
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FLAD1-PHYHD1 (FusionGDB2 ID:30553) |
Fusion Gene Summary for FLAD1-PHYHD1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FLAD1-PHYHD1 | Fusion gene ID: 30553 | Hgene | Tgene | Gene symbol | FLAD1 | PHYHD1 | Gene ID | 80308 | 254295 |
| Gene name | flavin adenine dinucleotide synthetase 1 | phytanoyl-CoA dioxygenase domain containing 1 | |
| Synonyms | FAD1|FADS|LSMFLAD|PP591 | - | |
| Cytomap | 1q21.3 | 9q34.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | FAD synthaseFAD pyrophosphorylaseFAD-synthetaseFMN adenylyltransferaseFad1, flavin adenine dinucleotide synthetase, homolog | phytanoyl-CoA dioxygenase domain-containing protein 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8NFF5 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000487371, ENST00000292180, ENST00000315144, ENST00000368432, ENST00000368433, ENST00000368431, ENST00000295530, ENST00000368428, ENST00000405236, | ENST00000308941, ENST00000353176, ENST00000372592, ENST00000421063, ENST00000487504, | |
| Fusion gene scores | * DoF score | 5 X 3 X 4=60 | 3 X 8 X 3=72 |
| # samples | 7 | 6 | |
| ** MAII score | log2(7/60*10)=0.222392421336448 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(6/72*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FLAD1 [Title/Abstract] AND PHYHD1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FLAD1(154956542)-PHYHD1(131696060), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across FLAD1 (5'-gene) Fusion gene breakpoints across FLAD1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
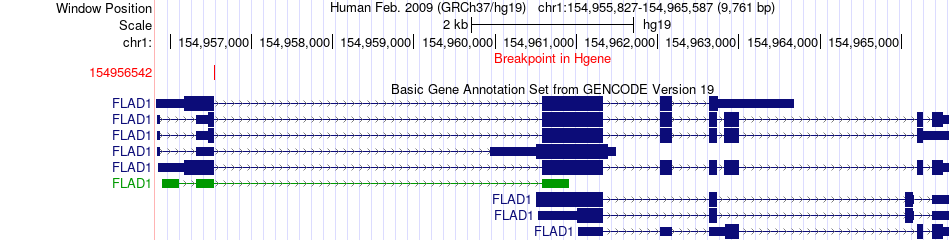 |
 Fusion gene breakpoints across PHYHD1 (3'-gene) Fusion gene breakpoints across PHYHD1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
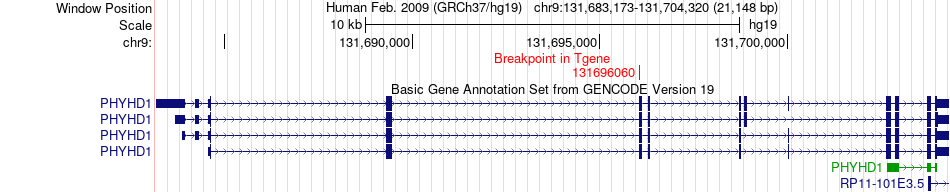 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-13-0762 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
Top |
Fusion Gene ORF analysis for FLAD1-PHYHD1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000487371 | ENST00000308941 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| 3UTR-3CDS | ENST00000487371 | ENST00000353176 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| 3UTR-3CDS | ENST00000487371 | ENST00000372592 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| 3UTR-3CDS | ENST00000487371 | ENST00000421063 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| 3UTR-intron | ENST00000487371 | ENST00000487504 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| 5CDS-intron | ENST00000292180 | ENST00000487504 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| 5CDS-intron | ENST00000315144 | ENST00000487504 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| 5CDS-intron | ENST00000368432 | ENST00000487504 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| 5CDS-intron | ENST00000368433 | ENST00000487504 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| 5UTR-3CDS | ENST00000368431 | ENST00000308941 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| 5UTR-3CDS | ENST00000368431 | ENST00000353176 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| 5UTR-3CDS | ENST00000368431 | ENST00000372592 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| 5UTR-3CDS | ENST00000368431 | ENST00000421063 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| 5UTR-intron | ENST00000368431 | ENST00000487504 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| In-frame | ENST00000292180 | ENST00000308941 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| In-frame | ENST00000292180 | ENST00000353176 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| In-frame | ENST00000292180 | ENST00000372592 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| In-frame | ENST00000292180 | ENST00000421063 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| In-frame | ENST00000315144 | ENST00000308941 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| In-frame | ENST00000315144 | ENST00000353176 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| In-frame | ENST00000315144 | ENST00000372592 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| In-frame | ENST00000315144 | ENST00000421063 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| In-frame | ENST00000368432 | ENST00000308941 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| In-frame | ENST00000368432 | ENST00000353176 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| In-frame | ENST00000368432 | ENST00000372592 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| In-frame | ENST00000368432 | ENST00000421063 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| In-frame | ENST00000368433 | ENST00000308941 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| In-frame | ENST00000368433 | ENST00000353176 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| In-frame | ENST00000368433 | ENST00000372592 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| In-frame | ENST00000368433 | ENST00000421063 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| intron-3CDS | ENST00000295530 | ENST00000308941 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| intron-3CDS | ENST00000295530 | ENST00000353176 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| intron-3CDS | ENST00000295530 | ENST00000372592 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| intron-3CDS | ENST00000295530 | ENST00000421063 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| intron-3CDS | ENST00000368428 | ENST00000308941 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| intron-3CDS | ENST00000368428 | ENST00000353176 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| intron-3CDS | ENST00000368428 | ENST00000372592 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| intron-3CDS | ENST00000368428 | ENST00000421063 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| intron-3CDS | ENST00000405236 | ENST00000308941 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| intron-3CDS | ENST00000405236 | ENST00000353176 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| intron-3CDS | ENST00000405236 | ENST00000372592 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| intron-3CDS | ENST00000405236 | ENST00000421063 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| intron-intron | ENST00000295530 | ENST00000487504 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| intron-intron | ENST00000368428 | ENST00000487504 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
| intron-intron | ENST00000405236 | ENST00000487504 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000368433 | FLAD1 | chr1 | 154956542 | + | ENST00000372592 | PHYHD1 | chr9 | 131696060 | + | 1723 | 715 | 343 | 1398 | 351 |
| ENST00000368433 | FLAD1 | chr1 | 154956542 | + | ENST00000308941 | PHYHD1 | chr9 | 131696060 | + | 1702 | 715 | 343 | 1416 | 357 |
