
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FGFR2-AP1M1 (FusionGDB2 ID:30275) |
Fusion Gene Summary for FGFR2-AP1M1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FGFR2-AP1M1 | Fusion gene ID: 30275 | Hgene | Tgene | Gene symbol | FGFR2 | AP1M1 | Gene ID | 2263 | 8907 |
| Gene name | fibroblast growth factor receptor 2 | adaptor related protein complex 1 subunit mu 1 | |
| Synonyms | BBDS|BEK|BFR-1|CD332|CEK3|CFD1|ECT1|JWS|K-SAM|KGFR|TK14|TK25 | AP47|CLAPM2|CLTNM|MU-1A | |
| Cytomap | 10q26.13 | 19p13.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | fibroblast growth factor receptor 2BEK fibroblast growth factor receptorbacteria-expressed kinasekeratinocyte growth factor receptorprotein tyrosine kinase, receptor like 14 | AP-1 complex subunit mu-1AP-mu chain family member mu1AHA1 47 kDa subunitadapter-related protein complex 1 subunit mu-1adaptor protein complex AP-1 mu-1 subunitadaptor protein complex AP-1 subunit mu-1adaptor related protein complex 1 mu 1 subunitc | |
| Modification date | 20200322 | 20200313 | |
| UniProtAcc | P21802 | Q9BXS5 | |
| Ensembl transtripts involved in fusion gene | ENST00000346997, ENST00000351936, ENST00000356226, ENST00000357555, ENST00000358487, ENST00000360144, ENST00000369056, ENST00000369059, ENST00000369060, ENST00000369061, ENST00000457416, ENST00000478859, ENST00000359354, ENST00000490349, | ENST00000429941, ENST00000444449, ENST00000541844, ENST00000590756, ENST00000291439, | |
| Fusion gene scores | * DoF score | 24 X 20 X 12=5760 | 5 X 5 X 4=100 |
| # samples | 38 | 6 | |
| ** MAII score | log2(38/5760*10)=-3.92199748799873 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/100*10)=-0.736965594166206 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FGFR2 [Title/Abstract] AND AP1M1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FGFR2(123256046)-AP1M1(16314270), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | FGFR2-AP1M1 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. FGFR2-AP1M1 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. FGFR2-AP1M1 seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | FGFR2 | GO:0008284 | positive regulation of cell proliferation | 8663044 |
| Hgene | FGFR2 | GO:0008543 | fibroblast growth factor receptor signaling pathway | 8663044|15629145 |
| Hgene | FGFR2 | GO:0018108 | peptidyl-tyrosine phosphorylation | 15629145|16844695 |
| Hgene | FGFR2 | GO:0046777 | protein autophosphorylation | 15629145 |
 Fusion gene breakpoints across FGFR2 (5'-gene) Fusion gene breakpoints across FGFR2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
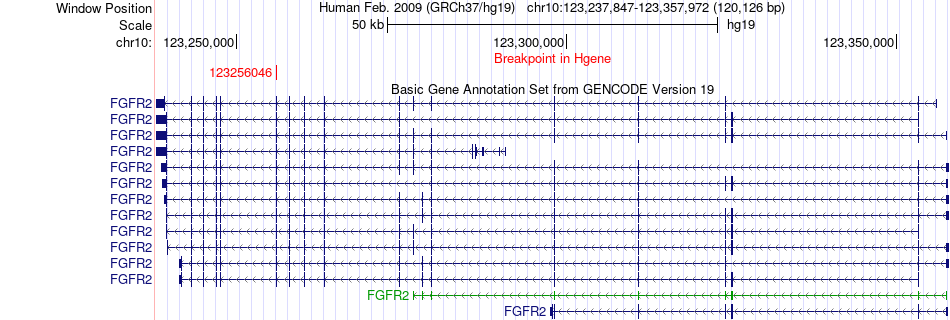 |
 Fusion gene breakpoints across AP1M1 (3'-gene) Fusion gene breakpoints across AP1M1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
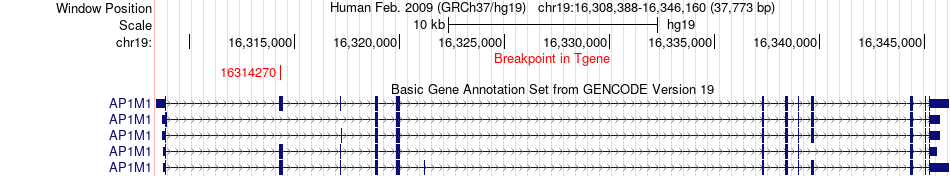 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-AQ-A04L-01B | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
Top |
Fusion Gene ORF analysis for FGFR2-AP1M1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000346997 | ENST00000429941 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000346997 | ENST00000444449 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000346997 | ENST00000541844 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000346997 | ENST00000590756 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000351936 | ENST00000429941 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000351936 | ENST00000444449 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000351936 | ENST00000541844 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000351936 | ENST00000590756 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000356226 | ENST00000429941 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000356226 | ENST00000444449 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000356226 | ENST00000541844 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000356226 | ENST00000590756 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000357555 | ENST00000429941 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000357555 | ENST00000444449 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000357555 | ENST00000541844 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000357555 | ENST00000590756 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000358487 | ENST00000429941 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000358487 | ENST00000444449 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000358487 | ENST00000541844 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000358487 | ENST00000590756 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000360144 | ENST00000429941 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000360144 | ENST00000444449 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000360144 | ENST00000541844 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000360144 | ENST00000590756 