
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FGFR2-AHCYL1 (FusionGDB2 ID:30273) |
Fusion Gene Summary for FGFR2-AHCYL1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FGFR2-AHCYL1 | Fusion gene ID: 30273 | Hgene | Tgene | Gene symbol | FGFR2 | AHCYL1 | Gene ID | 2263 | 10768 |
| Gene name | fibroblast growth factor receptor 2 | adenosylhomocysteinase like 1 | |
| Synonyms | BBDS|BEK|BFR-1|CD332|CEK3|CFD1|ECT1|JWS|K-SAM|KGFR|TK14|TK25 | DCAL|IRBIT|PPP1R78|PRO0233|XPVKONA | |
| Cytomap | 10q26.13 | 1p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | fibroblast growth factor receptor 2BEK fibroblast growth factor receptorbacteria-expressed kinasekeratinocyte growth factor receptorprotein tyrosine kinase, receptor like 14 | S-adenosylhomocysteine hydrolase-like protein 1DC-expressed AHCY-like moleculeIP(3)Rs binding protein released with IP(3)S-adenosyl homocysteine hydrolase homologS-adenosyl-L-homocysteine hydrolase 2adenosylhomocysteinase 2adoHcyase 2dendritic cell | |
| Modification date | 20200322 | 20200313 | |
| UniProtAcc | P21802 | O43865 | |
| Ensembl transtripts involved in fusion gene | ENST00000346997, ENST00000351936, ENST00000356226, ENST00000357555, ENST00000358487, ENST00000360144, ENST00000369056, ENST00000369059, ENST00000369060, ENST00000369061, ENST00000457416, ENST00000478859, ENST00000359354, ENST00000490349, | ENST00000475081, ENST00000359172, ENST00000393614, ENST00000369799, | |
| Fusion gene scores | * DoF score | 24 X 20 X 12=5760 | 11 X 13 X 4=572 |
| # samples | 38 | 12 | |
| ** MAII score | log2(38/5760*10)=-3.92199748799873 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/572*10)=-2.25298074116987 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FGFR2 [Title/Abstract] AND AHCYL1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FGFR2(123243212)-AHCYL1(110551654), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | FGFR2-AHCYL1 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. FGFR2-AHCYL1 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. FGFR2-AHCYL1 seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. FGFR2-AHCYL1 seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. FGFR2-AHCYL1 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. FGFR2-AHCYL1 seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | FGFR2 | GO:0008284 | positive regulation of cell proliferation | 8663044 |
| Hgene | FGFR2 | GO:0008543 | fibroblast growth factor receptor signaling pathway | 8663044|15629145 |
| Hgene | FGFR2 | GO:0018108 | peptidyl-tyrosine phosphorylation | 15629145|16844695 |
| Hgene | FGFR2 | GO:0046777 | protein autophosphorylation | 15629145 |
| Tgene | AHCYL1 | GO:0006378 | mRNA polyadenylation | 19224921 |
| Tgene | AHCYL1 | GO:0031440 | regulation of mRNA 3'-end processing | 19224921 |
| Tgene | AHCYL1 | GO:0038166 | angiotensin-activated signaling pathway | 20584908 |
| Tgene | AHCYL1 | GO:0051592 | response to calcium ion | 18829453 |
 Fusion gene breakpoints across FGFR2 (5'-gene) Fusion gene breakpoints across FGFR2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
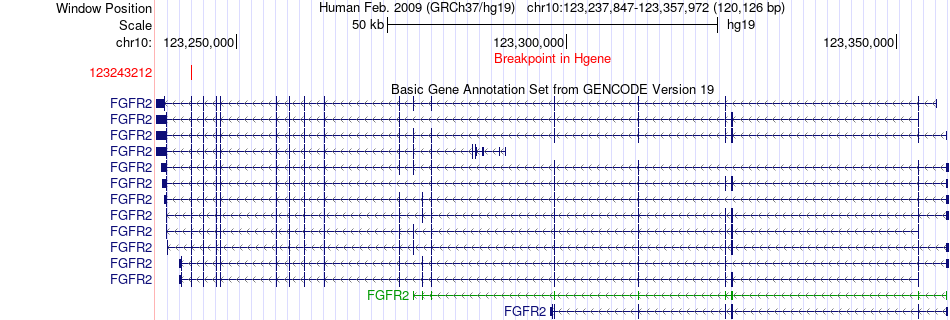 |
 Fusion gene breakpoints across AHCYL1 (3'-gene) Fusion gene breakpoints across AHCYL1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
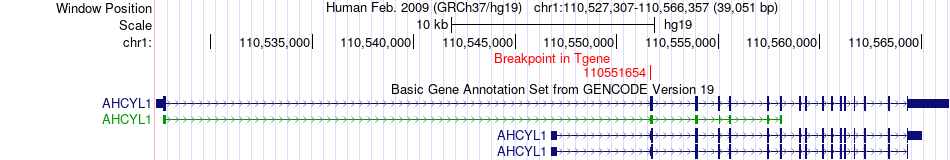 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChiTaRS5.0 | N/A | AB821309 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| ChiTaRS5.0 | N/A | LP131425 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
Top |
Fusion Gene ORF analysis for FGFR2-AHCYL1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000346997 | ENST00000475081 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-3UTR | ENST00000351936 | ENST00000475081 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-3UTR | ENST00000356226 | ENST00000475081 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-3UTR | ENST00000357555 | ENST00000475081 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-3UTR | ENST00000358487 | ENST00000475081 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-3UTR | ENST00000360144 | ENST00000475081 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-3UTR | ENST00000369056 | ENST00000475081 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-3UTR | ENST00000369059 | ENST00000475081 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-3UTR | ENST00000369060 | ENST00000475081 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-3UTR | ENST00000369061 | ENST00000475081 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-3UTR | ENST00000457416 | ENST00000475081 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-3UTR | ENST00000478859 | ENST00000475081 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000346997 | ENST00000359172 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000346997 | ENST00000393614 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000351936 | ENST00000359172 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000351936 | ENST00000393614 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000356226 | ENST00000359172 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000356226 | ENST00000393614 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000357555 | ENST00000359172 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000357555 | ENST00000393614 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000358487 | ENST00000359172 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000358487 | ENST00000393614 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000360144 | ENST00000359172 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000360144 | ENST00000393614 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000369056 | ENST00000359172 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000369056 | ENST00000393614 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000369059 | ENST00000359172 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000369059 | ENST00000393614 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000369060 | ENST00000359172 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000369060 | ENST00000393614 