| ENST00000368433 | FLAD1 | chr1 | 154956542 | + | ENST00000353176 | PHYHD1 | chr9 | 131696060 | + | 1661 | 715 | 343 | 1335 | 330 |
| ENST00000368433 | FLAD1 | chr1 | 154956542 | + | ENST00000421063 | PHYHD1 | chr9 | 131696060 | + | 1664 | 715 | 343 | 1335 | 330 |
| ENST00000315144 | FLAD1 | chr1 | 154956542 | + | ENST00000372592 | PHYHD1 | chr9 | 131696060 | + | 1272 | 264 | 60 | 947 | 295 |
| ENST00000315144 | FLAD1 | chr1 | 154956542 | + | ENST00000308941 | PHYHD1 | chr9 | 131696060 | + | 1251 | 264 | 60 | 965 | 301 |
| ENST00000315144 | FLAD1 | chr1 | 154956542 | + | ENST00000353176 | PHYHD1 | chr9 | 131696060 | + | 1210 | 264 | 60 | 884 | 274 |
| ENST00000315144 | FLAD1 | chr1 | 154956542 | + | ENST00000421063 | PHYHD1 | chr9 | 131696060 | + | 1213 | 264 | 60 | 884 | 274 |
| ENST00000368432 | FLAD1 | chr1 | 154956542 | + | ENST00000372592 | PHYHD1 | chr9 | 131696060 | + | 1266 | 258 | 54 | 941 | 295 |
| ENST00000368432 | FLAD1 | chr1 | 154956542 | + | ENST00000308941 | PHYHD1 | chr9 | 131696060 | + | 1245 | 258 | 54 | 959 | 301 |
| ENST00000368432 | FLAD1 | chr1 | 154956542 | + | ENST00000353176 | PHYHD1 | chr9 | 131696060 | + | 1204 | 258 | 54 | 878 | 274 |
| ENST00000368432 | FLAD1 | chr1 | 154956542 | + | ENST00000421063 | PHYHD1 | chr9 | 131696060 | + | 1207 | 258 | 54 | 878 | 274 |
| ENST00000292180 | FLAD1 | chr1 | 154956542 | + | ENST00000372592 | PHYHD1 | chr9 | 131696060 | + | 1702 | 694 | 322 | 1377 | 351 |
| ENST00000292180 | FLAD1 | chr1 | 154956542 | + | ENST00000308941 | PHYHD1 | chr9 | 131696060 | + | 1681 | 694 | 322 | 1395 | 357 |
| ENST00000292180 | FLAD1 | chr1 | 154956542 | + | ENST00000353176 | PHYHD1 | chr9 | 131696060 | + | 1640 | 694 | 322 | 1314 | 330 |
| ENST00000292180 | FLAD1 | chr1 | 154956542 | + | ENST00000421063 | PHYHD1 | chr9 | 131696060 | + | 1643 | 694 | 322 | 1314 | 330 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000368433 | ENST00000372592 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 0.20273376 | 0.79726624 |
| ENST00000368433 | ENST00000308941 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 0.30325285 | 0.6967472 |
| ENST00000368433 | ENST00000353176 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 0.26847664 | 0.73152333 |
| ENST00000368433 | ENST00000421063 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 0.26824844 | 0.73175156 |
| ENST00000315144 | ENST00000372592 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 0.046080675 | 0.9539193 |
| ENST00000315144 | ENST00000308941 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 0.41246137 | 0.58753866 |
| ENST00000315144 | ENST00000353176 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 0.053087417 | 0.94691265 |
| ENST00000315144 | ENST00000421063 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 0.053532094 | 0.94646794 |
| ENST00000368432 | ENST00000372592 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 0.045858294 | 0.95414174 |
| ENST00000368432 | ENST00000308941 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 0.44574976 | 0.5542503 |
| ENST00000368432 | ENST00000353176 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 0.059311137 | 0.94068885 |
| ENST00000368432 | ENST00000421063 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 0.059879735 | 0.94012034 |
| ENST00000292180 | ENST00000372592 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 0.19985308 | 0.800147 |
| ENST00000292180 | ENST00000308941 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 0.30084047 | 0.69915956 |
