
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FKBP8-HAUS8 (FusionGDB2 ID:30528) |
Fusion Gene Summary for FKBP8-HAUS8 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FKBP8-HAUS8 | Fusion gene ID: 30528 | Hgene | Tgene | Gene symbol | FKBP8 | HAUS8 | Gene ID | 23770 | 93323 |
| Gene name | FKBP prolyl isomerase 8 | HAUS augmin like complex subunit 8 | |
| Synonyms | FKBP38|FKBPr38 | DGT4|HICE1|NY-SAR-48 | |
| Cytomap | 19p13.11 | 19p13.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | peptidyl-prolyl cis-trans isomerase FKBP838 kDa FK506-binding protein38 kDa FKBPFK506 binding protein 8, 38kDaFK506-binding protein 8FKBP-38FKBP-8PPIase FKBP8hFKBP38rotamase | HAUS augmin-like complex subunit 8HEC1/NDC80 interacting, centrosome associated 1HEC1/NDC80-interacting centrosome-associated protein 1Hec1-interacting and centrosome-associated 1sarcoma antigen NY-SAR-48 | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | Q14318 | Q9BT25 | |
| Ensembl transtripts involved in fusion gene | ENST00000222308, ENST00000453489, ENST00000596558, ENST00000597960, ENST00000608443, ENST00000610101, | ENST00000593360, ENST00000253669, ENST00000448593, | |
| Fusion gene scores | * DoF score | 12 X 15 X 5=900 | 7 X 8 X 6=336 |
| # samples | 15 | 7 | |
| ** MAII score | log2(15/900*10)=-2.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/336*10)=-2.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FKBP8 [Title/Abstract] AND HAUS8 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FKBP8(18652489)-HAUS8(17184145), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | FKBP8-HAUS8 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across FKBP8 (5'-gene) Fusion gene breakpoints across FKBP8 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
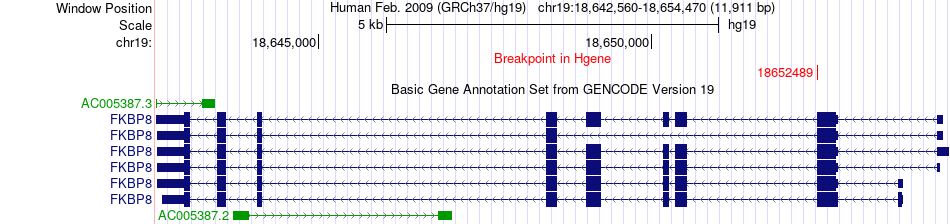 |
 Fusion gene breakpoints across HAUS8 (3'-gene) Fusion gene breakpoints across HAUS8 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
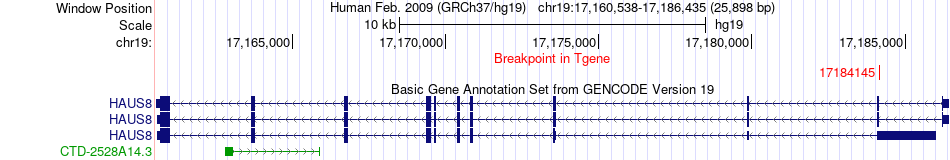 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ACC | TCGA-OR-A5KX-01A | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
Top |
Fusion Gene ORF analysis for FKBP8-HAUS8 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000222308 | ENST00000593360 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
| 5CDS-5UTR | ENST00000453489 | ENST00000593360 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
| 5CDS-5UTR | ENST00000596558 | ENST00000593360 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
| 5CDS-5UTR | ENST00000597960 | ENST00000593360 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
| 5CDS-5UTR | ENST00000608443 | ENST00000593360 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
| 5CDS-5UTR | ENST00000610101 | ENST00000593360 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
| Frame-shift | ENST00000453489 | ENST00000253669 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
| Frame-shift | ENST00000453489 | ENST00000448593 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