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000369061 | ENST00000359172 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000369061 | ENST00000393614 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000457416 | ENST00000359172 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000457416 | ENST00000393614 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000478859 | ENST00000359172 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| 5CDS-5UTR | ENST00000478859 | ENST00000393614 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| Frame-shift | ENST00000351936 | ENST00000369799 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| Frame-shift | ENST00000356226 | ENST00000369799 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| Frame-shift | ENST00000357555 | ENST00000369799 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| Frame-shift | ENST00000358487 | ENST00000369799 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| Frame-shift | ENST00000360144 | ENST00000369799 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| Frame-shift | ENST00000369059 | ENST00000369799 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| Frame-shift | ENST00000369060 | ENST00000369799 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| Frame-shift | ENST00000457416 | ENST00000369799 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| Frame-shift | ENST00000478859 | ENST00000369799 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| In-frame | ENST00000346997 | ENST00000369799 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| In-frame | ENST00000369056 | ENST00000369799 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| In-frame | ENST00000369061 | ENST00000369799 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| intron-3CDS | ENST00000359354 | ENST00000369799 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| intron-3CDS | ENST00000490349 | ENST00000369799 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| intron-3UTR | ENST00000359354 | ENST00000475081 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| intron-3UTR | ENST00000490349 | ENST00000475081 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| intron-5UTR | ENST00000359354 | ENST00000359172 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| intron-5UTR | ENST00000359354 | ENST00000393614 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| intron-5UTR | ENST00000490349 | ENST00000359172 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
| intron-5UTR | ENST00000490349 | ENST00000393614 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000369061 | FGFR2 | chr10 | 123243212 | - | ENST00000369799 | AHCYL1 | chr1 | 110551654 | + | 5643 | 2115 | 48 | 3587 | 1179 |
| ENST00000346997 | FGFR2 | chr10 | 123243212 | - | ENST00000369799 | AHCYL1 | chr1 | 110551654 | + | 5835 | 2307 | 12 | 3779 | 1255 |
| ENST00000369056 | FGFR2 | chr10 | 123243212 | - | ENST00000369799 | AHCYL1 | chr1 | 110551654 | + | 5856 | 2328 | 24 | 3800 | 1258 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000369061 | ENST00000369799 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + | 0.000258539 | 0.9997415 |
| ENST00000346997 | ENST00000369799 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + | 0.000199904 | 0.99980015 |
| ENST00000369056 | ENST00000369799 | FGFR2 | chr10 | 123243212 | - | AHCYL1 | chr1 | 110551654 | + | 0.000352547 | 0.9996475 |
Top |
Fusion Genomic Features for FGFR2-AHCYL1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| FGFR2 | chr10 | 123243211 | - | AHCYL1 | chr1 | 110551655 | + | 2.50E-06 | 0.9999975 |
| FGFR2 | chr10 | 123243211 | - | AHCYL1 | chr1 | 110551655 | + | 2.50E-06 | 0.9999975 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
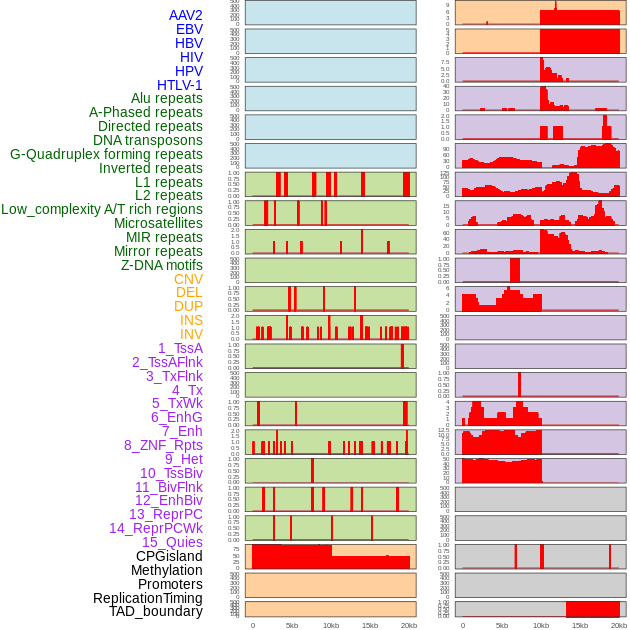 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
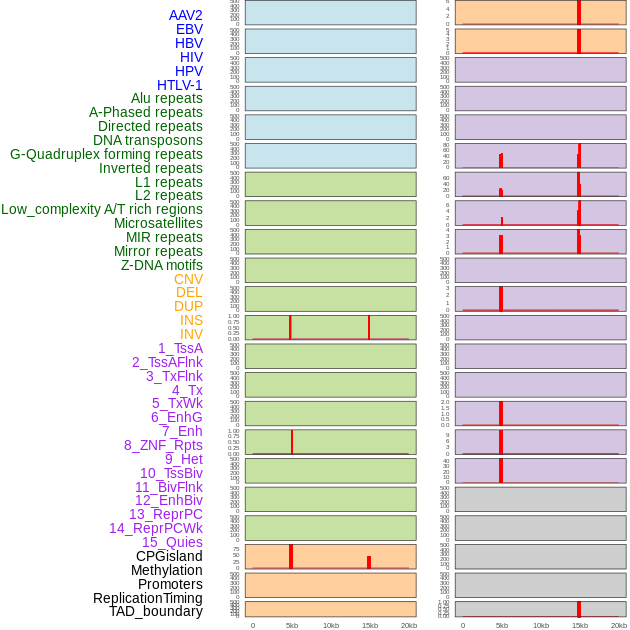 |
Top |
Fusion Protein Features for FGFR2-AHCYL1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:123243212/chr1:110551654) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FGFR2 | AHCYL1 |
| FUNCTION: Tyrosine-protein kinase that acts as cell-surface receptor for fibroblast growth factors and plays an essential role in the regulation of cell proliferation, differentiation, migration and apoptosis, and in the regulation of embryonic development. Required for normal embryonic patterning, trophoblast function, limb bud development, lung morphogenesis, osteogenesis and skin development. Plays an essential role in the regulation of osteoblast differentiation, proliferation and apoptosis, and is required for normal skeleton development. Promotes cell proliferation in keratinocytes and immature osteoblasts, but promotes apoptosis in differentiated osteoblasts. Phosphorylates PLCG1, FRS2 and PAK4. Ligand binding leads to the activation of several signaling cascades. Activation of PLCG1 leads to the production of the cellular signaling molecules diacylglycerol and inositol 1,4,5-trisphosphate. Phosphorylation of FRS2 triggers recruitment of GRB2, GAB1, PIK3R1 and SOS1, and mediates activation of RAS, MAPK1/ERK2, MAPK3/ERK1 and the MAP kinase signaling pathway, as well as of the AKT1 signaling pathway. FGFR2 signaling is down-regulated by ubiquitination, internalization and degradation. Mutations that lead to constitutive kinase activation or impair normal FGFR2 maturation, internalization and degradation lead to aberrant signaling. Over-expressed FGFR2 promotes activation of STAT1. {ECO:0000269|PubMed:12529371, ECO:0000269|PubMed:15190072, ECO:0000269|PubMed:15629145, ECO:0000269|PubMed:16384934, ECO:0000269|PubMed:16597617, ECO:0000269|PubMed:17311277, ECO:0000269|PubMed:17623664, ECO:0000269|PubMed:18374639, ECO:0000269|PubMed:19103595, ECO:0000269|PubMed:19387476, ECO:0000269|PubMed:19410646, ECO:0000269|PubMed:21596750, ECO:0000269|PubMed:8663044}. | FUNCTION: Multifaceted cellular regulator which coordinates several essential cellular functions including regulation of epithelial HCO3(-) and fluid secretion, mRNA processing and DNA replication. Regulates ITPR1 sensitivity to inositol 1,4,5-trisphosphate, competing for the common binding site and acting as endogenous 'pseudoligand' whose