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000369056 | ENST00000429941 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000369056 | ENST00000444449 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000369056 | ENST00000541844 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000369056 | ENST00000590756 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000369059 | ENST00000429941 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000369059 | ENST00000444449 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000369059 | ENST00000541844 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000369059 | ENST00000590756 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000369060 | ENST00000429941 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000369060 | ENST00000444449 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000369060 | ENST00000541844 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000369060 | ENST00000590756 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000369061 | ENST00000429941 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000369061 | ENST00000444449 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000369061 | ENST00000541844 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000369061 | ENST00000590756 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000457416 | ENST00000429941 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000457416 | ENST00000444449 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000457416 | ENST00000541844 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000457416 | ENST00000590756 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000478859 | ENST00000429941 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000478859 | ENST00000444449 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000478859 | ENST00000541844 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| 5CDS-intron | ENST00000478859 | ENST00000590756 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| Frame-shift | ENST00000351936 | ENST00000291439 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| Frame-shift | ENST00000356226 | ENST00000291439 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| Frame-shift | ENST00000357555 | ENST00000291439 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| Frame-shift | ENST00000358487 | ENST00000291439 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| Frame-shift | ENST00000360144 | ENST00000291439 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| Frame-shift | ENST00000369059 | ENST00000291439 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| Frame-shift | ENST00000369060 | ENST00000291439 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| Frame-shift | ENST00000457416 | ENST00000291439 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| Frame-shift | ENST00000478859 | ENST00000291439 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| In-frame | ENST00000346997 | ENST00000291439 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| In-frame | ENST00000369056 | ENST00000291439 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| In-frame | ENST00000369061 | ENST00000291439 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| intron-3CDS | ENST00000359354 | ENST00000291439 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| intron-3CDS | ENST00000490349 | ENST00000291439 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| intron-intron | ENST00000359354 | ENST00000429941 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| intron-intron | ENST00000359354 | ENST00000444449 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| intron-intron | ENST00000359354 | ENST00000541844 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| intron-intron | ENST00000359354 | ENST00000590756 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| intron-intron | ENST00000490349 | ENST00000429941 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| intron-intron | ENST00000490349 | ENST00000444449 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| intron-intron | ENST00000490349 | ENST00000541844 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
| intron-intron | ENST00000490349 | ENST00000590756 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000369061 | FGFR2 | chr10 | 123256046 | - | ENST00000291439 | AP1M1 | chr19 | 16314270 | + | 3821 | 1677 | 48 | 2906 | 952 |
| ENST00000346997 | FGFR2 | chr10 | 123256046 | - | ENST00000291439 | AP1M1 | chr19 | 16314270 | + | 4013 | 1869 | 12 | 3098 | 1028 |
| ENST00000369056 | FGFR2 | chr10 | 123256046 | - | ENST00000291439 | AP1M1 | chr19 | 16314270 | + | 4034 | 1890 | 24 | 3119 | 1031 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000369061 | ENST00000291439 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + | 0.000592621 | 0.9994074 |
| ENST00000346997 | ENST00000291439 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + | 0.000328161 | 0.9996718 |