| ENST00000292180 | ENST00000353176 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 0.265522 | 0.73447794 |
| ENST00000292180 | ENST00000421063 | FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 0.2672852 | 0.73271483 |
Top |
Fusion Genomic Features for FLAD1-PHYHD1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 1.27E-08 | 1 |
| FLAD1 | chr1 | 154956542 | + | PHYHD1 | chr9 | 131696060 | + | 1.27E-08 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
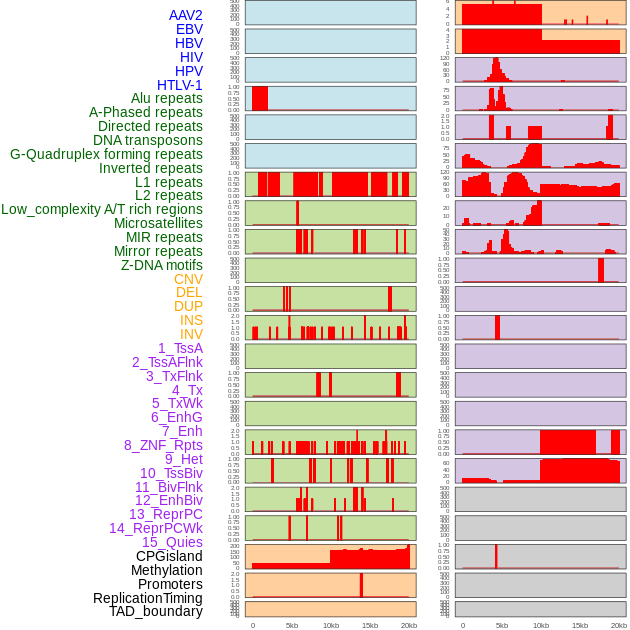 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
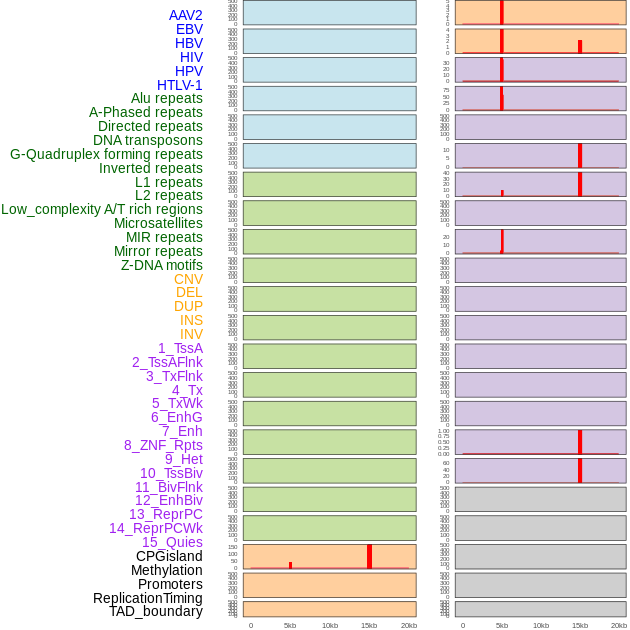 |
Top |
Fusion Protein Features for FLAD1-PHYHD1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:154956542/chr9:131696060) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FLAD1 | . |
| FUNCTION: Catalyzes the adenylation of flavin mononucleotide (FMN) to form flavin adenine dinucleotide (FAD) coenzyme. {ECO:0000269|PubMed:16643857, ECO:0000269|PubMed:27259049}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | PHYHD1 | chr1:154956542 | chr9:131696060 | ENST00000308941 | 3 | 12 | 156_158 | 64 | 298.0 | Region | Alpha-ketoglutarate binding | |
| Tgene | PHYHD1 | chr1:154956542 | chr9:131696060 | ENST00000353176 | 3 | 12 | 156_158 | 64 | 271.0 | Region | Alpha-ketoglutarate binding | |
| Tgene | PHYHD1 | chr1:154956542 | chr9:131696060 | ENST00000372592 | 3 | 13 | 156_158 | 64 | 292.0 | Region | Alpha-ketoglutarate binding | |
| Tgene | PHYHD1 | chr1:154956542 | chr9:131696060 | ENST00000421063 | 1 | 10 | 156_158 | 64 | 271.0 | Region | Alpha-ketoglutarate binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FLAD1 | chr1:154956542 | chr9:131696060 | ENST00000292180 | + | 1 | 7 | 114_205 | 124 | 588.0 | Region | Note=Molybdenum cofactor biosynthesis protein-like |
| Hgene | FLAD1 | chr1:154956542 | chr9:131696060 | ENST00000292180 | + | 1 | 7 | 398_555 | 124 | 588.0 | Region | Note=FAD synthase |
| Hgene | FLAD1 | chr1:154956542 | chr9:131696060 | ENST00000315144 | + | 2 | 8 | 114_205 | 27 | 491.0 | Region | Note=Molybdenum cofactor biosynthesis protein-like |
| Hgene | FLAD1 | chr1:154956542 | chr9:131696060 | ENST00000315144 | + | 2 | 8 | 398_555 | 27 | 491.0 | Region | Note=FAD synthase |
| Hgene | FLAD1 | chr1:154956542 | chr9:131696060 | ENST00000368431 | + | 2 | 3 | 114_205 | 0 | 295.0 | Region | Note=Molybdenum cofactor biosynthesis protein-like |
| Hgene | FLAD1 | chr1:154956542 | chr9:131696060 | ENST00000368431 | + | 2 | 3 | 398_555 | 0 | 295.0 | Region | Note=FAD synthase |
| Hgene | FLAD1 | chr1:154956542 | chr9:131696060 | ENST00000368432 | + | 2 | 7 | 114_205 | 27 | 447.0 | Region | Note=Molybdenum cofactor biosynthesis protein-like |
| Hgene | FLAD1 | chr1:154956542 | chr9:131696060 | ENST00000368432 | + | 2 | 7 | 398_555 | 27 | 447.0 | Region | Note=FAD synthase |
| Hgene | FLAD1 | chr1:154956542 | chr9:131696060 | ENST00000405236 | + | 1 | 4 | 114_205 | 0 | 135.66666666666666 | Region | Note=Molybdenum cofactor biosynthesis protein-like |
| Hgene | FLAD1 | chr1:154956542 | chr9:131696060 | ENST00000405236 | + | 1 | 4 | 398_555 | 0 | 135.66666666666666 | Region | Note=FAD synthase |
Top |