inhibitory activity can be modulated by its phosphorylation status. Promotes the formation of contact points between the endoplasmic reticulum (ER) and mitochondria, facilitating transfer of Ca(2+) from the ER to mitochondria (PubMed:27995898). Under normal cellular conditions, functions cooperatively with BCL2L10 to limit ITPR1-mediated Ca(2+) release but, under apoptotic stress conditions, dephosphorylated which promotes dissociation of both AHCYL1 and BCL2L10 from mitochondria-associated endoplasmic reticulum membranes, inhibits BCL2L10 interaction with ITPR1 and leads to increased Ca(2+) transfer to mitochondria which promotes apoptosis (PubMed:27995898). In the pancreatic and salivary ducts, at resting state, attenuates inositol 1,4,5-trisphosphate-induced calcium release by interacting with ITPR1 (PubMed:16793548). When extracellular stimuli induce ITPR1 phosphorylation or inositol 1,4,5-trisphosphate production, dissociates from ITPR1 to interact with CFTR and SLC26A6, mediating their synergistic activation by calcium and cAMP that stimulates the epithelial secretion of electrolytes and fluid (By similarity). Also activates basolateral SLC4A4 isoform 1 to coordinate fluid and HCO3(-) secretion (PubMed:16769890). Inhibits the effect of STK39 on SLC4A4 and CFTR by recruiting PP1 phosphatase which activates SLC4A4, SLC26A6 and CFTR through dephosphorylation (By similarity). Mediates the induction of SLC9A3 surface expression produced by Angiotensin-2 (PubMed:20584908). Depending on the cell type, activates SLC9A3 in response to calcium or reverses SLC9A3R2-dependent calcium inhibition (PubMed:18829453). May modulate the polyadenylation state of specific mRNAs, both by controlling the subcellular location of FIP1L1 and by inhibiting PAPOLA activity, in response to a stimulus that alters its phosphorylation state (PubMed:19224921). Acts as a (dATP)-dependent inhibitor of ribonucleotide reductase large subunit RRM1, controlling the endogenous dNTP pool and ensuring normal cell cycle progression (PubMed:25237103). In vitro does not exhibit any S-adenosyl-L-homocysteine hydrolase activity (By similarity). {ECO:0000250|UniProtKB:B5DFN2, ECO:0000250|UniProtKB:Q80SW1, ECO:0000269|PubMed:16769890, ECO:0000269|PubMed:16793548, ECO:0000269|PubMed:18829453, ECO:0000269|PubMed:19224921, ECO:0000269|PubMed:20584908, ECO:0000269|PubMed:25237103, ECO:0000269|PubMed:27995898}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000346997 | - | 16 | 17 | 154_247 | 765 | 820.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000346997 | - | 16 | 17 | 256_358 | 765 | 820.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000346997 | - | 16 | 17 | 25_125 | 765 | 820.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000351936 | - | 17 | 18 | 154_247 | 765 | 786.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000351936 | - | 17 | 18 | 256_358 | 765 | 786.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000351936 | - | 17 | 18 | 25_125 | 765 | 786.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000356226 | - | 15 | 16 | 154_247 | 650 | 705.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000356226 | - | 15 | 16 | 256_358 | 650 | 705.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000356226 | - | 15 | 16 | 25_125 | 650 | 705.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000357555 | - | 16 | 17 | 154_247 | 678 | 708.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000357555 | - | 16 | 17 | 256_358 | 678 | 708.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000357555 | - | 16 | 17 | 25_125 | 678 | 708.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000358487 | - | 17 | 18 | 154_247 | 767 | 822.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000358487 | - | 17 | 18 | 256_358 | 767 | 822.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000358487 | - | 17 | 18 | 25_125 | 767 | 822.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000358487 | - | 17 | 18 | 481_770 | 767 | 822.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000360144 | - | 16 | 17 | 154_247 | 679 | 681.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000360144 | - | 16 | 17 | 256_358 | 679 | 681.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000360144 | - | 16 | 17 | 25_125 | 679 | 681.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369056 | - | 16 | 17 | 154_247 | 768 | 770.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369056 | - | 16 | 17 | 256_358 | 768 | 770.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369056 | - | 16 | 17 | 25_125 | 768 | 770.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369056 | - | 16 | 17 | 481_770 | 768 | 770.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369060 | - | 15 | 16 | 154_247 | 651 | 706.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369060 | - | 15 | 16 | 256_358 | 651 | 706.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369060 | - | 15 | 16 | 25_125 | 651 | 706.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369061 | - | 14 | 15 | 154_247 | 655 | 710.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369061 | - | 14 | 15 | 256_358 | 655 | 710.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369061 | - | 14 | 15 | 25_125 | 655 | 710.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000457416 | - | 17 | 18 | 154_247 | 768 | 823.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000457416 | - | 17 | 18 | 256_358 | 768 | 823.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000457416 | - | 17 | 18 | 25_125 | 768 | 823.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000457416 | - | 17 | 18 | 481_770 | 768 | 823.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000346997 | - | 16 | 17 | 487_495 | 765 | 820.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000346997 | - | 16 | 17 | 565_567 | 765 | 820.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000351936 | - | 17 | 18 | 487_495 | 765 | 786.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000351936 | - | 17 | 18 | 565_567 | 765 | 786.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000356226 | - | 15 | 16 | 487_495 | 650 | 705.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000356226 | - | 15 | 16 | 565_567 | 650 | 705.