| ENST00000369056 | ENST00000291439 | FGFR2 | chr10 | 123256046 | - | AP1M1 | chr19 | 16314270 | + | 0.000445601 | 0.99955434 |
Top |
Fusion Genomic Features for FGFR2-AP1M1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| FGFR2 | chr10 | 123256045 | - | AP1M1 | chr19 | 16314269 | + | 3.36E-12 | 1 |
| FGFR2 | chr10 | 123256045 | - | AP1M1 | chr19 | 16314269 | + | 3.36E-12 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
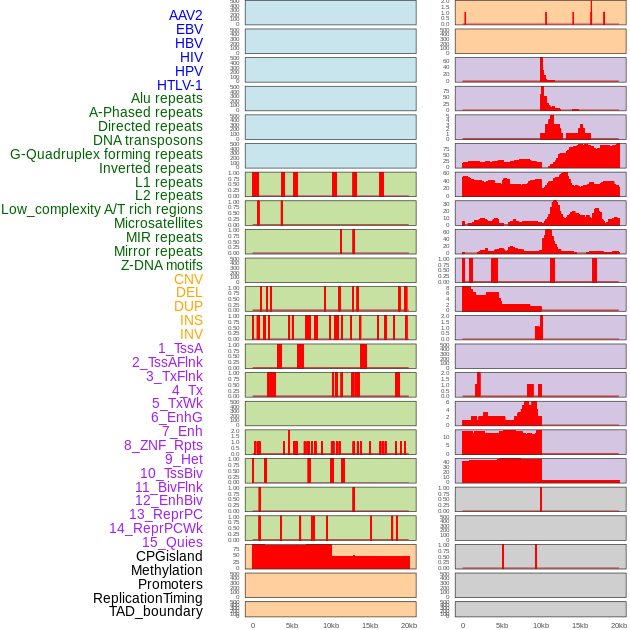 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
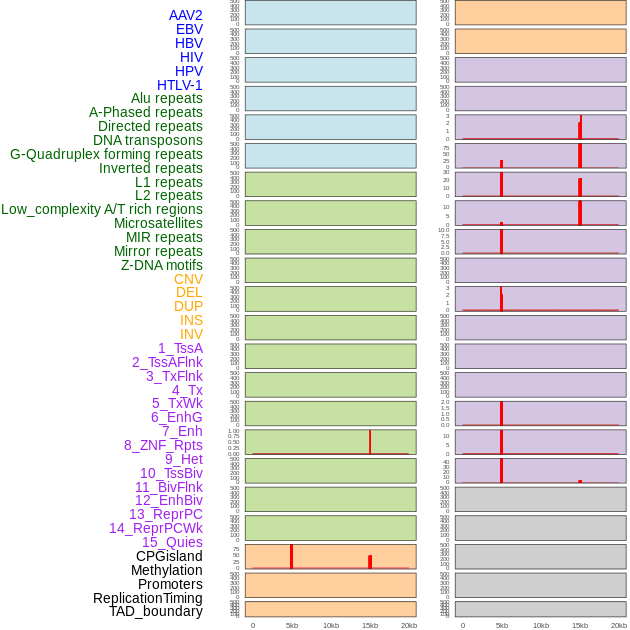 |
Top |
Fusion Protein Features for FGFR2-AP1M1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:123256046/chr19:16314270) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FGFR2 | AP1M1 |
| FUNCTION: Tyrosine-protein kinase that acts as cell-surface receptor for fibroblast growth factors and plays an essential role in the regulation of cell proliferation, differentiation, migration and apoptosis, and in the regulation of embryonic development. Required for normal embryonic patterning, trophoblast function, limb bud development, lung morphogenesis, osteogenesis and skin development. Plays an essential role in the regulation of osteoblast differentiation, proliferation and apoptosis, and is required for normal skeleton development. Promotes cell proliferation in keratinocytes and immature osteoblasts, but promotes apoptosis in differentiated osteoblasts. Phosphorylates PLCG1, FRS2 and PAK4. Ligand binding leads to the activation of several signaling cascades. Activation of PLCG1 leads to the production of the cellular signaling molecules diacylglycerol and inositol 1,4,5-trisphosphate. Phosphorylation of FRS2 triggers recruitment of GRB2, GAB1, PIK3R1 and SOS1, and mediates activation of RAS, MAPK1/ERK2, MAPK3/ERK1 and the MAP kinase signaling pathway, as well as of the AKT1 signaling pathway. FGFR2 signaling is down-regulated by ubiquitination, internalization and degradation. Mutations that lead to constitutive kinase activation or impair normal FGFR2 maturation, internalization and degradation lead to aberrant signaling. Over-expressed FGFR2 promotes activation of STAT1. {ECO:0000269|PubMed:12529371, ECO:0000269|PubMed:15190072, ECO:0000269|PubMed:15629145, ECO:0000269|PubMed:16384934, ECO:0000269|PubMed:16597617, ECO:0000269|PubMed:17311277, ECO:0000269|PubMed:17623664, ECO:0000269|PubMed:18374639, ECO:0000269|PubMed:19103595, ECO:0000269|PubMed:19387476, ECO:0000269|PubMed:19410646, ECO:0000269|PubMed:21596750, ECO:0000269|PubMed:8663044}. | FUNCTION: Subunit of clathrin-associated adaptor protein complex 1 that plays a role in protein sorting in the trans-Golgi network (TGN) and endosomes. The AP complexes mediate the recruitment of clathrin to membranes and the recognition of sorting signals within the cytosolic tails of transmembrane cargo molecules. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000346997 | - | 12 | 17 | 154_247 | 619 | 820.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000346997 | - | 12 | 17 | 256_358 | 619 | 820.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000346997 | - | 12 | 17 | 25_125 | 619 | 820.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000351936 | - | 13 | 18 | 154_247 | 619 | 786.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000351936 | - | 13 | 18 | 256_358 | 619 | 786.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000351936 | - | 13 | 18 | 25_125 | 619 | 786.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000356226 | - | 11 | 16 | 154_247 | 504 | 705.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000356226 | - | 11 | 16 | 256_358 | 504 | 705.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000356226 | - | 11 | 16 | 25_125 | 504 | 705.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000357555 | - | 12 | 17 | 154_247 | 532 | 708.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000357555 | - | 12 | 17 | 256_358 | 532 | 708.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000357555 | - | 12 | 17 | 25_125 | 532 | 708.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000358487 | - | 13 | 18 | 154_247 | 621 | 822.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000358487 | - | 13 | 18 | 256_358 | 621 | 822.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000358487 | - | 13 | 18 | 25_125 | 621 | 822.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000360144 | - | 12 | 17 | 154_247 | 533 | 681.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000360144 | - | 12 | 17 | 256_358 | 533 | 681.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000360144 | - | 12 | 17 | 25_125 | 533 | 681.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369056 | - | 12 | 17 | 154_247 | 622 | 770.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369056 | - | 12 | 17 | 256_358 | 622 | 770.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369056 | - | 12 | 17 | 25_125 | 622 | 770.