| Frame-shift | ENST00000596558 | ENST00000253669 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
| Frame-shift | ENST00000596558 | ENST00000448593 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
| Frame-shift | ENST00000597960 | ENST00000253669 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
| Frame-shift | ENST00000597960 | ENST00000448593 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
| In-frame | ENST00000222308 | ENST00000253669 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
| In-frame | ENST00000222308 | ENST00000448593 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
| In-frame | ENST00000608443 | ENST00000253669 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
| In-frame | ENST00000608443 | ENST00000448593 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
| In-frame | ENST00000610101 | ENST00000253669 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
| In-frame | ENST00000610101 | ENST00000448593 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000610101 | FKBP8 | chr19 | 18652489 | - | ENST00000253669 | HAUS8 | chr19 | 17184145 | - | 1753 | 405 | 493 | 1608 | 371 |
| ENST00000610101 | FKBP8 | chr19 | 18652489 | - | ENST00000448593 | HAUS8 | chr19 | 17184145 | - | 1718 | 405 | 493 | 1605 | 370 |
| ENST00000222308 | FKBP8 | chr19 | 18652489 | - | ENST00000253669 | HAUS8 | chr19 | 17184145 | - | 1704 | 356 | 444 | 1559 | 371 |
| ENST00000222308 | FKBP8 | chr19 | 18652489 | - | ENST00000448593 | HAUS8 | chr19 | 17184145 | - | 1669 | 356 | 444 | 1556 | 370 |
| ENST00000608443 | FKBP8 | chr19 | 18652489 | - | ENST00000253669 | HAUS8 | chr19 | 17184145 | - | 1840 | 492 | 580 | 1695 | 371 |
| ENST00000608443 | FKBP8 | chr19 | 18652489 | - | ENST00000448593 | HAUS8 | chr19 | 17184145 | - | 1805 | 492 | 580 | 1692 | 370 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000610101 | ENST00000253669 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - | 0.002263883 | 0.9977361 |
| ENST00000610101 | ENST00000448593 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - | 0.002698932 | 0.99730104 |
| ENST00000222308 | ENST00000253669 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - | 0.00239172 | 0.9976083 |
| ENST00000222308 | ENST00000448593 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - | 0.002793792 | 0.99720615 |
| ENST00000608443 | ENST00000253669 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - | 0.002207686 | 0.99779236 |
| ENST00000608443 | ENST00000448593 | FKBP8 | chr19 | 18652489 | - | HAUS8 | chr19 | 17184145 | - | 0.002623978 | 0.9973761 |
Top |
Fusion Genomic Features for FKBP8-HAUS8 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
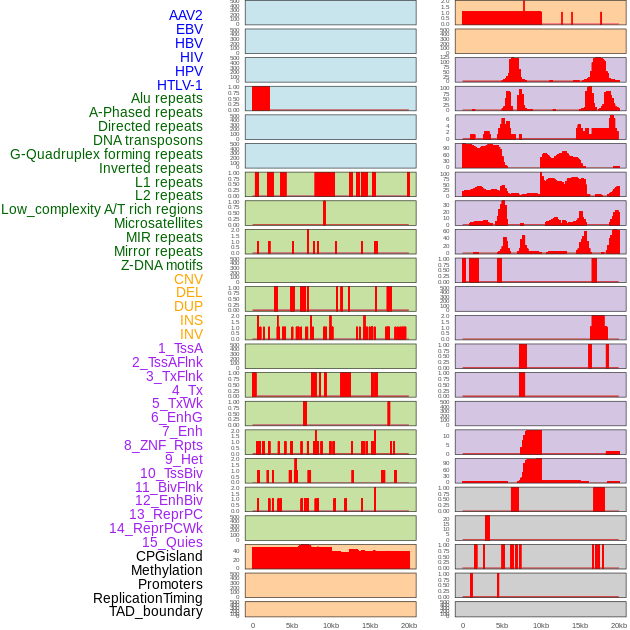 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for FKBP8-HAUS8 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:18652489/chr19:17184145) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FKBP8 | HAUS8 |