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000357555 | - | 16 | 17 | 487_495 | 678 | 708.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000357555 | - | 16 | 17 | 565_567 | 678 | 708.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000358487 | - | 17 | 18 | 487_495 | 767 | 822.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000358487 | - | 17 | 18 | 565_567 | 767 | 822.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000360144 | - | 16 | 17 | 487_495 | 679 | 681.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000360144 | - | 16 | 17 | 565_567 | 679 | 681.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369056 | - | 16 | 17 | 487_495 | 768 | 770.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369056 | - | 16 | 17 | 565_567 | 768 | 770.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369060 | - | 15 | 16 | 487_495 | 651 | 706.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369060 | - | 15 | 16 | 565_567 | 651 | 706.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369061 | - | 14 | 15 | 487_495 | 655 | 710.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369061 | - | 14 | 15 | 565_567 | 655 | 710.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000457416 | - | 17 | 18 | 487_495 | 768 | 823.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000457416 | - | 17 | 18 | 565_567 | 768 | 823.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000346997 | - | 16 | 17 | 161_178 | 765 | 820.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000351936 | - | 17 | 18 | 161_178 | 765 | 786.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000356226 | - | 15 | 16 | 161_178 | 650 | 705.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000357555 | - | 16 | 17 | 161_178 | 678 | 708.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000358487 | - | 17 | 18 | 161_178 | 767 | 822.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000360144 | - | 16 | 17 | 161_178 | 679 | 681.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369056 | - | 16 | 17 | 161_178 | 768 | 770.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369060 | - | 15 | 16 | 161_178 | 651 | 706.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369061 | - | 14 | 15 | 161_178 | 655 | 710.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000457416 | - | 17 | 18 | 161_178 | 768 | 823.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000346997 | - | 16 | 17 | 22_377 | 765 | 820.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000351936 | - | 17 | 18 | 22_377 | 765 | 786.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000356226 | - | 15 | 16 | 22_377 | 650 | 705.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000357555 | - | 16 | 17 | 22_377 | 678 | 708.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000358487 | - | 17 | 18 | 22_377 | 767 | 822.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000360144 | - | 16 | 17 | 22_377 | 679 | 681.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369056 | - | 16 | 17 | 22_377 | 768 | 770.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369060 | - | 15 | 16 | 22_377 | 651 | 706.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369061 | - | 14 | 15 | 22_377 | 655 | 710.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000457416 | - | 17 | 18 | 22_377 | 768 | 823.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000346997 | - | 16 | 17 | 378_398 | 765 | 820.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000351936 | - | 17 | 18 | 378_398 | 765 | 786.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000356226 | - | 15 | 16 | 378_398 | 650 | 705.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000357555 | - | 16 | 17 | 378_398 | 678 | 708.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000358487 | - | 17 | 18 | 378_398 | 767 | 822.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000360144 | - | 16 | 17 | 378_398 | 679 | 681.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369056 | - | 16 | 17 | 378_398 | 768 | 770.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369060 | - | 15 | 16 | 378_398 | 651 | 706.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369061 | - | 14 | 15 | 378_398 | 655 | 710.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000457416 | - | 17 | 18 | 378_398 | 768 | 823.0 | Transmembrane | Helical |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000359172 | 0 | 17 | 318_322 | 0 | 484.0 | Nucleotide binding | NAD | |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000359172 | 0 | 17 | 397_399 | 0 | 484.0 | Nucleotide binding | NAD | |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000369799 | 0 | 17 | 318_322 | 40 | 531.0 | Nucleotide binding | NAD | |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000369799 | 0 | 17 | 397_399 | 40 | 531.0 | Nucleotide binding | NAD | |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000393614 | 0 | 17 | 318_322 | 0 | 484.0 | Nucleotide binding | NAD | |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000393614 | 0 | 17 | 397_399 | 0 | 484.0 | Nucleotide binding | NAD | |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000359172 | 0 | 17 | 281_448 | 0 | 484.0 | Region | NAD binding | |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000359172 | 0 | 17 | 520_530 | 0 | 484.0 | Region | PDZ-binding | |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000359172 | 0 | 17 | 65_92 | 0 | 484.0 | Region | PEST | |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000369799 | 0 | 17 | 281_448 | 40 | 531.0 | Region | NAD binding | |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000369799 | 0 | 17 | 520_530 | 40 | 531.0 | Region | PDZ-binding | |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000369799 | 0 | 17 | 65_92 | 40 | 531.0 | Region | PEST | |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000393614 | 0 | 17 | 281_448 | 0 | 484.0 | Region | NAD binding | |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000393614 | 0 | 17 | 520_530 | 0 | 484.0 | Region | PDZ-binding | |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000393614 | 0 | 17 | 65_92 | 0 | 484.0 | Region | PEST |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000346997 | - | 16 | 17 | 481_770 | 765 | 820.