Fusion Gene Sequence for FLAD1-PHYHD1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >30553_30553_1_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000292180_PHYHD1_chr9_131696060_ENST00000308941_length(transcript)=1681nt_BP=694nt GCCGGTGAAGGGGCAGAACAGGCAGGTGAGAGTCTAAGAGGGCTCAGTAATCTGAAGCTTGGTGGGAAGAAGGGATTCTGGGCTAGAAAG GGTGCAGAAGCCTGAAGTAGAAAGAGACGGGATTTTGGTCCGGGGTGGAGAGCGAATGCATTGAAAAGGGCCAAGGCCCAGGATAAGGTA GACATTTAAAGGGGTACGGATGCCCAAGGTAGAGCAGACACTTGAGGAGACCAGCTCAGCAAACGGAAGACACTTAAAGTGGTAGGTTCT CAAGAGAGAAGAAGTTTTTAAGACTAGAGCTAAGCAAGACATTTAAAAGGACATGGGTTGGGATTTGGGAACACGTTTATTCCAGAGGCA GGAACAAAGGAGTCGCTTGTCAAGGATCTGGTTAGAGAAGACTAGGGTCTTCCTCGAAGGAAGCACGCGCACGCCTGCTCTCCCCCATTG TCTTTTCTGGCTTCTCCAGGTTCCCTCGACCCAGGACCCCCTGTTCCCAGGCTATGGCCCCCAGTGCCCTGTAGACCTGGCAGGCCCCCC GTGCTTGCGACCCCTATTTGGGGGTCTGGGTGGCTACTGGAGGGCCTTGCAGAGGGGCAGAGAAGGCAGGACCATGACATCTAGGGCCTC TGAACTTTCTCCGGGGCGCAGCGTGACGGCTGGCATCATCATTGTTGGAGATGAGATCCTTAAGGGCAGCACAGACTATTTCTTGAGCAG TGGTGACAAGATTCGATTCTTCTTTGAGAAAGGCGTTTTTGATGAGAAAGGAAATTTCCTGGTCCCTCCGGAGAAATCCATCAACAAAAT TGGCCACGCTCTGCACGCCCACGACCCCGTCTTCAAGAGCATCACACACTCCTTCAAGGTGCAGACCTTGGCCAGAAGTCTGGGCCTCCA GATGCCCGTGGTGGTGCAGAGCATGTACATCTTTAAGTCTCCCCTCATCAGGACGCCTCCTTCCTGTACACGGAGCCCCTGGGCCGGGTG CTGGGCGTGTGGATCGCAGTGGAGGATGCCACGCTGGAGAACGGCTGTCTCTGGTTCATCCCTGGCTCCCACACCAGTGGTGTGTCAAGA AGGATGGTCCGGGCCCCTGTTGGCTCAGCGCCTGGTACCAGCTTCCTTGGGTCAGAGCCAGCCCGGGATAACAGCCTCTTTGTGCCCACC CCAGTGCAGAGAGGGGCCCTGGTCCTCATCCATGGAGAAGTGGTACACAAGAGCAAGCAGAACCTCTCTGACCGCTCGCGCCAGGCCTAC ACTTTCCACCTCATGGAGGCCTCTGGCACCACCTGGAGCCCGGAGAACTGGCTCCAGCCAACAGCTGAACTGCCCTTTCCCCAACTGTAC ACCTAAAGGCTCTCGCAGGGCAGGAGCCCTCGCCCCTCCCGGGTGAAGCTGTGGGCTGTAAACACCAGTGCCTTGCTCAGCCTCCTGGTT GCAACAGGGAGGTCTTGTCTCCCCTCCTGGGCTTTCCTCCTGCCCTGTGGGCAGCAGCCTAGGCTGGGTCAGGGGCTTCCCTAAGATCTT CACCTCTCTGCCTCCCTACTGCCCCAACATAGCCTTGAGGAGGCTTCTCAGCCACCAAAGGGTTCTGGCCCCTTCTCACTCTCCTCTCCT >30553_30553_1_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000292180_PHYHD1_chr9_131696060_ENST00000308941_length(amino acids)=357AA_BP=124 MGWDLGTRLFQRQEQRSRLSRIWLEKTRVFLEGSTRTPALPHCLFWLLQVPSTQDPLFPGYGPQCPVDLAGPPCLRPLFGGLGGYWRALQ RGREGRTMTSRASELSPGRSVTAGIIIVGDEILKGSTDYFLSSGDKIRFFFEKGVFDEKGNFLVPPEKSINKIGHALHAHDPVFKSITHS FKVQTLARSLGLQMPVVVQSMYIFKSPLIRTPPSCTRSPWAGCWACGSQWRMPRWRTAVSGSSLAPTPVVCQEGWSGPLLAQRLVPASLG -------------------------------------------------------------- >30553_30553_2_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000292180_PHYHD1_chr9_131696060_ENST00000353176_length(transcript)=1640nt_BP=694nt GCCGGTGAAGGGGCAGAACAGGCAGGTGAGAGTCTAAGAGGGCTCAGTAATCTGAAGCTTGGTGGGAAGAAGGGATTCTGGGCTAGAAAG GGTGCAGAAGCCTGAAGTAGAAAGAGACGGGATTTTGGTCCGGGGTGGAGAGCGAATGCATTGAAAAGGGCCAAGGCCCAGGATAAGGTA GACATTTAAAGGGGTACGGATGCCCAAGGTAGAGCAGACACTTGAGGAGACCAGCTCAGCAAACGGAAGACACTTAAAGTGGTAGGTTCT CAAGAGAGAAGAAGTTTTTAAGACTAGAGCTAAGCAAGACATTTAAAAGGACATGGGTTGGGATTTGGGAACACGTTTATTCCAGAGGCA GGAACAAAGGAGTCGCTTGTCAAGGATCTGGTTAGAGAAGACTAGGGTCTTCCTCGAAGGAAGCACGCGCACGCCTGCTCTCCCCCATTG TCTTTTCTGGCTTCTCCAGGTTCCCTCGACCCAGGACCCCCTGTTCCCAGGCTATGGCCCCCAGTGCCCTGTAGACCTGGCAGGCCCCCC GTGCTTGCGACCCCTATTTGGGGGTCTGGGTGGCTACTGGAGGGCCTTGCAGAGGGGCAGAGAAGGCAGGACCATGACATCTAGGGCCTC TGAACTTTCTCCGGGGCGCAGCGTGACGGCTGGCATCATCATTGTTGGAGATGAGATCCTTAAGGGCAGCACAGACTATTTCTTGAGCAG TGGTGACAAGATTCGATTCTTCTTTGAGAAAGGCGTTTTTGATGAGAAAGGAAATTTCCTGGTCCCTCCGGAGAAATCCATCAACAAAAT TGGCCACGCTCTGCACGCCCACGACCCCGTCTTCAAGAGCATCACACACTCCTTCAAGGTGCAGCAACCTCACTTTGGCGGTGAAGTCTC CCCTCATCAGGACGCCTCCTTCCTGTACACGGAGCCCCTGGGCCGGGTGCTGGGCGTGTGGATCGCAGTGGAGGATGCCACGCTGGAGAA CGGCTGTCTCTGGTTCATCCCTGGCTCCCACACCAGTGGTGTGTCAAGAAGGATGGTCCGGGCCCCTGTTGGCTCAGCGCCTGGTACCAG CTTCCTTGGGTCAGAGCCAGCCCGGGATAACAGCCTCTTTGTGCCCACCCCAGTGCAGAGAGGGGCCCTGGTCCTCATCCATGGAGAAGT