| FUNCTION: Constitutively inactive PPiase, which becomes active when bound to calmodulin and calcium. Seems to act as a chaperone for BCL2, targets it to the mitochondria and modulates its phosphorylation state. The BCL2/FKBP8/calmodulin/calcium complex probably interferes with the binding of BCL2 to its targets. The active form of FKBP8 may therefore play a role in the regulation of apoptosis. {ECO:0000269|PubMed:12510191, ECO:0000269|PubMed:15757646, ECO:0000269|PubMed:16176796}. | FUNCTION: Contributes to mitotic spindle assembly, maintenance of centrosome integrity and completion of cytokinesis as part of the HAUS augmin-like complex. {ECO:0000269|PubMed:18362163, ECO:0000269|PubMed:19369198, ECO:0000269|PubMed:19427217}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000222308 | - | 2 | 9 | 22_92 | 97 | 413.0 | Compositional bias | Note=Glu-rich |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000596558 | - | 2 | 9 | 22_92 | 97 | 413.0 | Compositional bias | Note=Glu-rich |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000597960 | - | 2 | 9 | 22_92 | 97 | 414.0 | Compositional bias | Note=Glu-rich |
| Tgene | HAUS8 | chr19:18652489 | chr19:17184145 | ENST00000253669 | 0 | 11 | 156_208 | 9 | 411.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | HAUS8 | chr19:18652489 | chr19:17184145 | ENST00000448593 | 0 | 11 | 156_208 | 9 | 410.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | HAUS8 | chr19:18652489 | chr19:17184145 | ENST00000593360 | 0 | 10 | 156_208 | 0 | 350.0 | Coiled coil | Ontology_term=ECO:0000255 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000222308 | - | 2 | 9 | 120_204 | 97 | 413.0 | Domain | PPIase FKBP-type |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000596558 | - | 2 | 9 | 120_204 | 97 | 413.0 | Domain | PPIase FKBP-type |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000597960 | - | 2 | 9 | 120_204 | 97 | 414.0 | Domain | PPIase FKBP-type |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000222308 | - | 2 | 9 | 221_254 | 97 | 413.0 | Repeat | Note=TPR 1 |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000222308 | - | 2 | 9 | 272_305 | 97 | 413.0 | Repeat | Note=TPR 2 |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000222308 | - | 2 | 9 | 306_339 | 97 | 413.0 | Repeat | Note=TPR 3 |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000596558 | - | 2 | 9 | 221_254 | 97 | 413.0 | Repeat | Note=TPR 1 |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000596558 | - | 2 | 9 | 272_305 | 97 | 413.0 | Repeat | Note=TPR 2 |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000596558 | - | 2 | 9 | 306_339 | 97 | 413.0 | Repeat | Note=TPR 3 |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000597960 | - | 2 | 9 | 221_254 | 97 | 414.0 | Repeat | Note=TPR 1 |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000597960 | - | 2 | 9 | 272_305 | 97 | 414.0 | Repeat | Note=TPR 2 |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000597960 | - | 2 | 9 | 306_339 | 97 | 414.0 | Repeat | Note=TPR 3 |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000222308 | - | 2 | 9 | 390_410 | 97 | 413.0 | Transmembrane | Helical |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000596558 | - | 2 | 9 | 390_410 | 97 | 413.0 | Transmembrane | Helical |
