCCCCAGCAGCCAAGCCCCCTCTTCTCTCCCTCCC GCCCTGGCTCAGCTCCCGCGGCCCCGCCCGTCCCCCCTCCCAGGACTGGTCTGTTGTCTTTTCATCTGTTCAAGAGGAGATTGAAACTGA AAACAAAATGAGAACAACAAAAAAAATTGTATGGCAGTTTTTACTTTTTATCGCTCGTTTTTAACTTCACAAATAAATGATAACAAAACC >30059_30059_3_FDPS-KHSRP_FDPS_chr1_155282186_ENST00000447866_KHSRP_chr19_6422447_ENST00000398148_length(amino acids)=722AA_BP=94 MNGDQNSDVYAQEKQDFVQHFSQIVRVLTEDEMGHPEIGDAIARLKEVLEYNAIGGKYNRGLTVVVAFRELVEPRKQDADSLQRAWTVGW CVELIAAKIGGDAATTVNNSTPDFGFGGQKRQLEDGDQPESKKLASQGDSISSQLGPIHPPPRTSMTEEYRVPDGMVGLIIGRGGEQINK IQQDSGCKVQISPDSGGLPERSVSLTGAPESVQKAKMMLDDIVSRGRGGPPGQFHDNANGGQNGTVQEIMIPAGKAGLVIGKGGETIKQL QERAGVKMILIQDGSQNTNVDKPLRIIGDPYKVQQACEMVMDILRERDQGGFGDRNEYGSRIGGGIDVPVPRHSVGVVIGRSGEMIKKIQ NDAGVRIQFKQDDGTGPEKIAHIMGPPDRCEHAARIINDLLQSLRSGPPGPPGGPGMPPGGRGRGRGQGNWGPPGGEMTFSIPTHKCGLV IGRGGENVKAINQQTGAFVEISRQLPPNGDPNFKLFIIRGSPQQIDHAKQLIEEKIEGPLCPVGPGPGGPGPAGPMGPFNPGPFNQGPPG APPHAGGPPPHQYPPQGWGNTYPQWQPPAPHDPSKAAAAAADPNAAWAAYYSHYYQQPPGPVPGPAPAPAAPPAQGEPPQPPPTGQSDYT KAWEEYYKKIGQQPQQPGAPPQQDYTKAWEEYYKKQAQVATGGGPGAPPGSQPDYSAAWAEYYRQQAAYYGQTPGPGGPQPPPTQQGQQQ -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FDPS-KHSRP |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FDPS-KHSRP |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FDPS-KHSRP |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