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369060 | - | 11 | 16 | 154_247 | 505 | 706.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369060 | - | 11 | 16 | 256_358 | 505 | 706.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369060 | - | 11 | 16 | 25_125 | 505 | 706.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369061 | - | 10 | 15 | 154_247 | 509 | 710.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369061 | - | 10 | 15 | 256_358 | 509 | 710.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369061 | - | 10 | 15 | 25_125 | 509 | 710.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000457416 | - | 13 | 18 | 154_247 | 622 | 823.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000457416 | - | 13 | 18 | 256_358 | 622 | 823.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000457416 | - | 13 | 18 | 25_125 | 622 | 823.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000346997 | - | 12 | 17 | 487_495 | 619 | 820.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000346997 | - | 12 | 17 | 565_567 | 619 | 820.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000351936 | - | 13 | 18 | 487_495 | 619 | 786.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000351936 | - | 13 | 18 | 565_567 | 619 | 786.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000356226 | - | 11 | 16 | 487_495 | 504 | 705.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000357555 | - | 12 | 17 | 487_495 | 532 | 708.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000358487 | - | 13 | 18 | 487_495 | 621 | 822.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000358487 | - | 13 | 18 | 565_567 | 621 | 822.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000360144 | - | 12 | 17 | 487_495 | 533 | 681.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369056 | - | 12 | 17 | 487_495 | 622 | 770.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369056 | - | 12 | 17 | 565_567 | 622 | 770.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369060 | - | 11 | 16 | 487_495 | 505 | 706.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369061 | - | 10 | 15 | 487_495 | 509 | 710.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000457416 | - | 13 | 18 | 487_495 | 622 | 823.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000457416 | - | 13 | 18 | 565_567 | 622 | 823.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000346997 | - | 12 | 17 | 161_178 | 619 | 820.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000351936 | - | 13 | 18 | 161_178 | 619 | 786.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000356226 | - | 11 | 16 | 161_178 | 504 | 705.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000357555 | - | 12 | 17 | 161_178 | 532 | 708.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000358487 | - | 13 | 18 | 161_178 | 621 | 822.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000360144 | - | 12 | 17 | 161_178 | 533 | 681.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369056 | - | 12 | 17 | 161_178 | 622 | 770.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369060 | - | 11 | 16 | 161_178 | 505 | 706.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369061 | - | 10 | 15 | 161_178 | 509 | 710.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000457416 | - | 13 | 18 | 161_178 | 622 | 823.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000346997 | - | 12 | 17 | 22_377 | 619 | 820.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000351936 | - | 13 | 18 | 22_377 | 619 | 786.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000356226 | - | 11 | 16 | 22_377 | 504 | 705.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000357555 | - | 12 | 17 | 22_377 | 532 | 708.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000358487 | - | 13 | 18 | 22_377 | 621 | 822.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000360144 | - | 12 | 17 | 22_377 | 533 | 681.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369056 | - | 12 | 17 | 22_377 | 622 | 770.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369060 | - | 11 | 16 | 22_377 | 505 | 706.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369061 | - | 10 | 15 | 22_377 | 509 | 710.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000457416 | - | 13 | 18 | 22_377 | 622 | 823.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000346997 | - | 12 | 17 | 378_398 | 619 | 820.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000351936 | - | 13 | 18 | 378_398 | 619 | 786.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000356226 | - | 11 | 16 | 378_398 | 504 | 705.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000357555 | - | 12 | 17 | 378_398 | 532 | 708.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000358487 | - | 13 | 18 | 378_398 | 621 | 822.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000360144 | - | 12 | 17 | 378_398 | 533 | 681.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369056 | - | 12 | 17 | 378_398 | 622 | 770.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369060 | - | 11 | 16 | 378_398 | 505 | 706.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369061 | - | 10 | 15 | 378_398 | 509 | 710.0 | Transmembrane | Helical |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000457416 | - | 13 | 18 | 378_398 | 622 | 823.0 | Transmembrane | Helical |