GGTACACAAGAGCAAGCAGAACCTCTCTGACCGCTCGCGCCAGGCCTACACTTTCCACCTCATGGAGGCCTCTGGCACCACCTGGAGCCC GGAGAACTGGCTCCAGCCAACAGCTGAACTGCCCTTTCCCCAACTGTACACCTAAAGGCTCTCGCAGGGCAGGAGCCCTCGCCCCTCCCG GGTGAAGCTGTGGGCTGTAAACACCAGTGCCTTGCTCAGCCTCCTGGTTGCAACAGGGAGGTCTTGTCTCCCCTCCTGGGCTTTCCTCCT GCCCTGTGGGCAGCAGCCTAGGCTGGGTCAGGGGCTTCCCTAAGATCTTCACCTCTCTGCCTCCCTACTGCCCCAACATAGCCTTGAGGA GGCTTCTCAGCCACCAAAGGGTTCTGGCCCCTTCTCACTCTCCTCTCCTCTCAGATGGAACTCTGGTTATTATGGTGTTAGTTATCGAAT >30553_30553_2_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000292180_PHYHD1_chr9_131696060_ENST00000353176_length(amino acids)=330AA_BP=124 MGWDLGTRLFQRQEQRSRLSRIWLEKTRVFLEGSTRTPALPHCLFWLLQVPSTQDPLFPGYGPQCPVDLAGPPCLRPLFGGLGGYWRALQ RGREGRTMTSRASELSPGRSVTAGIIIVGDEILKGSTDYFLSSGDKIRFFFEKGVFDEKGNFLVPPEKSINKIGHALHAHDPVFKSITHS FKVQQPHFGGEVSPHQDASFLYTEPLGRVLGVWIAVEDATLENGCLWFIPGSHTSGVSRRMVRAPVGSAPGTSFLGSEPARDNSLFVPTP -------------------------------------------------------------- >30553_30553_3_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000292180_PHYHD1_chr9_131696060_ENST00000372592_length(transcript)=1702nt_BP=694nt GCCGGTGAAGGGGCAGAACAGGCAGGTGAGAGTCTAAGAGGGCTCAGTAATCTGAAGCTTGGTGGGAAGAAGGGATTCTGGGCTAGAAAG GGTGCAGAAGCCTGAAGTAGAAAGAGACGGGATTTTGGTCCGGGGTGGAGAGCGAATGCATTGAAAAGGGCCAAGGCCCAGGATAAGGTA GACATTTAAAGGGGTACGGATGCCCAAGGTAGAGCAGACACTTGAGGAGACCAGCTCAGCAAACGGAAGACACTTAAAGTGGTAGGTTCT CAAGAGAGAAGAAGTTTTTAAGACTAGAGCTAAGCAAGACATTTAAAAGGACATGGGTTGGGATTTGGGAACACGTTTATTCCAGAGGCA GGAACAAAGGAGTCGCTTGTCAAGGATCTGGTTAGAGAAGACTAGGGTCTTCCTCGAAGGAAGCACGCGCACGCCTGCTCTCCCCCATTG TCTTTTCTGGCTTCTCCAGGTTCCCTCGACCCAGGACCCCCTGTTCCCAGGCTATGGCCCCCAGTGCCCTGTAGACCTGGCAGGCCCCCC GTGCTTGCGACCCCTATTTGGGGGTCTGGGTGGCTACTGGAGGGCCTTGCAGAGGGGCAGAGAAGGCAGGACCATGACATCTAGGGCCTC TGAACTTTCTCCGGGGCGCAGCGTGACGGCTGGCATCATCATTGTTGGAGATGAGATCCTTAAGGGCAGCACAGACTATTTCTTGAGCAG TGGTGACAAGATTCGATTCTTCTTTGAGAAAGGCGTTTTTGATGAGAAAGGAAATTTCCTGGTCCCTCCGGAGAAATCCATCAACAAAAT TGGCCACGCTCTGCACGCCCACGACCCCGTCTTCAAGAGCATCACACACTCCTTCAAGGTGCAGACCTTGGCCAGAAGTCTGGGCCTCCA GATGCCCGTGGTGGTGCAGAGCATGTACATCTTTAAGCAACCTCACTTTGGCGGTGAAGTCTCCCCTCATCAGGACGCCTCCTTCCTGTA CACGGAGCCCCTGGGCCGGGTGCTGGGCGTGTGGATCGCAGTGGAGGATGCCACGCTGGAGAACGGCTGTCTCTGGTTCATCCCTGGCTC CCACACCAGTGGTGTGTCAAGAAGGATGGTCCGGGCCCCTGTTGGCTCAGCGCCTGGTACCAGCTTCCTTGGGTCAGAGCCAGCCCGGGA TAACAGCCTCTTTGTGCCCACCCCAGTGCAGAGAGGGGCCCTGGTCCTCATCCATGGAGAAGTGGTACACAAGAGCAAGCAGAACCTCTC TGACCGCTCGCGCCAGGCCTACACTTTCCACCTCATGGAGGCCTCTGGCACCACCTGGAGCCCGGAGAACTGGCTCCAGCCAACAGCTGA ACTGCCCTTTCCCCAACTGTACACCTAAAGGCTCTCGCAGGGCAGGAGCCCTCGCCCCTCCCGGGTGAAGCTGTGGGCTGTAAACACCAG TGCCTTGCTCAGCCTCCTGGTTGCAACAGGGAGGTCTTGTCTCCCCTCCTGGGCTTTCCTCCTGCCCTGTGGGCAGCAGCCTAGGCTGGG TCAGGGGCTTCCCTAAGATCTTCACCTCTCTGCCTCCCTACTGCCCCAACATAGCCTTGAGGAGGCTTCTCAGCCACCAAAGGGTTCTGG >30553_30553_3_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000292180_PHYHD1_chr9_131696060_ENST00000372592_length(amino acids)=351AA_BP=124 MGWDLGTRLFQRQEQRSRLSRIWLEKTRVFLEGSTRTPALPHCLFWLLQVPSTQDPLFPGYGPQCPVDLAGPPCLRPLFGGLGGYWRALQ RGREGRTMTSRASELSPGRSVTAGIIIVGDEILKGSTDYFLSSGDKIRFFFEKGVFDEKGNFLVPPEKSINKIGHALHAHDPVFKSITHS FKVQTLARSLGLQMPVVVQSMYIFKQPHFGGEVSPHQDASFLYTEPLGRVLGVWIAVEDATLENGCLWFIPGSHTSGVSRRMVRAPVGSA -------------------------------------------------------------- >30553_30553_4_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000292180_PHYHD1_chr9_131696060_ENST00000421063_length(transcript)=1643nt_BP=694nt GCCGGTGAAGGGGCAGAACAGGCAGGTGAGAGTCTAAGAGGGCTCAGTAATCTGAAGCTTGGTGGGAAGAAGGGATTCTGGGCTAGAAAG