| Hgene | FKBP8 | chr19:18652489 | chr19:17184145 | ENST00000597960 | - | 2 | 9 | 390_410 | 97 | 414.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for FKBP8-HAUS8 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >30528_30528_1_FKBP8-HAUS8_FKBP8_chr19_18652489_ENST00000222308_HAUS8_chr19_17184145_ENST00000253669_length(transcript)=1704nt_BP=356nt GGGGGCGCAGGACCCCAAGTGGGGGTCCCGGAGCCAGAGGGCCAATTCCTGTCCCCCCAGCAGCATGGCATCGTGTGCTGAACCCTCTGA GCCCTCTGCCCCACTGCCCGCCGGGGTCCCACCGCTCGAGGACTTCGAGGTACTGGATGGGGTTGAGGATGCAGAGGGTGAGGAGGAAGA GGAGGAGGAAGAGGAGGAAGAGGATGACCTGAGTGAGCTGCCACCGCTGGAGGACATGGGACAACCCCCGGCGGAGGAGGCTGAGCAGCC TGGGGCCCTGGCCCGAGAGTTCCTTGCTGCCATGGAGCCCGAGCCCGCCCCAGCCCCGGCCCCAGAAGAGTGGCTGGACATTCTGGGAAG CCTGCAACCGGCCCCACAAATTCTAGCAGTGCCAAGAAGAAGGATAAAAGAGTTCAAGGTGGAAGAGTGATTGAGTCCCGGTATCTGCAG TATGAAAAGAAGACAACCCAAAAGGCTCCTGCAGGAGATGGGTCACAGACCCGAGGGAAGATGTCTGAAGGTGGAAGGAAATCCAGCCTG CTCCAGAAAAGCAAAGCAGATAGCAGTGGGGTCGGAAAGGGTGACCTGCAGTCCACGTTGCTGGAAGGGCATGGCACAGCTCCACCTGAC CTGGATCTCTCTGCTATTAATGACAAAAGCATCGTCAAAAAGACGCCACAGTTAGCAAAAACAATATCAAAGAAACCTGAGTCAACATCA TTTTCTGCCCCTCGGAAAAAGAGCCCGGATTTATCTGAAGCAATGGAAATGATGGAGTCTCAGACACTACTGCTGACGCTACTATCCGTA AAGATGGAGAACAATCTTGCTGAGTTTGAAAGAAGGGCAGAAAAGAATTTATTAATAATGTGTAAGGAGAAGGAGAAGCTACAGAAAAAG GCCCACGAGCTGAAGCGCAGGCTTCTCCTCTCTCAGAGGAAGCGGGAGCTGGCAGATGTCCTGGATGCCCAGATCGAGATGCTCAGCCCC TTCGAGGCAGTGGCCACACGCTTCAAGGAGCAATACAGGACATTCGCCACGGCCCTGGACACTACCAGGCACGAGCTGCCCGTGAGGTCC ATCCACCTGGAGGGAGATGGGCAGCAGCTCTTAGACGCCCTGCAGCATGAACTGGTGACCACTCAGCGCCTCCTGGGAGAACTTGATGTT GGTGATTCGGAAGAAAATGTGCAGGTGCTGGACTTACTGAGCGAACTCAAGGACGTGACGGCGAAAAAGGACCTTGAGCTCCGAAGGAGC TTTGCCCAGGTGCTGGAACTCTCCGCAGAGGCAAGCAAAGAGGCAGCCTTGGCAAACCAGGAAGTCTGGGAAGAGACCCAGGGCATGGCG CCCCCCAGCCGGTGGTATTTCAATCAAGACAGTGCCTGCAGAGAATCTGGGGGAGCACCCAAGAACACGCCCCTGTCTGAGGACGACAAC CCGGGTGCCTCGTCAGCCCCCGCTCAGGCCACGTTCATCAGCCCAAGCGAAGATTTTTCTTCAAGCAGCCAGGCAGAAGTCCCGCCCTCT CTCTCTCGTTCAGGGAGGGACTTGTCATGACTCATGGTTACATTCAGGATACTTGAGCACTTTATATACTACCGTAGCACTGTAGCTATT >30528_30528_1_FKBP8-HAUS8_FKBP8_chr19_18652489_ENST00000222308_HAUS8_chr19_17184145_ENST00000253669_length(amino acids)=371AA_BP= MQYEKKTTQKAPAGDGSQTRGKMSEGGRKSSLLQKSKADSSGVGKGDLQSTLLEGHGTAPPDLDLSAINDKSIVKKTPQLAKTISKKPES TSFSAPRKKSPDLSEAMEMMESQTLLLTLLSVKMENNLAEFERRAEKNLLIMCKEKEKLQKKAHELKRRLLLSQRKRELADVLDAQIEML SPFEAVATRFKEQYRTFATALDTTRHELPVRSIHLEGDGQQLLDALQHELVTTQRLLGELDVGDSEENVQVLDLLSELKDVTAKKDLELR RSFAQVLELSAEASKEAALANQEVWEETQGMAPPSRWYFNQDSACRESGGAPKNTPLSEDDNPGASSAPAQATFISPSEDFSSSSQAEVP -------------------------------------------------------------- >30528_30528_2_FKBP8-HAUS8_FKBP8_chr19_18652489_ENST00000222308_HAUS8_chr19_17184145_ENST00000448593_length(transcript)=1669nt_BP=356nt GGGGGCGCAGGACCCCAAGTGGGGGTCCCGGAGCCAGAGGGCCAATTCCTGTCCCCCCAGCAGCATGGCATCGTGTGCTGAACCCTCTGA GCCCTCTGCCCCACTGCCCGCCGGGGTCCCACCGCTCGAGGACTTCGAGGTACTGGATGGGGTTGAGGATGCAGAGGGTGAGGAGGAAGA