| Tgene | AP1M1 | chr10:123256046 | chr19:16314270 | ENST00000291439 | 0 | 12 | 168_421 | 14 | 424.0 | Domain | MHD | |
| Tgene | AP1M1 | chr10:123256046 | chr19:16314270 | ENST00000444449 | 0 | 13 | 168_421 | 14 | 436.0 | Domain | MHD |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000346997 | - | 12 | 17 | 481_770 | 619 | 820.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000351936 | - | 13 | 18 | 481_770 | 619 | 786.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000356226 | - | 11 | 16 | 481_770 | 504 | 705.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000357555 | - | 12 | 17 | 481_770 | 532 | 708.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000358487 | - | 13 | 18 | 481_770 | 621 | 822.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000359354 | - | 1 | 7 | 154_247 | 0 | 255.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000359354 | - | 1 | 7 | 256_358 | 0 | 255.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000359354 | - | 1 | 7 | 25_125 | 0 | 255.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000359354 | - | 1 | 7 | 481_770 | 0 | 255.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000360144 | - | 12 | 17 | 481_770 | 533 | 681.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369056 | - | 12 | 17 | 481_770 | 622 | 770.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369060 | - | 11 | 16 | 481_770 | 505 | 706.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369061 | - | 10 | 15 | 481_770 | 509 | 710.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000457416 | - | 13 | 18 | 481_770 | 622 | 823.0 | Domain | Protein kinase |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000356226 | - | 11 | 16 | 565_567 | 504 | 705.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000357555 | - | 12 | 17 | 565_567 | 532 | 708.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000359354 | - | 1 | 7 | 487_495 | 0 | 255.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000359354 | - | 1 | 7 | 565_567 | 0 | 255.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000360144 | - | 12 | 17 | 565_567 | 533 | 681.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369060 | - | 11 | 16 | 565_567 | 505 | 706.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369061 | - | 10 | 15 | 565_567 | 509 | 710.0 | Nucleotide binding | ATP |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000359354 | - | 1 | 7 | 161_178 | 0 | 255.0 | Region | Note=Heparin-binding |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000346997 | - | 12 | 17 | 399_821 | 619 | 820.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000351936 | - | 13 | 18 | 399_821 | 619 | 786.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000356226 | - | 11 | 16 | 399_821 | 504 | 705.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000357555 | - | 12 | 17 | 399_821 | 532 | 708.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000358487 | - | 13 | 18 | 399_821 | 621 | 822.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000359354 | - | 1 | 7 | 22_377 | 0 | 255.0 | Topological domain | Extracellular |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000359354 | - | 1 | 7 | 399_821 | 0 | 255.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000360144 | - | 12 | 17 | 399_821 | 533 | 681.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369056 | - | 12 | 17 | 399_821 | 622 | 770.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369060 | - | 11 | 16 | 399_821 | 505 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000369061 | - | 10 | 15 | 399_821 | 509 | 710.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000457416 | - | 13 | 18 | 399_821 | 622 | 823.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR2 | chr10:123256046 | chr19:16314270 | ENST00000359354 | - | 1 | 7 | 378_398 | 0 | 255.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for FGFR2-AP1M1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >30275_30275_1_FGFR2-AP1M1_FGFR2_chr10_123256046_ENST00000346997_AP1M1_chr19_16314270_ENST00000291439_length(transcript)=4013nt_BP=1869nt GGTACCGTAACCATGGTCAGCTGGGGTCGTTTCATCTGCCTGGTCGTGGTCACCATGGCAACCTTGTCCCTGGCCCGGCCCTCCTTCAGT TTAGTTGAGGATACCACATTAGAGCCAGAAGAGCCACCAACCAAATACCAAATCTCTCAACCAGAAGTGTACGTGGCTGCGCCAGGGGAG TCGCTAGAGGTGCGCTGCCTGTTGAAAGATGCCGCCGTGATCAGTTGGACTAAGGATGGGGTGCACTTGGGGCCCAACAATAGGACAGTG CTTATTGGGGAGTACTTGCAGATAAAGGGCGCCACGCCTAGAGACTCCGGCCTCTATGCTTGTACTGCCAGTAGGACTGTAGACAGTGAA ACTTGGTACTTCATGGTGAATGTCACAGATGCCATCTCATCCGGAGATGATGAGGATGACACCGATGGTGCGGAAGATTTTGTCAGTGAG AACAGTAACAACAAGAGAGCACCATACTGGACCAACACAGAAAAGATGGAAAAGCGGCTCCATGCTGTGCCTGCGGCCAACACTGTCAAG