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000351936 | - | 17 | 18 | 481_770 | 765 | 786.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000356226 | - | 15 | 16 | 481_770 | 650 | 705.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000357555 | - | 16 | 17 | 481_770 | 678 | 708.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000359354 | - | 1 | 7 | 154_247 | 0 | 255.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000359354 | - | 1 | 7 | 256_358 | 0 | 255.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000359354 | - | 1 | 7 | 25_125 | 0 | 255.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000359354 | - | 1 | 7 | 481_770 | 0 | 255.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000360144 | - | 16 | 17 | 481_770 | 679 | 681.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369060 | - | 15 | 16 | 481_770 | 651 | 706.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369061 | - | 14 | 15 | 481_770 | 655 | 710.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000359354 | - | 1 | 7 | 487_495 | 0 | 255.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000359354 | - | 1 | 7 | 565_567 | 0 | 255.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000359354 | - | 1 | 7 | 161_178 | 0 | 255.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000346997 | - | 16 | 17 | 399_821 | 765 | 820.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000351936 | - | 17 | 18 | 399_821 | 765 | 786.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000356226 | - | 15 | 16 | 399_821 | 650 | 705.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000357555 | - | 16 | 17 | 399_821 | 678 | 708.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000358487 | - | 17 | 18 | 399_821 | 767 | 822.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000359354 | - | 1 | 7 | 22_377 | 0 | 255.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000359354 | - | 1 | 7 | 399_821 | 0 | 255.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000360144 | - | 16 | 17 | 399_821 | 679 | 681.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369056 | - | 16 | 17 | 399_821 | 768 | 770.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369060 | - | 15 | 16 | 399_821 | 651 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000369061 | - | 14 | 15 | 399_821 | 655 | 710.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000457416 | - | 17 | 18 | 399_821 | 768 | 823.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123243212 | chr1:110551654 | ENST00000359354 | - | 1 | 7 | 378_398 | 0 | 255.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for FGFR2-AHCYL1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >30273_30273_1_FGFR2-AHCYL1_FGFR2_chr10_123243212_ENST00000346997_AHCYL1_chr1_110551654_ENST00000369799_length(transcript)=5835nt_BP=2307nt GGTACCGTAACCATGGTCAGCTGGGGTCGTTTCATCTGCCTGGTCGTGGTCACCATGGCAACCTTGTCCCTGGCCCGGCCCTCCTTCAGT TTAGTTGAGGATACCACATTAGAGCCAGAAGAGCCACCAACCAAATACCAAATCTCTCAACCAGAAGTGTACGTGGCTGCGCCAGGGGAG TCGCTAGAGGTGCGCTGCCTGTTGAAAGATGCCGCCGTGATCAGTTGGACTAAGGATGGGGTGCACTTGGGGCCCAACAATAGGACAGTG CTTATTGGGGAGTACTTGCAGATAAAGGGCGCCACGCCTAGAGACTCCGGCCTCTATGCTTGTACTGCCAGTAGGACTGTAGACAGTGAA ACTTGGTACTTCATGGTGAATGTCACAGATGCCATCTCATCCGGAGATGATGAGGATGACACCGATGGTGCGGAAGATTTTGTCAGTGAG AACAGTAACAACAAGAGAGCACCATACTGGACCAACACAGAAAAGATGGAAAAGCGGCTCCATGCTGTGCCTGCGGCCAACACTGTCAAG TTTCGCTGCCCAGCCGGGGGGAACCCAATGCCAACCATGCGGTGGCTGAAAAACGGGAAGGAGTTTAAGCAGGAGCATCGCATTGGAGGC TACAAGGTACGAAACCAGCACTGGAGCCTCATTATGGAAAGTGTGGTCCCATCTGACAAGGGAAATTATACCTGTGTAGTGGAGAATGAA TACGGGTCCATCAATCACACGTACCACCTGGATGTTGTGGAGCGATCGCCTCACCGGCCCATCCTCCAAGCCGGACTGCCGGCAAATGCC TCCACAGTGGTCGGAGGAGACGTAGAGTTTGTCTGCAAGGTTTACAGTGATGCCCAGCCCCACATCCAGTGGATCAAGCACGTGGAAAAG AACGGCAGTAAATACGGGCCCGACGGGCTGCCCTACCTCAAGGTTCTCAAGGCCGCCGGTGTTAACACCACGGACAAAGAGATTGAGGTT CTCTATATTCGGAATGTAACTTTTGAGGACGCTGGGGAATATACGTGCTTGGCGGGTAATTCTATTGGGATATCCTTTCACTCTGCATGG TTGACAGTTCTGCCAGCGCCTGGAAGAGAAAAGGAGATTACAGCTTCCCCAGACTACCTGGAGATAGCCATTTACTGCATAGGGGTCTTC TTAATCGCCTGTATGGTGGTAACAGTCATCCTGTGCCGAATGAAGAACACGACCAAGAAGCCAGACTTCAGCAGCCAGCCGGCTGTGCAC AAGCTGACCAAACGTATCCCCCTGCGGAGACAGGTTTCGGCTGAGTCCAGCTCCTCCATGAACTCCAACACCCCGCTGGTGAGGATAACA ACACGCCTCTCTTCAACGGCAGACACCCCCATGCTGGCAGGGGTCTCCGAGTATGAACTTCCAGAGGACCCAAAATGGGAGTTTCCAAGA GATAAGCTGACACTGGGCAAGCCCCTGGGAGAAGGTTGCTTTGGGCAAGTGGTCATGGCGGAAGCAGTGGGAATTGACAAAGACAAGCCC AAGGAGGCGGTCACCGTGGCCGTGAAGATGTTGAAAGATGATGCCACAGAGAAAGACCTTTCTGATCTGGTGTCAGAGATGGAGATGATG AAGATGATTGGGAAACACAAGAATATCATAAATCTTCTTGGAGCCTGCACACAGGATGGGCCTCTCTATGTCATAGTTGAGTATGCCTCT AAAGGCAACCTCCGAGAATACCTCCGAGCCCGGAGGCCACCCGGGATGGAGTACTCCTATGACATTAACCGTGTTCCTGAGGAGCAGATG ACCTTCAAGGACTTGGTGTCATGCACCTACCAGCTGGCCAGAGGCATGGAGTACTTGGCTTCCCAAAAATGTATTCATCGAGATTTAGCA GCCAGAAATGTTTTGGTAACAGAAAACAATGTGATGAAAATAGCAGACTTTGGACTCGCCAGAGATATCAACAATATAGACTATTACAAA AAGACCACCAATGGGCGGCTTCCAGTCAAGTGGATGGCTCCAGAAGCCCTGTTTGATAGAGTATACACTCATCAGAGTGATGTCTGGTCC TTCGGGGTGTTAATGTGGGAGATCTTCACTTTAGGGGGCTCGCCCTACCCAGGGATTCCCGTGGAGGAACTTTTTAAGCTGCTGAAGGAA GGACACAGAATGGATAAGCCAGCCAACTGCACCAACGAACTGTACATGATGATGAGGGACTGTTGGCATGCAGTGCCCTCCCAGAGACCA ACGTTCAAGCAGTTGGTAGAAGACTTGGATCGAATTCTCACTCTCACAACCAATGAGCAAATCCAGTTTGCTGATGACATGCAGGAGTTC ACCAAATTCCCCACCAAAACTGGCCGAAGATCTTTGTCTCGCTCGATCTCACAGTCCTCCACTGACAGCTACAGTTCAGCTGCATCCTAC ACAGATAGCTCTGATGATGAGGTTTCTCCCCGAGAGAAGCAGCAAACCAACTCCAAGGGCAGCAGCAATTTCTGTGTGAAGAACATCAAG CAGGCAGAATTTGGACGCCGGGAGATTGAGATTGCAGAGCAAGACATGTCTGCTCTGATTTCACTCAGGAAACGTGCTCAGGGGGAGAAG CCCTTGGCTGGTGCTAAAATAGTGGGCTGTACACACATCACAGCCCAGACAGCGGTGTTGATTGAGACACTCTGTGCCCTGGGGGCTCAG TGCCGCTGGTCTGCTTGTAACATCTACTCAACTCAGAATGAAGTAGCTGCAGCACTGGCTGAGGCTGGAGTTGCAGTGTTCGCTTGGAAG GGCGAGTCAGAAGATGACTTCTGGTGGTGTATTGACCGCTGTGTGAACATGGATGGGTGGCAGGCCAACATGATCCTGGATGATGGGGGA GACTTAACCCACTGGGTTTATAAGAAGTATCCAAACGTGTTTAAGAAGATCCGAGGCATTGTGGAAGAGAGCGTGACTGGTGTTCACAGG CTGTATCAGCTCTCCAAAGCTGGGAAGCTCTGTGTTCCGGCCATGAACGTCAATGATTCTGTTACCAAACAGAAGTTTGATAACTTGTAC TGCTGCCGAGAATCCATTTTGGATGGCCTGAAGAGGACCACAGATGTGATGTTTGGTGGGAAACAAGTGGTGGTGTGTGGCTATGGTGAG GTAGGCAAGGGCTGCTGTGCTGCTCTCAAAGCTCTTGGAGCAATTGTCTACATTACCGAAATCGACCCCATCTGTGCTCTGCAGGCCTGC