GGTGCAGAAGCCTGAAGTAGAAAGAGACGGGATTTTGGTCCGGGGTGGAGAGCGAATGCATTGAAAAGGGCCAAGGCCCAGGATAAGGTA GACATTTAAAGGGGTACGGATGCCCAAGGTAGAGCAGACACTTGAGGAGACCAGCTCAGCAAACGGAAGACACTTAAAGTGGTAGGTTCT CAAGAGAGAAGAAGTTTTTAAGACTAGAGCTAAGCAAGACATTTAAAAGGACATGGGTTGGGATTTGGGAACACGTTTATTCCAGAGGCA GGAACAAAGGAGTCGCTTGTCAAGGATCTGGTTAGAGAAGACTAGGGTCTTCCTCGAAGGAAGCACGCGCACGCCTGCTCTCCCCCATTG TCTTTTCTGGCTTCTCCAGGTTCCCTCGACCCAGGACCCCCTGTTCCCAGGCTATGGCCCCCAGTGCCCTGTAGACCTGGCAGGCCCCCC GTGCTTGCGACCCCTATTTGGGGGTCTGGGTGGCTACTGGAGGGCCTTGCAGAGGGGCAGAGAAGGCAGGACCATGACATCTAGGGCCTC TGAACTTTCTCCGGGGCGCAGCGTGACGGCTGGCATCATCATTGTTGGAGATGAGATCCTTAAGGGCAGCACAGACTATTTCTTGAGCAG TGGTGACAAGATTCGATTCTTCTTTGAGAAAGGCGTTTTTGATGAGAAAGGAAATTTCCTGGTCCCTCCGGAGAAATCCATCAACAAAAT TGGCCACGCTCTGCACGCCCACGACCCCGTCTTCAAGAGCATCACACACTCCTTCAAGGTGCAGCAACCTCACTTTGGCGGTGAAGTCTC CCCTCATCAGGACGCCTCCTTCCTGTACACGGAGCCCCTGGGCCGGGTGCTGGGCGTGTGGATCGCAGTGGAGGATGCCACGCTGGAGAA CGGCTGTCTCTGGTTCATCCCTGGCTCCCACACCAGTGGTGTGTCAAGAAGGATGGTCCGGGCCCCTGTTGGCTCAGCGCCTGGTACCAG CTTCCTTGGGTCAGAGCCAGCCCGGGATAACAGCCTCTTTGTGCCCACCCCAGTGCAGAGAGGGGCCCTGGTCCTCATCCATGGAGAAGT GGTACACAAGAGCAAGCAGAACCTCTCTGACCGCTCGCGCCAGGCCTACACTTTCCACCTCATGGAGGCCTCTGGCACCACCTGGAGCCC GGAGAACTGGCTCCAGCCAACAGCTGAACTGCCCTTTCCCCAACTGTACACCTAAAGGCTCTCGCAGGGCAGGAGCCCTCGCCCCTCCCG GGTGAAGCTGTGGGCTGTAAACACCAGTGCCTTGCTCAGCCTCCTGGTTGCAACAGGGAGGTCTTGTCTCCCCTCCTGGGCTTTCCTCCT GCCCTGTGGGCAGCAGCCTAGGCTGGGTCAGGGGCTTCCCTAAGATCTTCACCTCTCTGCCTCCCTACTGCCCCAACATAGCCTTGAGGA GGCTTCTCAGCCACCAAAGGGTTCTGGCCCCTTCTCACTCTCCTCTCCTCTCAGATGGAACTCTGGTTATTATGGTGTTAGTTATCGAAT >30553_30553_4_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000292180_PHYHD1_chr9_131696060_ENST00000421063_length(amino acids)=330AA_BP=124 MGWDLGTRLFQRQEQRSRLSRIWLEKTRVFLEGSTRTPALPHCLFWLLQVPSTQDPLFPGYGPQCPVDLAGPPCLRPLFGGLGGYWRALQ RGREGRTMTSRASELSPGRSVTAGIIIVGDEILKGSTDYFLSSGDKIRFFFEKGVFDEKGNFLVPPEKSINKIGHALHAHDPVFKSITHS FKVQQPHFGGEVSPHQDASFLYTEPLGRVLGVWIAVEDATLENGCLWFIPGSHTSGVSRRMVRAPVGSAPGTSFLGSEPARDNSLFVPTP -------------------------------------------------------------- >30553_30553_5_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000315144_PHYHD1_chr9_131696060_ENST00000308941_length(transcript)=1251nt_BP=264nt GAACAAGGGAAAGAGCCGGTGAAGGGGCAGAACAGGCAGGTTCCCTCGACCCAGGACCCCCTGTTCCCAGGCTATGGCCCCCAGTGCCCT GTAGACCTGGCAGGCCCCCCGTGCTTGCGACCCCTATTTGGGGGTCTGGGTGGCTACTGGAGGGCCTTGCAGAGGGGCAGAGAAGGCAGG ACCATGACATCTAGGGCCTCTGAACTTTCTCCGGGGCGCAGCGTGACGGCTGGCATCATCATTGTTGGAGATGAGATCCTTAAGGGCAGC ACAGACTATTTCTTGAGCAGTGGTGACAAGATTCGATTCTTCTTTGAGAAAGGCGTTTTTGATGAGAAAGGAAATTTCCTGGTCCCTCCG GAGAAATCCATCAACAAAATTGGCCACGCTCTGCACGCCCACGACCCCGTCTTCAAGAGCATCACACACTCCTTCAAGGTGCAGACCTTG GCCAGAAGTCTGGGCCTCCAGATGCCCGTGGTGGTGCAGAGCATGTACATCTTTAAGTCTCCCCTCATCAGGACGCCTCCTTCCTGTACA CGGAGCCCCTGGGCCGGGTGCTGGGCGTGTGGATCGCAGTGGAGGATGCCACGCTGGAGAACGGCTGTCTCTGGTTCATCCCTGGCTCCC ACACCAGTGGTGTGTCAAGAAGGATGGTCCGGGCCCCTGTTGGCTCAGCGCCTGGTACCAGCTTCCTTGGGTCAGAGCCAGCCCGGGATA ACAGCCTCTTTGTGCCCACCCCAGTGCAGAGAGGGGCCCTGGTCCTCATCCATGGAGAAGTGGTACACAAGAGCAAGCAGAACCTCTCTG ACCGCTCGCGCCAGGCCTACACTTTCCACCTCATGGAGGCCTCTGGCACCACCTGGAGCCCGGAGAACTGGCTCCAGCCAACAGCTGAAC TGCCCTTTCCCCAACTGTACACCTAAAGGCTCTCGCAGGGCAGGAGCCCTCGCCCCTCCCGGGTGAAGCTGTGGGCTGTAAACACCAGTG CCTTGCTCAGCCTCCTGGTTGCAACAGGGAGGTCTTGTCTCCCCTCCTGGGCTTTCCTCCTGCCCTGTGGGCAGCAGCCTAGGCTGGGTC AGGGGCTTCCCTAAGATCTTCACCTCTCTGCCTCCCTACTGCCCCAACATAGCCTTGAGGAGGCTTCTCAGCCACCAAAGGGTTCTGGCC >30553_30553_5_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000315144_PHYHD1_chr9_131696060_ENST00000308941_length(amino