TTTCGCTGCCCAGCCGGGGGGAACCCAATGCCAACCATGCGGTGGCTGAAAAACGGGAAGGAGTTTAAGCAGGAGCATCGCATTGGAGGC TACAAGGTACGAAACCAGCACTGGAGCCTCATTATGGAAAGTGTGGTCCCATCTGACAAGGGAAATTATACCTGTGTAGTGGAGAATGAA TACGGGTCCATCAATCACACGTACCACCTGGATGTTGTGGAGCGATCGCCTCACCGGCCCATCCTCCAAGCCGGACTGCCGGCAAATGCC TCCACAGTGGTCGGAGGAGACGTAGAGTTTGTCTGCAAGGTTTACAGTGATGCCCAGCCCCACATCCAGTGGATCAAGCACGTGGAAAAG AACGGCAGTAAATACGGGCCCGACGGGCTGCCCTACCTCAAGGTTCTCAAGGCCGCCGGTGTTAACACCACGGACAAAGAGATTGAGGTT CTCTATATTCGGAATGTAACTTTTGAGGACGCTGGGGAATATACGTGCTTGGCGGGTAATTCTATTGGGATATCCTTTCACTCTGCATGG TTGACAGTTCTGCCAGCGCCTGGAAGAGAAAAGGAGATTACAGCTTCCCCAGACTACCTGGAGATAGCCATTTACTGCATAGGGGTCTTC TTAATCGCCTGTATGGTGGTAACAGTCATCCTGTGCCGAATGAAGAACACGACCAAGAAGCCAGACTTCAGCAGCCAGCCGGCTGTGCAC AAGCTGACCAAACGTATCCCCCTGCGGAGACAGGTTTCGGCTGAGTCCAGCTCCTCCATGAACTCCAACACCCCGCTGGTGAGGATAACA ACACGCCTCTCTTCAACGGCAGACACCCCCATGCTGGCAGGGGTCTCCGAGTATGAACTTCCAGAGGACCCAAAATGGGAGTTTCCAAGA GATAAGCTGACACTGGGCAAGCCCCTGGGAGAAGGTTGCTTTGGGCAAGTGGTCATGGCGGAAGCAGTGGGAATTGACAAAGACAAGCCC AAGGAGGCGGTCACCGTGGCCGTGAAGATGTTGAAAGATGATGCCACAGAGAAAGACCTTTCTGATCTGGTGTCAGAGATGGAGATGATG AAGATGATTGGGAAACACAAGAATATCATAAATCTTCTTGGAGCCTGCACACAGGATGGGCCTCTCTATGTCATAGTTGAGTATGCCTCT AAAGGCAACCTCCGAGAATACCTCCGAGCCCGGAGGCCACCCGGGATGGAGTACTCCTATGACATTAACCGTGTTCCTGAGGAGCAGATG ACCTTCAAGGACTTGGTGTCATGCACCTACCAGCTGGCCAGAGGCATGGAGTACTTGGCTTCCCAAAAAGTGCTCATCTGCCGGAACTAC CGTGGCGACGTGGACATGTCAGAGGTGGAGCACTTCATGCCCATCCTGATGGAGAAGGAGGAGGAGGGGATGCTGTCGCCCATCCTGGCC CACGGGGGGGTCCGTTTCATGTGGATCAAACACAACAACCTGTATCTGGTTGCCACATCCAAGAAGAACGCGTGCGTGTCGCTGGTCTTT TCTTTCCTCTATAAGGTGGTGCAGGTGTTTTCCGAGTACTTCAAGGAGCTGGAGGAGGAGAGCATCCGGGACAACTTTGTTATCATCTAC GAGCTGCTGGACGAGCTCATGGACTTCGGCTACCCCCAGACCACCGACAGCAAGATCCTGCAGGAGTACATCACTCAGGAAGGCCACAAG CTGGAAACAGGGGCCCCGCGGCCACCAGCCACCGTCACCAACGCGGTGTCCTGGCGGTCCGAAGGCATCAAGTATCGGAAGAATGAGGTG TTCTTGGACGTCATCGAGTCTGTCAACCTCTTGGTCAGCGCCAACGGCAATGTCCTGCGCAGCGAGATCGTGGGCTCCATCAAGATGCGA GTCTTCCTCTCGGGCATGCCCGAGCTGCGCCTGGGCCTCAACGACAAGGTCCTCTTTGACAACACGGGCCGCGGCAAAAGCAAATCCGTG GAGCTGGAGGATGTGAAGTTCCACCAGTGTGTGCGGCTATCACGCTTCGAGAATGACCGCACCATCTCCTTCATCCCACCCGACGGCGAG TTCGAGCTCATGTCCTACCGTCTCAACACCCACGTCAAGCCTTTGATATGGATCGAGTCGGTGATCGAGAAGCACTCCCACAGCCGCATC GAGTACATGATCAAGGCCAAAAGCCAGTTCAAGCGGCGGTCAACAGCCAACAACGTGGAGATCCACATTCCCGTGCCCAATGATGCCGAC TCACCCAAGTTCAAGACGACGGTGGGGAGCGTTAAGTGGGTCCCCGAGAACAGCGAGATCGTGTGGTCCATCAAGTCCTTCCCGGGCGGC AAGGAGTACCTGATGCGGGCCCACTTCGGCCTGCCTAGTGTGGAGGCCGAAGACAAGGAGGGCAAGCCCCCGATCAGTGTCAAGTTCGAG ATCCCTTACTTCACTACCTCCGGCATCCAGGTGCGCTACCTGAAGATCATTGAGAAGAGTGGGTACCAGGCCCTGCCCTGGGTGCGTTAT ATCACGCAGAATGGAGATTACCAGCTCCGGACCCAGTGAGGGGCTGTCGCAGCCAACACCCCGGCCTCGGGGCTCCTGGTGGCAGCACCA GGGGACACACCTGCCAAACCCACCAGATGGAGGGGCCCTCCCTGGTCTCTGGCCACCCTCCCAGCCTCTGCCCAGGGACCCCTGCCTTCC CCAGGCCATCTGCTCTGCCGTCGACACTCGTCTCAGAAGCCCCTTTCCCAGAAGAGGCTGGTCTTCAAGAAGTCTCGTTTCTTTGCCCCT GAAGTCAGTTTCAGGGGAAGGATGTGAAATTTTTCCGTGTAGAGGTTACAGCCTTTTATGCTGTTGAGCTCCCAGGTACCAAAAAGCTTG GCCAACGCTTGCCAGCCAGCCAGCTGCAGGTGGCATCTGCCACGAAGGAAGCGCCAGCCTCGCCAGGCCAGCAGGGGCGTCGTTTTGTTG CCATTTTGTTGAACGTTATGGGTTTATGGGTGTTCCTGGAACTTGTCTTTGTGCATTCGTTGCTGTTTGTGTTACCCTCACTGTCCCCAT GTCCCACCCACGTCCTACGGCACTCAGGAAGCACTTGGTGAGGACGAGCCCTCACCCTTCTTGTCTTCCTTCCCGGCAGCGCCCGCAGCG GGCCATTTACACGTCGAGGCTGGCACCTGGCGCGCTCGGGGGCCACTGTAGCGTCTGCCTGCTCCCTGGACTCGCAGGCCTCGCCTGTGG CGCCTTCCCAGGGCCAGCCTGGGTCACGAGATGCTGTCACTCAGCCAGATCAGTATTGACCCACCAGGGGAGGTGGGGTTTGGTGAGAGA CGCCAGCCTCAGACTTTTTCCCACTGAGGGTCCAGAGAGCGGGGCCACGTGTCACCCACGTCTGCGCTTGGTCACCCGTCCTCCCCACCC >30275_30275_1_FGFR2-AP1M1_FGFR2_chr10_123256046_ENST00000346997_AP1M1_chr19_16314270_ENST00000291439_length(amino acids)=1028AA_BP=619 MVSWGRFICLVVVTMATLSLARPSFSLVEDTTLEPEEPPTKYQISQPEVYVAAPGESLEVRCLLKDAAVISWTKDGVHLGPNNRTVLIGE YLQIKGATPRDSGLYACTASRTVDSETWYFMVNVTDAISSGDDEDDTDGAEDFVSENSNNKRAPYWTNTEKMEKRLHAVPAANTVKFRCP AGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVV GGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVL PAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSAESSSSMNSNTPLVRITTRLS STADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIG KHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKVLICRNYRGDV DMSEVEHFMPILMEKEEEGMLSPILAHGGVRFMWIKHNNLYLVATSKKNACVSLVFSFLYKVVQVFSEYFKELEEESIRDNFVIIYELLD ELMDFGYPQTTDSKILQEYITQEGHKLETGAPRPPATVTNAVSWRSEGIKYRKNEVFLDVIESVNLLVSANGNVLRSEIVGSIKMRVFLS GMPELRLGLNDKVLFDNTGRGKSKSVELEDVKFHQCVRLSRFENDRTISFIPPDGEFELMSYRLNTHVKPLIWIESVIEKHSHSRIEYMI KAKSQFKRRSTANNVEIHIPVPNDADSPKFKTTVGSVKWVPENSEIVWSIKSFPGGKEYLMRAHFGLPSVEAEDKEGKPPISVKFEIPYF -------------------------------------------------------------- >30275_30275_2_FGFR2-AP1M1_FGFR2_chr10_123256046_ENST00000369056_AP1M1_chr19_16314270_ENST00000291439_length(transcript)=4034nt_BP=1890nt GGACCGGGGATTGGTACCGTAACCATGGTCAGCTGGGGTCGTTTCATCTGCCTGGTCGTGGTCACCATGGCAACCTTGTCCCTGGCCCGG CCCTCCTTCAGTTTAGTTGAGGATACCACATTAGAGCCAGAAGAGCCACCAACCAAATACCAAATCTCTCAACCAGAAGTGTACGTGGCT GCGCCAGGGGAGTCGCTAGAGGTGCGCTGCCTGTTGAAAGATGCCGCCGTGATCAGTTGGACTAAGGATGGGGTGCACTTGGGGCCCAAC