GGAGGAGGAAGAGGAGGAAGAGGATGACCTGAGTGAGCTGCCACCGCTGGAGGACATGGGACAACCCCCGGCGGAGGAGGCTGAGCAGCC TGGGGCCCTGGCCCGAGAGTTCCTTGCTGCCATGGAGCCCGAGCCCGCCCCAGCCCCGGCCCCAGAAGAGTGGCTGGACATTCTGGGAAG CCTGCAACCGGCCCCACAAATTCTAGCAGTGCCAAGAAGAAGGATAAAAGAGTTCAAGGTGGAAGAGTGATTGAGTCCCGGTATCTGCAG TATGAAAAGAAGACAACCCAAAAGGCTCCTGCAGGAGATGGGTCACAGACCCGAGGGAAGATGTCTGAAGGTGGAAGGAAATCCAGCCTG CTCCAGAAAAGCAAAGATAGCAGTGGGGTCGGAAAGGGTGACCTGCAGTCCACGTTGCTGGAAGGGCATGGCACAGCTCCACCTGACCTG GATCTCTCTGCTATTAATGACAAAAGCATCGTCAAAAAGACGCCACAGTTAGCAAAAACAATATCAAAGAAACCTGAGTCAACATCATTT TCTGCCCCTCGGAAAAAGAGCCCGGATTTATCTGAAGCAATGGAAATGATGGAGTCTCAGACACTACTGCTGACGCTACTATCCGTAAAG ATGGAGAACAATCTTGCTGAGTTTGAAAGAAGGGCAGAAAAGAATTTATTAATAATGTGTAAGGAGAAGGAGAAGCTACAGAAAAAGGCC CACGAGCTGAAGCGCAGGCTTCTCCTCTCTCAGAGGAAGCGGGAGCTGGCAGATGTCCTGGATGCCCAGATCGAGATGCTCAGCCCCTTC GAGGCAGTGGCCACACGCTTCAAGGAGCAATACAGGACATTCGCCACGGCCCTGGACACTACCAGGCACGAGCTGCCCGTGAGGTCCATC CACCTGGAGGGAGATGGGCAGCAGCTCTTAGACGCCCTGCAGCATGAACTGGTGACCACTCAGCGCCTCCTGGGAGAACTTGATGTTGGT GATTCGGAAGAAAATGTGCAGGTGCTGGACTTACTGAGCGAACTCAAGGACGTGACGGCGAAAAAGGACCTTGAGCTCCGAAGGAGCTTT GCCCAGGTGCTGGAACTCTCCGCAGAGGCAAGCAAAGAGGCAGCCTTGGCAAACCAGGAAGTCTGGGAAGAGACCCAGGGCATGGCGCCC CCCAGCCGGTGGTATTTCAATCAAGACAGTGCCTGCAGAGAATCTGGGGGAGCACCCAAGAACACGCCCCTGTCTGAGGACGACAACCCG GGTGCCTCGTCAGCCCCCGCTCAGGCCACGTTCATCAGCCCAAGCGAAGATTTTTCTTCAAGCAGCCAGGCAGAAGTCCCGCCCTCTCTC TCTCGTTCAGGGAGGGACTTGTCATGACTCATGGTTACATTCAGGATACTTGAGCACTTTATATACTACCGTAGCACTGTAGCTATTTTT >30528_30528_2_FKBP8-HAUS8_FKBP8_chr19_18652489_ENST00000222308_HAUS8_chr19_17184145_ENST00000448593_length(amino acids)=370AA_BP= MQYEKKTTQKAPAGDGSQTRGKMSEGGRKSSLLQKSKDSSGVGKGDLQSTLLEGHGTAPPDLDLSAINDKSIVKKTPQLAKTISKKPEST SFSAPRKKSPDLSEAMEMMESQTLLLTLLSVKMENNLAEFERRAEKNLLIMCKEKEKLQKKAHELKRRLLLSQRKRELADVLDAQIEMLS PFEAVATRFKEQYRTFATALDTTRHELPVRSIHLEGDGQQLLDALQHELVTTQRLLGELDVGDSEENVQVLDLLSELKDVTAKKDLELRR SFAQVLELSAEASKEAALANQEVWEETQGMAPPSRWYFNQDSACRESGGAPKNTPLSEDDNPGASSAPAQATFISPSEDFSSSSQAEVPP -------------------------------------------------------------- >30528_30528_3_FKBP8-HAUS8_FKBP8_chr19_18652489_ENST00000608443_HAUS8_chr19_17184145_ENST00000253669_length(transcript)=1840nt_BP=492nt ACGCCCGGGACGGACGTCTGCATGAGCCAACGGGTGCGGCAGTCGCGGCAGCGGCGGCCAATGGGCACGGCAGGGGCGGGGCCAAGCGGC CGAACCTGGTTCCCGAGGCGCGGCGGCCGCGGCTGGGGGCGGGGAGGGGGGCGCAGGACCCCAAGTGGGGGTCCCGGAGCCAGAGGGCCA ATTCCTGTCCCCCCAGCAGCATGGCATCGTGTGCTGAACCCTCTGAGCCCTCTGCCCCACTGCCCGCCGGGGTCCCACCGCTCGAGGACT TCGAGGTACTGGATGGGGTTGAGGATGCAGAGGGTGAGGAGGAAGAGGAGGAGGAAGAGGAGGAAGAGGATGACCTGAGTGAGCTGCCAC CGCTGGAGGACATGGGACAACCCCCGGCGGAGGAGGCTGAGCAGCCTGGGGCCCTGGCCCGAGAGTTCCTTGCTGCCATGGAGCCCGAGC CCGCCCCAGCCCCGGCCCCAGAAGAGTGGCTGGACATTCTGGGAAGCCTGCAACCGGCCCCACAAATTCTAGCAGTGCCAAGAAGAAGGA TAAAAGAGTTCAAGGTGGAAGAGTGATTGAGTCCCGGTATCTGCAGTATGAAAAGAAGACAACCCAAAAGGCTCCTGCAGGAGATGGGTC ACAGACCCGAGGGAAGATGTCTGAAGGTGGAAGGAAATCCAGCCTGCTCCAGAAAAGCAAAGCAGATAGCAGTGGGGTCGGAAAGGGTGA CCTGCAGTCCACGTTGCTGGAAGGGCATGGCACAGCTCCACCTGACCTGGATCTCTCTGCTATTAATGACAAAAGCATCGTCAAAAAGAC