ATGGATGGGTTCAGGGTGGTAAAGCTAAATGAAGTCATCCGGCAAGTCGATGTCGTAATAACTTGCACAGGAAATAAGAATGTAGTGACA CGGGAGCACTTGGATCGCATGAAAAACAGTTGTATCGTATGCAATATGGGCCACTCCAACACAGAAATCGATGTGACCAGCCTCCGCACT CCGGAGCTGACGTGGGAGCGAGTACGTTCTCAGGTGGACCATGTCATCTGGCCAGATGGCAAACGAGTTGTCCTCCTGGCAGAGGGTCGT CTACTCAATTTGAGCTGCTCCACAGTTCCCACCTTTGTTCTGTCCATCACAGCCACAACACAGGCTTTGGCACTGATAGAACTCTATAAT GCACCCGAGGGGCGATACAAGCAGGATGTGTACTTGCTTCCTAAGAAAATGGATGAATACGTTGCCAGCTTGCATCTGCCATCATTTGAT GCCCACCTTACAGAGCTGACAGATGACCAAGCAAAATATCTGGGACTCAACAAAAATGGGCCATTCAAACCTAATTATTACAGATACTAA TGGACCATACTACCAAGGACCAGTCCACCTGAACCACACACTCTAAAGAAATATTTTTTAAGATAACTTTTATTTTCTTCTTACTCCTTT CCTCTTGATTTTTTTCCTATAATTTCATTCTTGTTTTTTCATCTCATTATCCAAGTTCTGCAGACCACACAGGAACTTGCTTCATGGCTC TTTAGATGAAATAGAAGTTCAGGGTTCCTCACTCTAGTCACTAAAGAAGGATTTTACTCTCCCAGCCCAGAAAGGTGATTCTTTCTTTAC CATTTCTGGGGACTTTAGTCTTAATTAGGTACCTTATTAACAGGAAATGCTAAGGTACCTTCTCTGTGGAACAATCTGCAATGTCTAAAT CGCCTTAAAAGAGCCCATTTCTTAGCTGCTGAAATCAGTGCTCTTTCACTTCTTCAGAGAAGCAGGGATGGTACCTACCCGGCAGGTAGG TTAGATGTGGGTGGTGCATGTTAATTTCCCTTAGAAGTTCCAAGCCCTGTTTCCTGCGTAAAGGTGGTATGTCCAGTTCAGAGATGTGTA TAATGAGCATGGCTTGTTAAGATCAGGAGGCCCACTTGGATTTATAGTATAGCCCTTCCTCCACTCCCACCAGACTTGCTCATTTTTCGA GTTTTTAACTAGACTACACTCTATTGAGTTTAATTTTGTCCTCTAGGATTTATTTCTGTTGTCCAAAAAAAAAAAAAAAGAAAAGAAAAA TTAAGGAGAATTTTTGGTGTTAATGCTGAGGAATTGCTTGAGTGGTTAGTTGTTACCAATTTCTCTTTTGAACCTTTGGAGCTAAGGATG CTGAGTCTAGAGAAATGCTAGTCTCAAGCCCTGTTAAGTCCCTCTGTTTCTAGCCCGTAGTTCATAGCATCAGTGAACTGGAGCCACAAC AGCAAATTCTATCAGCTGTGTACCATACAGCTTGTGCTGAAGGCGAATTTCTTGAGCCATTACTCAGTATAAAGCACTGAGTTCTATCTT TAGGATTTATCTTTAAGAGCAAATTTCTGGTCAGCTGTGCTTCTGCAACCTAAAATATTTAAAGGGAGGTAGGTGTGGGCAGGAGGAGGA ATGATAAATTGGGCCAGGGCAAGAAAAATCTAGCTTCATATAATTTGTCTGGGACTATACACCCTATATAATGTTAGTTTTACAGAAGTA ATATGACTTTTGATTGCTACATACCACAAAGAGTTTATGAACTGAGATCATAAAGGGCAACTGATGTGTGAAGAAAGTAGTCAGTACATC CTGGCTCATGCTCTGAAAGAATATCCAGAGAGGCTCTCTCAAAGATCAGGGAGATGTATTCCCATGCCATGCACCCTGCTTCCCAGCATT TCTGCATGGTCAAGTGAGCTTTATGCTCATGAGCTTTAAGTATATAATTATCCAGGATTTTAAATCCTCAACTTGTTCTAGCTTGTGATC CCTCAAAGTTGGGTCATACGTTAGTGCTAGATACTAGAAATTTTCACTTTTCCACTGATCAGAGAGACAGACATTAAAAACAAAAATAGA AGAAAGGAAAGCTTTCACCCTGCAGCTTCTTAGCAGGGAACAATTGTCTTGCCAAAACTTTTTTCCCTTTTCTCTCCCATTTTCTTTTAC CCAATCCCTTCTTACTCCTTGCCAGTGTGACCATGCTTTCTTCTCTGTAGATGTTAACAGTTAAGGCCTATTTTCCTCGGGCACTTAACC AACCAATCAGAACACCACATCTGTTAGGGGAGGTAACCTGGCCAACAGTGTATCCATCACGTTAGCCCTGCTGGAGGGAAGGGACCCACA TTCACCTGCCCTCTGACCTGCCCCTTGATCCCATATCTATTACCGTGTCCATAGGAATAATAGGTAAGGGCTCTGTCTCTGTCAAGCCAT GTAACAAAGGACACTGTTAAAAAAAAAAAAAAGTCTGGCATCAGAGGGAGCATGTGGAGAGCAACTTGGGAAGAACAAGTTCATTTTGTA >30273_30273_1_FGFR2-AHCYL1_FGFR2_chr10_123243212_ENST00000346997_AHCYL1_chr1_110551654_ENST00000369799_length(amino acids)=1255AA_BP=765 MVSWGRFICLVVVTMATLSLARPSFSLVEDTTLEPEEPPTKYQISQPEVYVAAPGESLEVRCLLKDAAVISWTKDGVHLGPNNRTVLIGE YLQIKGATPRDSGLYACTASRTVDSETWYFMVNVTDAISSGDDEDDTDGAEDFVSENSNNKRAPYWTNTEKMEKRLHAVPAANTVKFRCP AGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVV GGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVL PAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSAESSSSMNSNTPLVRITTRLS STADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIG KHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNV LVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRM DKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEQIQFADDMQEFTKFPTKTGRRSLSRSISQSSTDSYSSAASYTDSS DDEVSPREKQQTNSKGSSNFCVKNIKQAEFGRREIEIAEQDMSALISLRKRAQGEKPLAGAKIVGCTHITAQTAVLIETLCALGAQCRWS ACNIYSTQNEVAAALAEAGVAVFAWKGESEDDFWWCIDRCVNMDGWQANMILDDGGDLTHWVYKKYPNVFKKIRGIVEESVTGVHRLYQL SKAGKLCVPAMNVNDSVTKQKFDNLYCCRESILDGLKRTTDVMFGGKQVVVCGYGEVGKGCCAALKALGAIVYITEIDPICALQACMDGF RVVKLNEVIRQVDVVITCTGNKNVVTREHLDRMKNSCIVCNMGHSNTEIDVTSLRTPELTWERVRSQVDHVIWPDGKRVVLLAEGRLLNL -------------------------------------------------------------- >30273_30273_2_FGFR2-AHCYL1_FGFR2_chr10_123243212_ENST00000369056_AHCYL1_chr1_110551654_ENST00000369799_length(transcript)=5856nt_BP=2328nt GGACCGGGGATTGGTACCGTAACCATGGTCAGCTGGGGTCGTTTCATCTGCCTGGTCGTGGTCACCATGGCAACCTTGTCCCTGGCCCGG CCCTCCTTCAGTTTAGTTGAGGATACCACATTAGAGCCAGAAGAGCCACCAACCAAATACCAAATCTCTCAACCAGAAGTGTACGTGGCT GCGCCAGGGGAGTCGCTAGAGGTGCGCTGCCTGTTGAAAGATGCCGCCGTGATCAGTTGGACTAAGGATGGGGTGCACTTGGGGCCCAAC AATAGGACAGTGCTTATTGGGGAGTACTTGCAGATAAAGGGCGCCACGCCTAGAGACTCCGGCCTCTATGCTTGTACTGCCAGTAGGACT GTAGACAGTGAAACTTGGTACTTCATGGTGAATGTCACAGATGCCATCTCATCCGGAGATGATGAGGATGACACCGATGGTGCGGAAGAT TTTGTCAGTGAGAACAGTAACAACAAGAGAGCACCATACTGGACCAACACAGAAAAGATGGAAAAGCGGCTCCATGCTGTGCCTGCGGCC AACACTGTCAAGTTTCGCTGCCCAGCCGGGGGGAACCCAATGCCAACCATGCGGTGGCTGAAAAACGGGAAGGAGTTTAAGCAGGAGCAT CGCATTGGAGGCTACAAGGTACGAAACCAGCACTGGAGCCTCATTATGGAAAGTGTGGTCCCATCTGACAAGGGAAATTATACCTGTGTA GTGGAGAATGAATACGGGTCCATCAATCACACGTACCACCTGGATGTTGTGGAGCGATCGCCTCACCGGCCCATCCTCCAAGCCGGACTG CCGGCAAATGCCTCCACAGTGGTCGGAGGAGACGTAGAGTTTGTCTGCAAGGTTTACAGTGATGCCCAGCCCCACATCCAGTGGATCAAG CACGTGGAAAAGAACGGCAGTAAATACGGGCCCGACGGGCTGCCCTACCTCAAGGTTCTCAAGCACTCGGGGATAAATAGTTCCAATGCA GAAGTGCTGGCTCTGTTCAATGTGACCGAGGCGGATGCTGGGGAATATATATGTAAGGTCTCCAATTATATAGGGCAGGCCAACCAGTCT GCCTGGCTCACTGTCCTGCCAAAACAGCAAGCGCCTGGAAGAGAAAAGGAGATTACAGCTTCCCCAGACTACCTGGAGATAGCCATTTAC TGCATAGGGGTCTTCTTAATCGCCTGTATGGTGGTAACAGTCATCCTGTGCCGAATGAAGAACACGACCAAGAAGCCAGACTTCAGCAGC CAGCCGGCTGTGCACAAGCTGACCAAACGTATCCCCCTGCGGAGACAGGTAACAGTTTCGGCTGAGTCCAGCTCCTCCATGAACTCCAAC ACCCCGCTGGTGAGGATAACAACACGCCTCTCTTCAACGGCAGACACCCCCATGCTGGCAGGGGTCTCCGAGTATGAACTTCCAGAGGAC CCAAAATGGGAGTTTCCAAGAGATAAGCTGACACTGGGCAAGCCCCTGGGAGAAGGTTGCTTTGGGCAAGTGGTCATGGCGGAAGCAGTG GGAATTGACAAAGACAAGCCCAAGGAGGCGGTCACCGTGGCCGTGAAGATGTTGAAAGATGATGCCACAGAGAAAGACCTTTCTGATCTG GTGTCAGAGATGGAGATGATGAAGATGATTGGGAAACACAAGAATATCATAAATCTTCTTGGAGCCTGCACACAGGATGGGCCTCTCTAT GTCATAGTTGAGTATGCCTCTAAAGGCAACCTCCGAGAATACCTCCGAGCCCGGAGGCCACCCGGGATGGAGTACTCCTATGACATTAAC CGTGTTCCTGAGGAGCAGATGACCTTCAAGGACTTGGTGTCATGCACCTACCAGCTGGCCAGAGGCATGGAGTACTTGGCTTCCCAAAAA TGTATTCATCGAGATTTAGCAGCCAGAAATGTTTTGGTAACAGAAAACAATGTGATGAAAATAGCAGACTTTGGACTCGCCAGAGATATC AACAATATAGACTATTACAAAAAGACCACCAATGGGCGGCTTCCAGTCAAGTGGATGGCTCCAGAAGCCCTGTTTGATAGAGTATACACT CATCAGAGTGATGTCTGGTCCTTCGGGGTGTTAATGTGGGAGATCTTCACTTTAGGGGGCTCGCCCTACCCAGGGATTCCCGTGGAGGAA CTTTTTAAGCTGCTGAAGGAAGGACACAGAATGGATAAGCCAGCCAACTGCACCAACGAACTGTACATGATGATGAGGGACTGTTGGCAT