AATAGGACAGTGCTTATTGGGGAGTACTTGCAGATAAAGGGCGCCACGCCTAGAGACTCCGGCCTCTATGCTTGTACTGCCAGTAGGACT GTAGACAGTGAAACTTGGTACTTCATGGTGAATGTCACAGATGCCATCTCATCCGGAGATGATGAGGATGACACCGATGGTGCGGAAGAT TTTGTCAGTGAGAACAGTAACAACAAGAGAGCACCATACTGGACCAACACAGAAAAGATGGAAAAGCGGCTCCATGCTGTGCCTGCGGCC AACACTGTCAAGTTTCGCTGCCCAGCCGGGGGGAACCCAATGCCAACCATGCGGTGGCTGAAAAACGGGAAGGAGTTTAAGCAGGAGCAT CGCATTGGAGGCTACAAGGTACGAAACCAGCACTGGAGCCTCATTATGGAAAGTGTGGTCCCATCTGACAAGGGAAATTATACCTGTGTA GTGGAGAATGAATACGGGTCCATCAATCACACGTACCACCTGGATGTTGTGGAGCGATCGCCTCACCGGCCCATCCTCCAAGCCGGACTG CCGGCAAATGCCTCCACAGTGGTCGGAGGAGACGTAGAGTTTGTCTGCAAGGTTTACAGTGATGCCCAGCCCCACATCCAGTGGATCAAG CACGTGGAAAAGAACGGCAGTAAATACGGGCCCGACGGGCTGCCCTACCTCAAGGTTCTCAAGCACTCGGGGATAAATAGTTCCAATGCA GAAGTGCTGGCTCTGTTCAATGTGACCGAGGCGGATGCTGGGGAATATATATGTAAGGTCTCCAATTATATAGGGCAGGCCAACCAGTCT GCCTGGCTCACTGTCCTGCCAAAACAGCAAGCGCCTGGAAGAGAAAAGGAGATTACAGCTTCCCCAGACTACCTGGAGATAGCCATTTAC TGCATAGGGGTCTTCTTAATCGCCTGTATGGTGGTAACAGTCATCCTGTGCCGAATGAAGAACACGACCAAGAAGCCAGACTTCAGCAGC CAGCCGGCTGTGCACAAGCTGACCAAACGTATCCCCCTGCGGAGACAGGTAACAGTTTCGGCTGAGTCCAGCTCCTCCATGAACTCCAAC ACCCCGCTGGTGAGGATAACAACACGCCTCTCTTCAACGGCAGACACCCCCATGCTGGCAGGGGTCTCCGAGTATGAACTTCCAGAGGAC CCAAAATGGGAGTTTCCAAGAGATAAGCTGACACTGGGCAAGCCCCTGGGAGAAGGTTGCTTTGGGCAAGTGGTCATGGCGGAAGCAGTG GGAATTGACAAAGACAAGCCCAAGGAGGCGGTCACCGTGGCCGTGAAGATGTTGAAAGATGATGCCACAGAGAAAGACCTTTCTGATCTG GTGTCAGAGATGGAGATGATGAAGATGATTGGGAAACACAAGAATATCATAAATCTTCTTGGAGCCTGCACACAGGATGGGCCTCTCTAT GTCATAGTTGAGTATGCCTCTAAAGGCAACCTCCGAGAATACCTCCGAGCCCGGAGGCCACCCGGGATGGAGTACTCCTATGACATTAAC CGTGTTCCTGAGGAGCAGATGACCTTCAAGGACTTGGTGTCATGCACCTACCAGCTGGCCAGAGGCATGGAGTACTTGGCTTCCCAAAAA GTGCTCATCTGCCGGAACTACCGTGGCGACGTGGACATGTCAGAGGTGGAGCACTTCATGCCCATCCTGATGGAGAAGGAGGAGGAGGGG ATGCTGTCGCCCATCCTGGCCCACGGGGGGGTCCGTTTCATGTGGATCAAACACAACAACCTGTATCTGGTTGCCACATCCAAGAAGAAC GCGTGCGTGTCGCTGGTCTTTTCTTTCCTCTATAAGGTGGTGCAGGTGTTTTCCGAGTACTTCAAGGAGCTGGAGGAGGAGAGCATCCGG GACAACTTTGTTATCATCTACGAGCTGCTGGACGAGCTCATGGACTTCGGCTACCCCCAGACCACCGACAGCAAGATCCTGCAGGAGTAC ATCACTCAGGAAGGCCACAAGCTGGAAACAGGGGCCCCGCGGCCACCAGCCACCGTCACCAACGCGGTGTCCTGGCGGTCCGAAGGCATC AAGTATCGGAAGAATGAGGTGTTCTTGGACGTCATCGAGTCTGTCAACCTCTTGGTCAGCGCCAACGGCAATGTCCTGCGCAGCGAGATC GTGGGCTCCATCAAGATGCGAGTCTTCCTCTCGGGCATGCCCGAGCTGCGCCTGGGCCTCAACGACAAGGTCCTCTTTGACAACACGGGC CGCGGCAAAAGCAAATCCGTGGAGCTGGAGGATGTGAAGTTCCACCAGTGTGTGCGGCTATCACGCTTCGAGAATGACCGCACCATCTCC TTCATCCCACCCGACGGCGAGTTCGAGCTCATGTCCTACCGTCTCAACACCCACGTCAAGCCTTTGATATGGATCGAGTCGGTGATCGAG AAGCACTCCCACAGCCGCATCGAGTACATGATCAAGGCCAAAAGCCAGTTCAAGCGGCGGTCAACAGCCAACAACGTGGAGATCCACATT CCCGTGCCCAATGATGCCGACTCACCCAAGTTCAAGACGACGGTGGGGAGCGTTAAGTGGGTCCCCGAGAACAGCGAGATCGTGTGGTCC ATCAAGTCCTTCCCGGGCGGCAAGGAGTACCTGATGCGGGCCCACTTCGGCCTGCCTAGTGTGGAGGCCGAAGACAAGGAGGGCAAGCCC CCGATCAGTGTCAAGTTCGAGATCCCTTACTTCACTACCTCCGGCATCCAGGTGCGCTACCTGAAGATCATTGAGAAGAGTGGGTACCAG GCCCTGCCCTGGGTGCGTTATATCACGCAGAATGGAGATTACCAGCTCCGGACCCAGTGAGGGGCTGTCGCAGCCAACACCCCGGCCTCG GGGCTCCTGGTGGCAGCACCAGGGGACACACCTGCCAAACCCACCAGATGGAGGGGCCCTCCCTGGTCTCTGGCCACCCTCCCAGCCTCT GCCCAGGGACCCCTGCCTTCCCCAGGCCATCTGCTCTGCCGTCGACACTCGTCTCAGAAGCCCCTTTCCCAGAAGAGGCTGGTCTTCAAG AAGTCTCGTTTCTTTGCCCCTGAAGTCAGTTTCAGGGGAAGGATGTGAAATTTTTCCGTGTAGAGGTTACAGCCTTTTATGCTGTTGAGC TCCCAGGTACCAAAAAGCTTGGCCAACGCTTGCCAGCCAGCCAGCTGCAGGTGGCATCTGCCACGAAGGAAGCGCCAGCCTCGCCAGGCC AGCAGGGGCGTCGTTTTGTTGCCATTTTGTTGAACGTTATGGGTTTATGGGTGTTCCTGGAACTTGTCTTTGTGCATTCGTTGCTGTTTG TGTTACCCTCACTGTCCCCATGTCCCACCCACGTCCTACGGCACTCAGGAAGCACTTGGTGAGGACGAGCCCTCACCCTTCTTGTCTTCC TTCCCGGCAGCGCCCGCAGCGGGCCATTTACACGTCGAGGCTGGCACCTGGCGCGCTCGGGGGCCACTGTAGCGTCTGCCTGCTCCCTGG ACTCGCAGGCCTCGCCTGTGGCGCCTTCCCAGGGCCAGCCTGGGTCACGAGATGCTGTCACTCAGCCAGATCAGTATTGACCCACCAGGG GAGGTGGGGTTTGGTGAGAGACGCCAGCCTCAGACTTTTTCCCACTGAGGGTCCAGAGAGCGGGGCCACGTGTCACCCACGTCTGCGCTT >30275_30275_2_FGFR2-AP1M1_FGFR2_chr10_123256046_ENST00000369056_AP1M1_chr19_16314270_ENST00000291439_length(amino acids)=1031AA_BP=622 MVSWGRFICLVVVTMATLSLARPSFSLVEDTTLEPEEPPTKYQISQPEVYVAAPGESLEVRCLLKDAAVISWTKDGVHLGPNNRTVLIGE YLQIKGATPRDSGLYACTASRTVDSETWYFMVNVTDAISSGDDEDDTDGAEDFVSENSNNKRAPYWTNTEKMEKRLHAVPAANTVKFRCP AGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVV GGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYICKVSNYIGQANQSAWLTVLPK QQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITT RLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMK MIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKVLICRNYR GDVDMSEVEHFMPILMEKEEEGMLSPILAHGGVRFMWIKHNNLYLVATSKKNACVSLVFSFLYKVVQVFSEYFKELEEESIRDNFVIIYE LLDELMDFGYPQTTDSKILQEYITQEGHKLETGAPRPPATVTNAVSWRSEGIKYRKNEVFLDVIESVNLLVSANGNVLRSEIVGSIKMRV FLSGMPELRLGLNDKVLFDNTGRGKSKSVELEDVKFHQCVRLSRFENDRTISFIPPDGEFELMSYRLNTHVKPLIWIESVIEKHSHSRIE YMIKAKSQFKRRSTANNVEIHIPVPNDADSPKFKTTVGSVKWVPENSEIVWSIKSFPGGKEYLMRAHFGLPSVEAEDKEGKPPISVKFEI -------------------------------------------------------------- >30275_30275_3_FGFR2-AP1M1_FGFR2_chr10_123256046_ENST00000369061_AP1M1_chr19_16314270_ENST00000291439_length(transcript)=3821nt_BP=1677nt