GCCACAGTTAGCAAAAACAATATCAAAGAAACCTGAGTCAACATCATTTTCTGCCCCTCGGAAAAAGAGCCCGGATTTATCTGAAGCAAT GGAAATGATGGAGTCTCAGACACTACTGCTGACGCTACTATCCGTAAAGATGGAGAACAATCTTGCTGAGTTTGAAAGAAGGGCAGAAAA GAATTTATTAATAATGTGTAAGGAGAAGGAGAAGCTACAGAAAAAGGCCCACGAGCTGAAGCGCAGGCTTCTCCTCTCTCAGAGGAAGCG GGAGCTGGCAGATGTCCTGGATGCCCAGATCGAGATGCTCAGCCCCTTCGAGGCAGTGGCCACACGCTTCAAGGAGCAATACAGGACATT CGCCACGGCCCTGGACACTACCAGGCACGAGCTGCCCGTGAGGTCCATCCACCTGGAGGGAGATGGGCAGCAGCTCTTAGACGCCCTGCA GCATGAACTGGTGACCACTCAGCGCCTCCTGGGAGAACTTGATGTTGGTGATTCGGAAGAAAATGTGCAGGTGCTGGACTTACTGAGCGA ACTCAAGGACGTGACGGCGAAAAAGGACCTTGAGCTCCGAAGGAGCTTTGCCCAGGTGCTGGAACTCTCCGCAGAGGCAAGCAAAGAGGC AGCCTTGGCAAACCAGGAAGTCTGGGAAGAGACCCAGGGCATGGCGCCCCCCAGCCGGTGGTATTTCAATCAAGACAGTGCCTGCAGAGA ATCTGGGGGAGCACCCAAGAACACGCCCCTGTCTGAGGACGACAACCCGGGTGCCTCGTCAGCCCCCGCTCAGGCCACGTTCATCAGCCC AAGCGAAGATTTTTCTTCAAGCAGCCAGGCAGAAGTCCCGCCCTCTCTCTCTCGTTCAGGGAGGGACTTGTCATGACTCATGGTTACATT CAGGATACTTGAGCACTTTATATACTACCGTAGCACTGTAGCTATTTTTTATGTGATAATCTTGTTTGATAAAAAAGTAAACCTGTTTTG >30528_30528_3_FKBP8-HAUS8_FKBP8_chr19_18652489_ENST00000608443_HAUS8_chr19_17184145_ENST00000253669_length(amino acids)=371AA_BP= MQYEKKTTQKAPAGDGSQTRGKMSEGGRKSSLLQKSKADSSGVGKGDLQSTLLEGHGTAPPDLDLSAINDKSIVKKTPQLAKTISKKPES TSFSAPRKKSPDLSEAMEMMESQTLLLTLLSVKMENNLAEFERRAEKNLLIMCKEKEKLQKKAHELKRRLLLSQRKRELADVLDAQIEML SPFEAVATRFKEQYRTFATALDTTRHELPVRSIHLEGDGQQLLDALQHELVTTQRLLGELDVGDSEENVQVLDLLSELKDVTAKKDLELR RSFAQVLELSAEASKEAALANQEVWEETQGMAPPSRWYFNQDSACRESGGAPKNTPLSEDDNPGASSAPAQATFISPSEDFSSSSQAEVP -------------------------------------------------------------- >30528_30528_4_FKBP8-HAUS8_FKBP8_chr19_18652489_ENST00000608443_HAUS8_chr19_17184145_ENST00000448593_length(transcript)=1805nt_BP=492nt ACGCCCGGGACGGACGTCTGCATGAGCCAACGGGTGCGGCAGTCGCGGCAGCGGCGGCCAATGGGCACGGCAGGGGCGGGGCCAAGCGGC CGAACCTGGTTCCCGAGGCGCGGCGGCCGCGGCTGGGGGCGGGGAGGGGGGCGCAGGACCCCAAGTGGGGGTCCCGGAGCCAGAGGGCCA ATTCCTGTCCCCCCAGCAGCATGGCATCGTGTGCTGAACCCTCTGAGCCCTCTGCCCCACTGCCCGCCGGGGTCCCACCGCTCGAGGACT TCGAGGTACTGGATGGGGTTGAGGATGCAGAGGGTGAGGAGGAAGAGGAGGAGGAAGAGGAGGAAGAGGATGACCTGAGTGAGCTGCCAC CGCTGGAGGACATGGGACAACCCCCGGCGGAGGAGGCTGAGCAGCCTGGGGCCCTGGCCCGAGAGTTCCTTGCTGCCATGGAGCCCGAGC CCGCCCCAGCCCCGGCCCCAGAAGAGTGGCTGGACATTCTGGGAAGCCTGCAACCGGCCCCACAAATTCTAGCAGTGCCAAGAAGAAGGA TAAAAGAGTTCAAGGTGGAAGAGTGATTGAGTCCCGGTATCTGCAGTATGAAAAGAAGACAACCCAAAAGGCTCCTGCAGGAGATGGGTC ACAGACCCGAGGGAAGATGTCTGAAGGTGGAAGGAAATCCAGCCTGCTCCAGAAAAGCAAAGATAGCAGTGGGGTCGGAAAGGGTGACCT GCAGTCCACGTTGCTGGAAGGGCATGGCACAGCTCCACCTGACCTGGATCTCTCTGCTATTAATGACAAAAGCATCGTCAAAAAGACGCC ACAGTTAGCAAAAACAATATCAAAGAAACCTGAGTCAACATCATTTTCTGCCCCTCGGAAAAAGAGCCCGGATTTATCTGAAGCAATGGA AATGATGGAGTCTCAGACACTACTGCTGACGCTACTATCCGTAAAGATGGAGAACAATCTTGCTGAGTTTGAAAGAAGGGCAGAAAAGAA TTTATTAATAATGTGTAAGGAGAAGGAGAAGCTACAGAAAAAGGCCCACGAGCTGAAGCGCAGGCTTCTCCTCTCTCAGAGGAAGCGGGA GCTGGCAGATGTCCTGGATGCCCAGATCGAGATGCTCAGCCCCTTCGAGGCAGTGGCCACACGCTTCAAGGAGCAATACAGGACATTCGC CACGGCCCTGGACACTACCAGGCACGAGCTGCCCGTGAGGTCCATCCACCTGGAGGGAGATGGGCAGCAGCTCTTAGACGCCCTGCAGCA TGAACTGGTGACCACTCAGCGCCTCCTGGGAGAACTTGATGTTGGTGATTCGGAAGAAAATGTGCAGGTGCTGGACTTACTGAGCGAACT