GCAGTGCCCTCCCAGAGACCAACGTTCAAGCAGTTGGTAGAAGACTTGGATCGAATTCTCACTCTCACAACCAATGAGCAAATCCAGTTT GCTGATGACATGCAGGAGTTCACCAAATTCCCCACCAAAACTGGCCGAAGATCTTTGTCTCGCTCGATCTCACAGTCCTCCACTGACAGC TACAGTTCAGCTGCATCCTACACAGATAGCTCTGATGATGAGGTTTCTCCCCGAGAGAAGCAGCAAACCAACTCCAAGGGCAGCAGCAAT TTCTGTGTGAAGAACATCAAGCAGGCAGAATTTGGACGCCGGGAGATTGAGATTGCAGAGCAAGACATGTCTGCTCTGATTTCACTCAGG AAACGTGCTCAGGGGGAGAAGCCCTTGGCTGGTGCTAAAATAGTGGGCTGTACACACATCACAGCCCAGACAGCGGTGTTGATTGAGACA CTCTGTGCCCTGGGGGCTCAGTGCCGCTGGTCTGCTTGTAACATCTACTCAACTCAGAATGAAGTAGCTGCAGCACTGGCTGAGGCTGGA GTTGCAGTGTTCGCTTGGAAGGGCGAGTCAGAAGATGACTTCTGGTGGTGTATTGACCGCTGTGTGAACATGGATGGGTGGCAGGCCAAC ATGATCCTGGATGATGGGGGAGACTTAACCCACTGGGTTTATAAGAAGTATCCAAACGTGTTTAAGAAGATCCGAGGCATTGTGGAAGAG AGCGTGACTGGTGTTCACAGGCTGTATCAGCTCTCCAAAGCTGGGAAGCTCTGTGTTCCGGCCATGAACGTCAATGATTCTGTTACCAAA CAGAAGTTTGATAACTTGTACTGCTGCCGAGAATCCATTTTGGATGGCCTGAAGAGGACCACAGATGTGATGTTTGGTGGGAAACAAGTG GTGGTGTGTGGCTATGGTGAGGTAGGCAAGGGCTGCTGTGCTGCTCTCAAAGCTCTTGGAGCAATTGTCTACATTACCGAAATCGACCCC ATCTGTGCTCTGCAGGCCTGCATGGATGGGTTCAGGGTGGTAAAGCTAAATGAAGTCATCCGGCAAGTCGATGTCGTAATAACTTGCACA GGAAATAAGAATGTAGTGACACGGGAGCACTTGGATCGCATGAAAAACAGTTGTATCGTATGCAATATGGGCCACTCCAACACAGAAATC GATGTGACCAGCCTCCGCACTCCGGAGCTGACGTGGGAGCGAGTACGTTCTCAGGTGGACCATGTCATCTGGCCAGATGGCAAACGAGTT GTCCTCCTGGCAGAGGGTCGTCTACTCAATTTGAGCTGCTCCACAGTTCCCACCTTTGTTCTGTCCATCACAGCCACAACACAGGCTTTG GCACTGATAGAACTCTATAATGCACCCGAGGGGCGATACAAGCAGGATGTGTACTTGCTTCCTAAGAAAATGGATGAATACGTTGCCAGC TTGCATCTGCCATCATTTGATGCCCACCTTACAGAGCTGACAGATGACCAAGCAAAATATCTGGGACTCAACAAAAATGGGCCATTCAAA CCTAATTATTACAGATACTAATGGACCATACTACCAAGGACCAGTCCACCTGAACCACACACTCTAAAGAAATATTTTTTAAGATAACTT TTATTTTCTTCTTACTCCTTTCCTCTTGATTTTTTTCCTATAATTTCATTCTTGTTTTTTCATCTCATTATCCAAGTTCTGCAGACCACA CAGGAACTTGCTTCATGGCTCTTTAGATGAAATAGAAGTTCAGGGTTCCTCACTCTAGTCACTAAAGAAGGATTTTACTCTCCCAGCCCA GAAAGGTGATTCTTTCTTTACCATTTCTGGGGACTTTAGTCTTAATTAGGTACCTTATTAACAGGAAATGCTAAGGTACCTTCTCTGTGG AACAATCTGCAATGTCTAAATCGCCTTAAAAGAGCCCATTTCTTAGCTGCTGAAATCAGTGCTCTTTCACTTCTTCAGAGAAGCAGGGAT GGTACCTACCCGGCAGGTAGGTTAGATGTGGGTGGTGCATGTTAATTTCCCTTAGAAGTTCCAAGCCCTGTTTCCTGCGTAAAGGTGGTA TGTCCAGTTCAGAGATGTGTATAATGAGCATGGCTTGTTAAGATCAGGAGGCCCACTTGGATTTATAGTATAGCCCTTCCTCCACTCCCA CCAGACTTGCTCATTTTTCGAGTTTTTAACTAGACTACACTCTATTGAGTTTAATTTTGTCCTCTAGGATTTATTTCTGTTGTCCAAAAA AAAAAAAAAAGAAAAGAAAAATTAAGGAGAATTTTTGGTGTTAATGCTGAGGAATTGCTTGAGTGGTTAGTTGTTACCAATTTCTCTTTT GAACCTTTGGAGCTAAGGATGCTGAGTCTAGAGAAATGCTAGTCTCAAGCCCTGTTAAGTCCCTCTGTTTCTAGCCCGTAGTTCATAGCA TCAGTGAACTGGAGCCACAACAGCAAATTCTATCAGCTGTGTACCATACAGCTTGTGCTGAAGGCGAATTTCTTGAGCCATTACTCAGTA TAAAGCACTGAGTTCTATCTTTAGGATTTATCTTTAAGAGCAAATTTCTGGTCAGCTGTGCTTCTGCAACCTAAAATATTTAAAGGGAGG TAGGTGTGGGCAGGAGGAGGAATGATAAATTGGGCCAGGGCAAGAAAAATCTAGCTTCATATAATTTGTCTGGGACTATACACCCTATAT AATGTTAGTTTTACAGAAGTAATATGACTTTTGATTGCTACATACCACAAAGAGTTTATGAACTGAGATCATAAAGGGCAACTGATGTGT GAAGAAAGTAGTCAGTACATCCTGGCTCATGCTCTGAAAGAATATCCAGAGAGGCTCTCTCAAAGATCAGGGAGATGTATTCCCATGCCA TGCACCCTGCTTCCCAGCATTTCTGCATGGTCAAGTGAGCTTTATGCTCATGAGCTTTAAGTATATAATTATCCAGGATTTTAAATCCTC AACTTGTTCTAGCTTGTGATCCCTCAAAGTTGGGTCATACGTTAGTGCTAGATACTAGAAATTTTCACTTTTCCACTGATCAGAGAGACA GACATTAAAAACAAAAATAGAAGAAAGGAAAGCTTTCACCCTGCAGCTTCTTAGCAGGGAACAATTGTCTTGCCAAAACTTTTTTCCCTT TTCTCTCCCATTTTCTTTTACCCAATCCCTTCTTACTCCTTGCCAGTGTGACCATGCTTTCTTCTCTGTAGATGTTAACAGTTAAGGCCT ATTTTCCTCGGGCACTTAACCAACCAATCAGAACACCACATCTGTTAGGGGAGGTAACCTGGCCAACAGTGTATCCATCACGTTAGCCCT GCTGGAGGGAAGGGACCCACATTCACCTGCCCTCTGACCTGCCCCTTGATCCCATATCTATTACCGTGTCCATAGGAATAATAGGTAAGG GCTCTGTCTCTGTCAAGCCATGTAACAAAGGACACTGTTAAAAAAAAAAAAAAGTCTGGCATCAGAGGGAGCATGTGGAGAGCAACTTGG GAAGAACAAGTTCATTTTGTATTGAATGATTTTTAATGAATGCAATATTAATCCTTGCAGATGAGCAATAATCATTAAAATCGATTAAAA >30273_30273_2_FGFR2-AHCYL1_FGFR2_chr10_123243212_ENST00000369056_AHCYL1_chr1_110551654_ENST00000369799_length(amino acids)=1258AA_BP=768 MVSWGRFICLVVVTMATLSLARPSFSLVEDTTLEPEEPPTKYQISQPEVYVAAPGESLEVRCLLKDAAVISWTKDGVHLGPNNRTVLIGE YLQIKGATPRDSGLYACTASRTVDSETWYFMVNVTDAISSGDDEDDTDGAEDFVSENSNNKRAPYWTNTEKMEKRLHAVPAANTVKFRCP AGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVV GGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYICKVSNYIGQANQSAWLTVLPK QQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITT RLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMK MIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAA RNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEG HRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEQIQFADDMQEFTKFPTKTGRRSLSRSISQSSTDSYSSAASYT DSSDDEVSPREKQQTNSKGSSNFCVKNIKQAEFGRREIEIAEQDMSALISLRKRAQGEKPLAGAKIVGCTHITAQTAVLIETLCALGAQC RWSACNIYSTQNEVAAALAEAGVAVFAWKGESEDDFWWCIDRCVNMDGWQANMILDDGGDLTHWVYKKYPNVFKKIRGIVEESVTGVHRL YQLSKAGKLCVPAMNVNDSVTKQKFDNLYCCRESILDGLKRTTDVMFGGKQVVVCGYGEVGKGCCAALKALGAIVYITEIDPICALQACM DGFRVVKLNEVIRQVDVVITCTGNKNVVTREHLDRMKNSCIVCNMGHSNTEIDVTSLRTPELTWERVRSQVDHVIWPDGKRVVLLAEGRL -------------------------------------------------------------- >30273_30273_3_FGFR2-AHCYL1_FGFR2_chr10_123243212_ENST00000369061_AHCYL1_chr1_110551654_ENST00000369799_length(transcript)=5643nt_BP=2115nt TGACTGCAGCAGCAGCGGCAGCGCCTCGGTTCCTGAGCCCACCGCAGGCTGAAGGCATTGCGCGTAGTCCATGCCCGTAGAGGAAGTGTG CAGATGGGATTAACGTCCACATGGAGATATGGAAGAGGACCGGGGATTGGTACCGTAACCATGGTCAGCTGGGGTCGTTTCATCTGCCTG GTCGTGGTCACCATGGCAACCTTGTCCCTGGCCCGGCCCTCCTTCAGTTTAGTTGAGGATACCACATTAGAGCCAGAAGAGCCACCAACC AAATACCAAATCTCTCAACCAGAAGTGTACGTGGCTGCGCCAGGGGAGTCGCTAGAGGTGCGCTGCCTGTTGAAAGATGCCGCCGTGATC AGTTGGACTAAGGATGGGGTGCACTTGGGGCCCAACAATAGGACAGTGCTTATTGGGGAGTACTTGCAGATAAAGGGCGCCACGCCTAGA GACTCCGGCCTCTATGCTTGTACTGCCAGTAGGACTGTAGACAGTGAAACTTGGTACTTCATGGTGAATGTCACAGATGCCATCTCATCC GGAGATGATGAGGATGACACCGATGGTGCGGAAGATTTTGTCAGTGAGAACAGTAACAACAAGAGAGCACCATACTGGACCAACACAGAA AAGATGGAAAAGCGGCTCCATGCTGTGCCTGCGGCCAACACTGTCAAGTTTCGCTGCCCAGCCGGGGGGAACCCAATGCCAACCATGCGG TGGCTGAAAAACGGGAAGGAGTTTAAGCAGGAGCATCGCATTGGAGGCTACAAGGTACGAAACCAGCACTGGAGCCTCATTATGGAAAGT GTGGTCCCATCTGACAAGGGAAATTATACCTGTGTAGTGGAGAATGAATACGGGTCCATCAATCACACGTACCACCTGGATGTTGTGGCG CCTGGAAGAGAAAAGGAGATTACAGCTTCCCCAGACTACCTGGAGATAGCCATTTACTGCATAGGGGTCTTCTTAATCGCCTGTATGGTG GTAACAGTCATCCTGTGCCGAATGAAGAACACGACCAAGAAGCCAGACTTCAGCAGCCAGCCGGCTGTGCACAAGCTGACCAAACGTATC CCCCTGCGGAGACAGGTAACAGTTTCGGCTGAGTCCAGCTCCTCCATGAACTCCAACACCCCGCTGGTGAGGATAACAACACGCCTCTCT