TGACTGCAGCAGCAGCGGCAGCGCCTCGGTTCCTGAGCCCACCGCAGGCTGAAGGCATTGCGCGTAGTCCATGCCCGTAGAGGAAGTGTG CAGATGGGATTAACGTCCACATGGAGATATGGAAGAGGACCGGGGATTGGTACCGTAACCATGGTCAGCTGGGGTCGTTTCATCTGCCTG GTCGTGGTCACCATGGCAACCTTGTCCCTGGCCCGGCCCTCCTTCAGTTTAGTTGAGGATACCACATTAGAGCCAGAAGAGCCACCAACC AAATACCAAATCTCTCAACCAGAAGTGTACGTGGCTGCGCCAGGGGAGTCGCTAGAGGTGCGCTGCCTGTTGAAAGATGCCGCCGTGATC AGTTGGACTAAGGATGGGGTGCACTTGGGGCCCAACAATAGGACAGTGCTTATTGGGGAGTACTTGCAGATAAAGGGCGCCACGCCTAGA GACTCCGGCCTCTATGCTTGTACTGCCAGTAGGACTGTAGACAGTGAAACTTGGTACTTCATGGTGAATGTCACAGATGCCATCTCATCC GGAGATGATGAGGATGACACCGATGGTGCGGAAGATTTTGTCAGTGAGAACAGTAACAACAAGAGAGCACCATACTGGACCAACACAGAA AAGATGGAAAAGCGGCTCCATGCTGTGCCTGCGGCCAACACTGTCAAGTTTCGCTGCCCAGCCGGGGGGAACCCAATGCCAACCATGCGG TGGCTGAAAAACGGGAAGGAGTTTAAGCAGGAGCATCGCATTGGAGGCTACAAGGTACGAAACCAGCACTGGAGCCTCATTATGGAAAGT GTGGTCCCATCTGACAAGGGAAATTATACCTGTGTAGTGGAGAATGAATACGGGTCCATCAATCACACGTACCACCTGGATGTTGTGGCG CCTGGAAGAGAAAAGGAGATTACAGCTTCCCCAGACTACCTGGAGATAGCCATTTACTGCATAGGGGTCTTCTTAATCGCCTGTATGGTG GTAACAGTCATCCTGTGCCGAATGAAGAACACGACCAAGAAGCCAGACTTCAGCAGCCAGCCGGCTGTGCACAAGCTGACCAAACGTATC CCCCTGCGGAGACAGGTAACAGTTTCGGCTGAGTCCAGCTCCTCCATGAACTCCAACACCCCGCTGGTGAGGATAACAACACGCCTCTCT TCAACGGCAGACACCCCCATGCTGGCAGGGGTCTCCGAGTATGAACTTCCAGAGGACCCAAAATGGGAGTTTCCAAGAGATAAGCTGACA CTGGGCAAGCCCCTGGGAGAAGGTTGCTTTGGGCAAGTGGTCATGGCGGAAGCAGTGGGAATTGACAAAGACAAGCCCAAGGAGGCGGTC ACCGTGGCCGTGAAGATGTTGAAAGATGATGCCACAGAGAAAGACCTTTCTGATCTGGTGTCAGAGATGGAGATGATGAAGATGATTGGG AAACACAAGAATATCATAAATCTTCTTGGAGCCTGCACACAGGATGGGCCTCTCTATGTCATAGTTGAGTATGCCTCTAAAGGCAACCTC CGAGAATACCTCCGAGCCCGGAGGCCACCCGGGATGGAGTACTCCTATGACATTAACCGTGTTCCTGAGGAGCAGATGACCTTCAAGGAC TTGGTGTCATGCACCTACCAGCTGGCCAGAGGCATGGAGTACTTGGCTTCCCAAAAAGTGCTCATCTGCCGGAACTACCGTGGCGACGTG GACATGTCAGAGGTGGAGCACTTCATGCCCATCCTGATGGAGAAGGAGGAGGAGGGGATGCTGTCGCCCATCCTGGCCCACGGGGGGGTC CGTTTCATGTGGATCAAACACAACAACCTGTATCTGGTTGCCACATCCAAGAAGAACGCGTGCGTGTCGCTGGTCTTTTCTTTCCTCTAT AAGGTGGTGCAGGTGTTTTCCGAGTACTTCAAGGAGCTGGAGGAGGAGAGCATCCGGGACAACTTTGTTATCATCTACGAGCTGCTGGAC GAGCTCATGGACTTCGGCTACCCCCAGACCACCGACAGCAAGATCCTGCAGGAGTACATCACTCAGGAAGGCCACAAGCTGGAAACAGGG GCCCCGCGGCCACCAGCCACCGTCACCAACGCGGTGTCCTGGCGGTCCGAAGGCATCAAGTATCGGAAGAATGAGGTGTTCTTGGACGTC ATCGAGTCTGTCAACCTCTTGGTCAGCGCCAACGGCAATGTCCTGCGCAGCGAGATCGTGGGCTCCATCAAGATGCGAGTCTTCCTCTCG GGCATGCCCGAGCTGCGCCTGGGCCTCAACGACAAGGTCCTCTTTGACAACACGGGCCGCGGCAAAAGCAAATCCGTGGAGCTGGAGGAT GTGAAGTTCCACCAGTGTGTGCGGCTATCACGCTTCGAGAATGACCGCACCATCTCCTTCATCCCACCCGACGGCGAGTTCGAGCTCATG TCCTACCGTCTCAACACCCACGTCAAGCCTTTGATATGGATCGAGTCGGTGATCGAGAAGCACTCCCACAGCCGCATCGAGTACATGATC AAGGCCAAAAGCCAGTTCAAGCGGCGGTCAACAGCCAACAACGTGGAGATCCACATTCCCGTGCCCAATGATGCCGACTCACCCAAGTTC AAGACGACGGTGGGGAGCGTTAAGTGGGTCCCCGAGAACAGCGAGATCGTGTGGTCCATCAAGTCCTTCCCGGGCGGCAAGGAGTACCTG ATGCGGGCCCACTTCGGCCTGCCTAGTGTGGAGGCCGAAGACAAGGAGGGCAAGCCCCCGATCAGTGTCAAGTTCGAGATCCCTTACTTC ACTACCTCCGGCATCCAGGTGCGCTACCTGAAGATCATTGAGAAGAGTGGGTACCAGGCCCTGCCCTGGGTGCGTTATATCACGCAGAAT GGAGATTACCAGCTCCGGACCCAGTGAGGGGCTGTCGCAGCCAACACCCCGGCCTCGGGGCTCCTGGTGGCAGCACCAGGGGACACACCT GCCAAACCCACCAGATGGAGGGGCCCTCCCTGGTCTCTGGCCACCCTCCCAGCCTCTGCCCAGGGACCCCTGCCTTCCCCAGGCCATCTG CTCTGCCGTCGACACTCGTCTCAGAAGCCCCTTTCCCAGAAGAGGCTGGTCTTCAAGAAGTCTCGTTTCTTTGCCCCTGAAGTCAGTTTC AGGGGAAGGATGTGAAATTTTTCCGTGTAGAGGTTACAGCCTTTTATGCTGTTGAGCTCCCAGGTACCAAAAAGCTTGGCCAACGCTTGC CAGCCAGCCAGCTGCAGGTGGCATCTGCCACGAAGGAAGCGCCAGCCTCGCCAGGCCAGCAGGGGCGTCGTTTTGTTGCCATTTTGTTGA ACGTTATGGGTTTATGGGTGTTCCTGGAACTTGTCTTTGTGCATTCGTTGCTGTTTGTGTTACCCTCACTGTCCCCATGTCCCACCCACG TCCTACGGCACTCAGGAAGCACTTGGTGAGGACGAGCCCTCACCCTTCTTGTCTTCCTTCCCGGCAGCGCCCGCAGCGGGCCATTTACAC GTCGAGGCTGGCACCTGGCGCGCTCGGGGGCCACTGTAGCGTCTGCCTGCTCCCTGGACTCGCAGGCCTCGCCTGTGGCGCCTTCCCAGG GCCAGCCTGGGTCACGAGATGCTGTCACTCAGCCAGATCAGTATTGACCCACCAGGGGAGGTGGGGTTTGGTGAGAGACGCCAGCCTCAG ACTTTTTCCCACTGAGGGTCCAGAGAGCGGGGCCACGTGTCACCCACGTCTGCGCTTGGTCACCCGTCCTCCCCACCCTGTGTGTGTTTA >30275_30275_3_FGFR2-AP1M1_FGFR2_chr10_123256046_ENST00000369061_AP1M1_chr19_16314270_ENST00000291439_length(amino acids)=952AA_BP=543 MKALRVVHARRGSVQMGLTSTWRYGRGPGIGTVTMVSWGRFICLVVVTMATLSLARPSFSLVEDTTLEPEEPPTKYQISQPEVYVAAPGE SLEVRCLLKDAAVISWTKDGVHLGPNNRTVLIGEYLQIKGATPRDSGLYACTASRTVDSETWYFMVNVTDAISSGDDEDDTDGAEDFVSE NSNNKRAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENE YGSINHTYHLDVVAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSM NSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDL SDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLA SQKVLICRNYRGDVDMSEVEHFMPILMEKEEEGMLSPILAHGGVRFMWIKHNNLYLVATSKKNACVSLVFSFLYKVVQVFSEYFKELEEE SIRDNFVIIYELLDELMDFGYPQTTDSKILQEYITQEGHKLETGAPRPPATVTNAVSWRSEGIKYRKNEVFLDVIESVNLLVSANGNVLR SEIVGSIKMRVFLSGMPELRLGLNDKVLFDNTGRGKSKSVELEDVKFHQCVRLSRFENDRTISFIPPDGEFELMSYRLNTHVKPLIWIES VIEKHSHSRIEYMIKAKSQFKRRSTANNVEIHIPVPNDADSPKFKTTVGSVKWVPENSEIVWSIKSFPGGKEYLMRAHFGLPSVEAEDKE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FGFR2-AP1M1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FGFR2-AP1M1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FGFR2-AP1M1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