CAAGGACGTGACGGCGAAAAAGGACCTTGAGCTCCGAAGGAGCTTTGCCCAGGTGCTGGAACTCTCCGCAGAGGCAAGCAAAGAGGCAGC CTTGGCAAACCAGGAAGTCTGGGAAGAGACCCAGGGCATGGCGCCCCCCAGCCGGTGGTATTTCAATCAAGACAGTGCCTGCAGAGAATC TGGGGGAGCACCCAAGAACACGCCCCTGTCTGAGGACGACAACCCGGGTGCCTCGTCAGCCCCCGCTCAGGCCACGTTCATCAGCCCAAG CGAAGATTTTTCTTCAAGCAGCCAGGCAGAAGTCCCGCCCTCTCTCTCTCGTTCAGGGAGGGACTTGTCATGACTCATGGTTACATTCAG GATACTTGAGCACTTTATATACTACCGTAGCACTGTAGCTATTTTTTATGTGATAATCTTGTTTGATAAAAAAGTAAACCTGTTTTGCAA >30528_30528_4_FKBP8-HAUS8_FKBP8_chr19_18652489_ENST00000608443_HAUS8_chr19_17184145_ENST00000448593_length(amino acids)=370AA_BP= MQYEKKTTQKAPAGDGSQTRGKMSEGGRKSSLLQKSKDSSGVGKGDLQSTLLEGHGTAPPDLDLSAINDKSIVKKTPQLAKTISKKPEST SFSAPRKKSPDLSEAMEMMESQTLLLTLLSVKMENNLAEFERRAEKNLLIMCKEKEKLQKKAHELKRRLLLSQRKRELADVLDAQIEMLS PFEAVATRFKEQYRTFATALDTTRHELPVRSIHLEGDGQQLLDALQHELVTTQRLLGELDVGDSEENVQVLDLLSELKDVTAKKDLELRR SFAQVLELSAEASKEAALANQEVWEETQGMAPPSRWYFNQDSACRESGGAPKNTPLSEDDNPGASSAPAQATFISPSEDFSSSSQAEVPP -------------------------------------------------------------- >30528_30528_5_FKBP8-HAUS8_FKBP8_chr19_18652489_ENST00000610101_HAUS8_chr19_17184145_ENST00000253669_length(transcript)=1753nt_BP=405nt GGCCGAACCTGGTTCCCGAGGCGCGGCGGCCGCGGCTGGGGGCGGGGAGGGGGGCGCAGGACCCCAAGTGGGGGTCCCGGAGCCAGAGGG CCAATTCCTGTCCCCCCAGCAGCATGGCATCGTGTGCTGAACCCTCTGAGCCCTCTGCCCCACTGCCCGCCGGGGTCCCACCGCTCGAGG ACTTCGAGGTACTGGATGGGGTTGAGGATGCAGAGGGTGAGGAGGAAGAGGAGGAGGAAGAGGAGGAAGAGGATGACCTGAGTGAGCTGC CACCGCTGGAGGACATGGGACAACCCCCGGCGGAGGAGGCTGAGCAGCCTGGGGCCCTGGCCCGAGAGTTCCTTGCTGCCATGGAGCCCG AGCCCGCCCCAGCCCCGGCCCCAGAAGAGTGGCTGGACATTCTGGGAAGCCTGCAACCGGCCCCACAAATTCTAGCAGTGCCAAGAAGAA GGATAAAAGAGTTCAAGGTGGAAGAGTGATTGAGTCCCGGTATCTGCAGTATGAAAAGAAGACAACCCAAAAGGCTCCTGCAGGAGATGG GTCACAGACCCGAGGGAAGATGTCTGAAGGTGGAAGGAAATCCAGCCTGCTCCAGAAAAGCAAAGCAGATAGCAGTGGGGTCGGAAAGGG TGACCTGCAGTCCACGTTGCTGGAAGGGCATGGCACAGCTCCACCTGACCTGGATCTCTCTGCTATTAATGACAAAAGCATCGTCAAAAA GACGCCACAGTTAGCAAAAACAATATCAAAGAAACCTGAGTCAACATCATTTTCTGCCCCTCGGAAAAAGAGCCCGGATTTATCTGAAGC AATGGAAATGATGGAGTCTCAGACACTACTGCTGACGCTACTATCCGTAAAGATGGAGAACAATCTTGCTGAGTTTGAAAGAAGGGCAGA AAAGAATTTATTAATAATGTGTAAGGAGAAGGAGAAGCTACAGAAAAAGGCCCACGAGCTGAAGCGCAGGCTTCTCCTCTCTCAGAGGAA GCGGGAGCTGGCAGATGTCCTGGATGCCCAGATCGAGATGCTCAGCCCCTTCGAGGCAGTGGCCACACGCTTCAAGGAGCAATACAGGAC ATTCGCCACGGCCCTGGACACTACCAGGCACGAGCTGCCCGTGAGGTCCATCCACCTGGAGGGAGATGGGCAGCAGCTCTTAGACGCCCT GCAGCATGAACTGGTGACCACTCAGCGCCTCCTGGGAGAACTTGATGTTGGTGATTCGGAAGAAAATGTGCAGGTGCTGGACTTACTGAG CGAACTCAAGGACGTGACGGCGAAAAAGGACCTTGAGCTCCGAAGGAGCTTTGCCCAGGTGCTGGAACTCTCCGCAGAGGCAAGCAAAGA GGCAGCCTTGGCAAACCAGGAAGTCTGGGAAGAGACCCAGGGCATGGCGCCCCCCAGCCGGTGGTATTTCAATCAAGACAGTGCCTGCAG AGAATCTGGGGGAGCACCCAAGAACACGCCCCTGTCTGAGGACGACAACCCGGGTGCCTCGTCAGCCCCCGCTCAGGCCACGTTCATCAG CCCAAGCGAAGATTTTTCTTCAAGCAGCCAGGCAGAAGTCCCGCCCTCTCTCTCTCGTTCAGGGAGGGACTTGTCATGACTCATGGTTAC ATTCAGGATACTTGAGCACTTTATATACTACCGTAGCACTGTAGCTATTTTTTATGTGATAATCTTGTTTGATAAAAAAGTAAACCTGTT >30528_30528_5_FKBP8-HAUS8_FKBP8_chr19_18652489_ENST00000610101_HAUS8_chr19_17184145_ENST00000253669_length(amino