TCAACGGCAGACACCCCCATGCTGGCAGGGGTCTCCGAGTATGAACTTCCAGAGGACCCAAAATGGGAGTTTCCAAGAGATAAGCTGACA CTGGGCAAGCCCCTGGGAGAAGGTTGCTTTGGGCAAGTGGTCATGGCGGAAGCAGTGGGAATTGACAAAGACAAGCCCAAGGAGGCGGTC ACCGTGGCCGTGAAGATGTTGAAAGATGATGCCACAGAGAAAGACCTTTCTGATCTGGTGTCAGAGATGGAGATGATGAAGATGATTGGG AAACACAAGAATATCATAAATCTTCTTGGAGCCTGCACACAGGATGGGCCTCTCTATGTCATAGTTGAGTATGCCTCTAAAGGCAACCTC CGAGAATACCTCCGAGCCCGGAGGCCACCCGGGATGGAGTACTCCTATGACATTAACCGTGTTCCTGAGGAGCAGATGACCTTCAAGGAC TTGGTGTCATGCACCTACCAGCTGGCCAGAGGCATGGAGTACTTGGCTTCCCAAAAATGTATTCATCGAGATTTAGCAGCCAGAAATGTT TTGGTAACAGAAAACAATGTGATGAAAATAGCAGACTTTGGACTCGCCAGAGATATCAACAATATAGACTATTACAAAAAGACCACCAAT GGGCGGCTTCCAGTCAAGTGGATGGCTCCAGAAGCCCTGTTTGATAGAGTATACACTCATCAGAGTGATGTCTGGTCCTTCGGGGTGTTA ATGTGGGAGATCTTCACTTTAGGGGGCTCGCCCTACCCAGGGATTCCCGTGGAGGAACTTTTTAAGCTGCTGAAGGAAGGACACAGAATG GATAAGCCAGCCAACTGCACCAACGAACTGTACATGATGATGAGGGACTGTTGGCATGCAGTGCCCTCCCAGAGACCAACGTTCAAGCAG TTGGTAGAAGACTTGGATCGAATTCTCACTCTCACAACCAATGAGCAAATCCAGTTTGCTGATGACATGCAGGAGTTCACCAAATTCCCC ACCAAAACTGGCCGAAGATCTTTGTCTCGCTCGATCTCACAGTCCTCCACTGACAGCTACAGTTCAGCTGCATCCTACACAGATAGCTCT GATGATGAGGTTTCTCCCCGAGAGAAGCAGCAAACCAACTCCAAGGGCAGCAGCAATTTCTGTGTGAAGAACATCAAGCAGGCAGAATTT GGACGCCGGGAGATTGAGATTGCAGAGCAAGACATGTCTGCTCTGATTTCACTCAGGAAACGTGCTCAGGGGGAGAAGCCCTTGGCTGGT GCTAAAATAGTGGGCTGTACACACATCACAGCCCAGACAGCGGTGTTGATTGAGACACTCTGTGCCCTGGGGGCTCAGTGCCGCTGGTCT GCTTGTAACATCTACTCAACTCAGAATGAAGTAGCTGCAGCACTGGCTGAGGCTGGAGTTGCAGTGTTCGCTTGGAAGGGCGAGTCAGAA GATGACTTCTGGTGGTGTATTGACCGCTGTGTGAACATGGATGGGTGGCAGGCCAACATGATCCTGGATGATGGGGGAGACTTAACCCAC TGGGTTTATAAGAAGTATCCAAACGTGTTTAAGAAGATCCGAGGCATTGTGGAAGAGAGCGTGACTGGTGTTCACAGGCTGTATCAGCTC TCCAAAGCTGGGAAGCTCTGTGTTCCGGCCATGAACGTCAATGATTCTGTTACCAAACAGAAGTTTGATAACTTGTACTGCTGCCGAGAA TCCATTTTGGATGGCCTGAAGAGGACCACAGATGTGATGTTTGGTGGGAAACAAGTGGTGGTGTGTGGCTATGGTGAGGTAGGCAAGGGC TGCTGTGCTGCTCTCAAAGCTCTTGGAGCAATTGTCTACATTACCGAAATCGACCCCATCTGTGCTCTGCAGGCCTGCATGGATGGGTTC AGGGTGGTAAAGCTAAATGAAGTCATCCGGCAAGTCGATGTCGTAATAACTTGCACAGGAAATAAGAATGTAGTGACACGGGAGCACTTG GATCGCATGAAAAACAGTTGTATCGTATGCAATATGGGCCACTCCAACACAGAAATCGATGTGACCAGCCTCCGCACTCCGGAGCTGACG TGGGAGCGAGTACGTTCTCAGGTGGACCATGTCATCTGGCCAGATGGCAAACGAGTTGTCCTCCTGGCAGAGGGTCGTCTACTCAATTTG AGCTGCTCCACAGTTCCCACCTTTGTTCTGTCCATCACAGCCACAACACAGGCTTTGGCACTGATAGAACTCTATAATGCACCCGAGGGG CGATACAAGCAGGATGTGTACTTGCTTCCTAAGAAAATGGATGAATACGTTGCCAGCTTGCATCTGCCATCATTTGATGCCCACCTTACA GAGCTGACAGATGACCAAGCAAAATATCTGGGACTCAACAAAAATGGGCCATTCAAACCTAATTATTACAGATACTAATGGACCATACTA CCAAGGACCAGTCCACCTGAACCACACACTCTAAAGAAATATTTTTTAAGATAACTTTTATTTTCTTCTTACTCCTTTCCTCTTGATTTT TTTCCTATAATTTCATTCTTGTTTTTTCATCTCATTATCCAAGTTCTGCAGACCACACAGGAACTTGCTTCATGGCTCTTTAGATGAAAT AGAAGTTCAGGGTTCCTCACTCTAGTCACTAAAGAAGGATTTTACTCTCCCAGCCCAGAAAGGTGATTCTTTCTTTACCATTTCTGGGGA CTTTAGTCTTAATTAGGTACCTTATTAACAGGAAATGCTAAGGTACCTTCTCTGTGGAACAATCTGCAATGTCTAAATCGCCTTAAAAGA GCCCATTTCTTAGCTGCTGAAATCAGTGCTCTTTCACTTCTTCAGAGAAGCAGGGATGGTACCTACCCGGCAGGTAGGTTAGATGTGGGT GGTGCATGTTAATTTCCCTTAGAAGTTCCAAGCCCTGTTTCCTGCGTAAAGGTGGTATGTCCAGTTCAGAGATGTGTATAATGAGCATGG CTTGTTAAGATCAGGAGGCCCACTTGGATTTATAGTATAGCCCTTCCTCCACTCCCACCAGACTTGCTCATTTTTCGAGTTTTTAACTAG ACTACACTCTATTGAGTTTAATTTTGTCCTCTAGGATTTATTTCTGTTGTCCAAAAAAAAAAAAAAAGAAAAGAAAAATTAAGGAGAATT TTTGGTGTTAATGCTGAGGAATTGCTTGAGTGGTTAGTTGTTACCAATTTCTCTTTTGAACCTTTGGAGCTAAGGATGCTGAGTCTAGAG AAATGCTAGTCTCAAGCCCTGTTAAGTCCCTCTGTTTCTAGCCCGTAGTTCATAGCATCAGTGAACTGGAGCCACAACAGCAAATTCTAT CAGCTGTGTACCATACAGCTTGTGCTGAAGGCGAATTTCTTGAGCCATTACTCAGTATAAAGCACTGAGTTCTATCTTTAGGATTTATCT TTAAGAGCAAATTTCTGGTCAGCTGTGCTTCTGCAACCTAAAATATTTAAAGGGAGGTAGGTGTGGGCAGGAGGAGGAATGATAAATTGG GCCAGGGCAAGAAAAATCTAGCTTCATATAATTTGTCTGGGACTATACACCCTATATAATGTTAGTTTTACAGAAGTAATATGACTTTTG ATTGCTACATACCACAAAGAGTTTATGAACTGAGATCATAAAGGGCAACTGATGTGTGAAGAAAGTAGTCAGTACATCCTGGCTCATGCT CTGAAAGAATATCCAGAGAGGCTCTCTCAAAGATCAGGGAGATGTATTCCCATGCCATGCACCCTGCTTCCCAGCATTTCTGCATGGTCA AGTGAGCTTTATGCTCATGAGCTTTAAGTATATAATTATCCAGGATTTTAAATCCTCAACTTGTTCTAGCTTGTGATCCCTCAAAGTTGG GTCATACGTTAGTGCTAGATACTAGAAATTTTCACTTTTCCACTGATCAGAGAGACAGACATTAAAAACAAAAATAGAAGAAAGGAAAGC TTTCACCCTGCAGCTTCTTAGCAGGGAACAATTGTCTTGCCAAAACTTTTTTCCCTTTTCTCTCCCATTTTCTTTTACCCAATCCCTTCT TACTCCTTGCCAGTGTGACCATGCTTTCTTCTCTGTAGATGTTAACAGTTAAGGCCTATTTTCCTCGGGCACTTAACCAACCAATCAGAA CACCACATCTGTTAGGGGAGGTAACCTGGCCAACAGTGTATCCATCACGTTAGCCCTGCTGGAGGGAAGGGACCCACATTCACCTGCCCT CTGACCTGCCCCTTGATCCCATATCTATTACCGTGTCCATAGGAATAATAGGTAAGGGCTCTGTCTCTGTCAAGCCATGTAACAAAGGAC ACTGTTAAAAAAAAAAAAAAGTCTGGCATCAGAGGGAGCATGTGGAGAGCAACTTGGGAAGAACAAGTTCATTTTGTATTGAATGATTTT >30273_30273_3_FGFR2-AHCYL1_FGFR2_chr10_123243212_ENST00000369061_AHCYL1_chr1_110551654_ENST00000369799_length(amino acids)=1179AA_BP=689 MKALRVVHARRGSVQMGLTSTWRYGRGPGIGTVTMVSWGRFICLVVVTMATLSLARPSFSLVEDTTLEPEEPPTKYQISQPEVYVAAPGE SLEVRCLLKDAAVISWTKDGVHLGPNNRTVLIGEYLQIKGATPRDSGLYACTASRTVDSETWYFMVNVTDAISSGDDEDDTDGAEDFVSE NSNNKRAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENE YGSINHTYHLDVVAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSM NSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDL SDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLA SQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIP VEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEQIQFADDMQEFTKFPTKTGRRSLSRSISQSS TDSYSSAASYTDSSDDEVSPREKQQTNSKGSSNFCVKNIKQAEFGRREIEIAEQDMSALISLRKRAQGEKPLAGAKIVGCTHITAQTAVL IETLCALGAQCRWSACNIYSTQNEVAAALAEAGVAVFAWKGESEDDFWWCIDRCVNMDGWQANMILDDGGDLTHWVYKKYPNVFKKIRGI VEESVTGVHRLYQLSKAGKLCVPAMNVNDSVTKQKFDNLYCCRESILDGLKRTTDVMFGGKQVVVCGYGEVGKGCCAALKALGAIVYITE IDPICALQACMDGFRVVKLNEVIRQVDVVITCTGNKNVVTREHLDRMKNSCIVCNMGHSNTEIDVTSLRTPELTWERVRSQVDHVIWPDG KRVVLLAEGRLLNLSCSTVPTFVLSITATTQALALIELYNAPEGRYKQDVYLLPKKMDEYVASLHLPSFDAHLTELTDDQAKYLGLNKNG -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FGFR2-AHCYL1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000359172 | 0 | 17 | 138_201 | 0 | 484.0 | BCL2L10 | |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000369799 | 0 | 17 | 138_201 | 40.0 | 531.0 | BCL2L10 | |
| Tgene | AHCYL1 | chr10:123243212 | chr1:110551654 | ENST00000393614 | 0 | 17 | 138_201 | 0 | 484.0 | BCL2L10 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FGFR2-AHCYL1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FGFR2-AHCYL1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