acids)=371AA_BP= MQYEKKTTQKAPAGDGSQTRGKMSEGGRKSSLLQKSKADSSGVGKGDLQSTLLEGHGTAPPDLDLSAINDKSIVKKTPQLAKTISKKPES TSFSAPRKKSPDLSEAMEMMESQTLLLTLLSVKMENNLAEFERRAEKNLLIMCKEKEKLQKKAHELKRRLLLSQRKRELADVLDAQIEML SPFEAVATRFKEQYRTFATALDTTRHELPVRSIHLEGDGQQLLDALQHELVTTQRLLGELDVGDSEENVQVLDLLSELKDVTAKKDLELR RSFAQVLELSAEASKEAALANQEVWEETQGMAPPSRWYFNQDSACRESGGAPKNTPLSEDDNPGASSAPAQATFISPSEDFSSSSQAEVP -------------------------------------------------------------- >30528_30528_6_FKBP8-HAUS8_FKBP8_chr19_18652489_ENST00000610101_HAUS8_chr19_17184145_ENST00000448593_length(transcript)=1718nt_BP=405nt GGCCGAACCTGGTTCCCGAGGCGCGGCGGCCGCGGCTGGGGGCGGGGAGGGGGGCGCAGGACCCCAAGTGGGGGTCCCGGAGCCAGAGGG CCAATTCCTGTCCCCCCAGCAGCATGGCATCGTGTGCTGAACCCTCTGAGCCCTCTGCCCCACTGCCCGCCGGGGTCCCACCGCTCGAGG ACTTCGAGGTACTGGATGGGGTTGAGGATGCAGAGGGTGAGGAGGAAGAGGAGGAGGAAGAGGAGGAAGAGGATGACCTGAGTGAGCTGC CACCGCTGGAGGACATGGGACAACCCCCGGCGGAGGAGGCTGAGCAGCCTGGGGCCCTGGCCCGAGAGTTCCTTGCTGCCATGGAGCCCG AGCCCGCCCCAGCCCCGGCCCCAGAAGAGTGGCTGGACATTCTGGGAAGCCTGCAACCGGCCCCACAAATTCTAGCAGTGCCAAGAAGAA GGATAAAAGAGTTCAAGGTGGAAGAGTGATTGAGTCCCGGTATCTGCAGTATGAAAAGAAGACAACCCAAAAGGCTCCTGCAGGAGATGG GTCACAGACCCGAGGGAAGATGTCTGAAGGTGGAAGGAAATCCAGCCTGCTCCAGAAAAGCAAAGATAGCAGTGGGGTCGGAAAGGGTGA CCTGCAGTCCACGTTGCTGGAAGGGCATGGCACAGCTCCACCTGACCTGGATCTCTCTGCTATTAATGACAAAAGCATCGTCAAAAAGAC GCCACAGTTAGCAAAAACAATATCAAAGAAACCTGAGTCAACATCATTTTCTGCCCCTCGGAAAAAGAGCCCGGATTTATCTGAAGCAAT GGAAATGATGGAGTCTCAGACACTACTGCTGACGCTACTATCCGTAAAGATGGAGAACAATCTTGCTGAGTTTGAAAGAAGGGCAGAAAA GAATTTATTAATAATGTGTAAGGAGAAGGAGAAGCTACAGAAAAAGGCCCACGAGCTGAAGCGCAGGCTTCTCCTCTCTCAGAGGAAGCG GGAGCTGGCAGATGTCCTGGATGCCCAGATCGAGATGCTCAGCCCCTTCGAGGCAGTGGCCACACGCTTCAAGGAGCAATACAGGACATT CGCCACGGCCCTGGACACTACCAGGCACGAGCTGCCCGTGAGGTCCATCCACCTGGAGGGAGATGGGCAGCAGCTCTTAGACGCCCTGCA GCATGAACTGGTGACCACTCAGCGCCTCCTGGGAGAACTTGATGTTGGTGATTCGGAAGAAAATGTGCAGGTGCTGGACTTACTGAGCGA ACTCAAGGACGTGACGGCGAAAAAGGACCTTGAGCTCCGAAGGAGCTTTGCCCAGGTGCTGGAACTCTCCGCAGAGGCAAGCAAAGAGGC AGCCTTGGCAAACCAGGAAGTCTGGGAAGAGACCCAGGGCATGGCGCCCCCCAGCCGGTGGTATTTCAATCAAGACAGTGCCTGCAGAGA ATCTGGGGGAGCACCCAAGAACACGCCCCTGTCTGAGGACGACAACCCGGGTGCCTCGTCAGCCCCCGCTCAGGCCACGTTCATCAGCCC AAGCGAAGATTTTTCTTCAAGCAGCCAGGCAGAAGTCCCGCCCTCTCTCTCTCGTTCAGGGAGGGACTTGTCATGACTCATGGTTACATT CAGGATACTTGAGCACTTTATATACTACCGTAGCACTGTAGCTATTTTTTATGTGATAATCTTGTTTGATAAAAAAGTAAACCTGTTTTG >30528_30528_6_FKBP8-HAUS8_FKBP8_chr19_18652489_ENST00000610101_HAUS8_chr19_17184145_ENST00000448593_length(amino acids)=370AA_BP= MQYEKKTTQKAPAGDGSQTRGKMSEGGRKSSLLQKSKDSSGVGKGDLQSTLLEGHGTAPPDLDLSAINDKSIVKKTPQLAKTISKKPEST SFSAPRKKSPDLSEAMEMMESQTLLLTLLSVKMENNLAEFERRAEKNLLIMCKEKEKLQKKAHELKRRLLLSQRKRELADVLDAQIEMLS PFEAVATRFKEQYRTFATALDTTRHELPVRSIHLEGDGQQLLDALQHELVTTQRLLGELDVGDSEENVQVLDLLSELKDVTAKKDLELRR SFAQVLELSAEASKEAALANQEVWEETQGMAPPSRWYFNQDSACRESGGAPKNTPLSEDDNPGASSAPAQATFISPSEDFSSSSQAEVPP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FKBP8-HAUS8 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FKBP8-HAUS8 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FKBP8-HAUS8 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