acids)=301AA_BP=68 MFPGYGPQCPVDLAGPPCLRPLFGGLGGYWRALQRGREGRTMTSRASELSPGRSVTAGIIIVGDEILKGSTDYFLSSGDKIRFFFEKGVF DEKGNFLVPPEKSINKIGHALHAHDPVFKSITHSFKVQTLARSLGLQMPVVVQSMYIFKSPLIRTPPSCTRSPWAGCWACGSQWRMPRWR TAVSGSSLAPTPVVCQEGWSGPLLAQRLVPASLGQSQPGITASLCPPQCREGPWSSSMEKWYTRASRTSLTARARPTLSTSWRPLAPPGA -------------------------------------------------------------- >30553_30553_6_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000315144_PHYHD1_chr9_131696060_ENST00000353176_length(transcript)=1210nt_BP=264nt GAACAAGGGAAAGAGCCGGTGAAGGGGCAGAACAGGCAGGTTCCCTCGACCCAGGACCCCCTGTTCCCAGGCTATGGCCCCCAGTGCCCT GTAGACCTGGCAGGCCCCCCGTGCTTGCGACCCCTATTTGGGGGTCTGGGTGGCTACTGGAGGGCCTTGCAGAGGGGCAGAGAAGGCAGG ACCATGACATCTAGGGCCTCTGAACTTTCTCCGGGGCGCAGCGTGACGGCTGGCATCATCATTGTTGGAGATGAGATCCTTAAGGGCAGC ACAGACTATTTCTTGAGCAGTGGTGACAAGATTCGATTCTTCTTTGAGAAAGGCGTTTTTGATGAGAAAGGAAATTTCCTGGTCCCTCCG GAGAAATCCATCAACAAAATTGGCCACGCTCTGCACGCCCACGACCCCGTCTTCAAGAGCATCACACACTCCTTCAAGGTGCAGCAACCT CACTTTGGCGGTGAAGTCTCCCCTCATCAGGACGCCTCCTTCCTGTACACGGAGCCCCTGGGCCGGGTGCTGGGCGTGTGGATCGCAGTG GAGGATGCCACGCTGGAGAACGGCTGTCTCTGGTTCATCCCTGGCTCCCACACCAGTGGTGTGTCAAGAAGGATGGTCCGGGCCCCTGTT GGCTCAGCGCCTGGTACCAGCTTCCTTGGGTCAGAGCCAGCCCGGGATAACAGCCTCTTTGTGCCCACCCCAGTGCAGAGAGGGGCCCTG GTCCTCATCCATGGAGAAGTGGTACACAAGAGCAAGCAGAACCTCTCTGACCGCTCGCGCCAGGCCTACACTTTCCACCTCATGGAGGCC TCTGGCACCACCTGGAGCCCGGAGAACTGGCTCCAGCCAACAGCTGAACTGCCCTTTCCCCAACTGTACACCTAAAGGCTCTCGCAGGGC AGGAGCCCTCGCCCCTCCCGGGTGAAGCTGTGGGCTGTAAACACCAGTGCCTTGCTCAGCCTCCTGGTTGCAACAGGGAGGTCTTGTCTC CCCTCCTGGGCTTTCCTCCTGCCCTGTGGGCAGCAGCCTAGGCTGGGTCAGGGGCTTCCCTAAGATCTTCACCTCTCTGCCTCCCTACTG CCCCAACATAGCCTTGAGGAGGCTTCTCAGCCACCAAAGGGTTCTGGCCCCTTCTCACTCTCCTCTCCTCTCAGATGGAACTCTGGTTAT >30553_30553_6_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000315144_PHYHD1_chr9_131696060_ENST00000353176_length(amino acids)=274AA_BP=68 MFPGYGPQCPVDLAGPPCLRPLFGGLGGYWRALQRGREGRTMTSRASELSPGRSVTAGIIIVGDEILKGSTDYFLSSGDKIRFFFEKGVF DEKGNFLVPPEKSINKIGHALHAHDPVFKSITHSFKVQQPHFGGEVSPHQDASFLYTEPLGRVLGVWIAVEDATLENGCLWFIPGSHTSG VSRRMVRAPVGSAPGTSFLGSEPARDNSLFVPTPVQRGALVLIHGEVVHKSKQNLSDRSRQAYTFHLMEASGTTWSPENWLQPTAELPFP -------------------------------------------------------------- >30553_30553_7_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000315144_PHYHD1_chr9_131696060_ENST00000372592_length(transcript)=1272nt_BP=264nt GAACAAGGGAAAGAGCCGGTGAAGGGGCAGAACAGGCAGGTTCCCTCGACCCAGGACCCCCTGTTCCCAGGCTATGGCCCCCAGTGCCCT GTAGACCTGGCAGGCCCCCCGTGCTTGCGACCCCTATTTGGGGGTCTGGGTGGCTACTGGAGGGCCTTGCAGAGGGGCAGAGAAGGCAGG ACCATGACATCTAGGGCCTCTGAACTTTCTCCGGGGCGCAGCGTGACGGCTGGCATCATCATTGTTGGAGATGAGATCCTTAAGGGCAGC ACAGACTATTTCTTGAGCAGTGGTGACAAGATTCGATTCTTCTTTGAGAAAGGCGTTTTTGATGAGAAAGGAAATTTCCTGGTCCCTCCG GAGAAATCCATCAACAAAATTGGCCACGCTCTGCACGCCCACGACCCCGTCTTCAAGAGCATCACACACTCCTTCAAGGTGCAGACCTTG GCCAGAAGTCTGGGCCTCCAGATGCCCGTGGTGGTGCAGAGCATGTACATCTTTAAGCAACCTCACTTTGGCGGTGAAGTCTCCCCTCAT CAGGACGCCTCCTTCCTGTACACGGAGCCCCTGGGCCGGGTGCTGGGCGTGTGGATCGCAGTGGAGGATGCCACGCTGGAGAACGGCTGT CTCTGGTTCATCCCTGGCTCCCACACCAGTGGTGTGTCAAGAAGGATGGTCCGGGCCCCTGTTGGCTCAGCGCCTGGTACCAGCTTCCTT GGGTCAGAGCCAGCCCGGGATAACAGCCTCTTTGTGCCCACCCCAGTGCAGAGAGGGGCCCTGGTCCTCATCCATGGAGAAGTGGTACAC AAGAGCAAGCAGAACCTCTCTGACCGCTCGCGCCAGGCCTACACTTTCCACCTCATGGAGGCCTCTGGCACCACCTGGAGCCCGGAGAAC TGGCTCCAGCCAACAGCTGAACTGCCCTTTCCCCAACTGTACACCTAAAGGCTCTCGCAGGGCAGGAGCCCTCGCCCCTCCCGGGTGAAG CTGTGGGCTGTAAACACCAGTGCCTTGCTCAGCCTCCTGGTTGCAACAGGGAGGTCTTGTCTCCCCTCCTGGGCTTTCCTCCTGCCCTGT GGGCAGCAGCCTAGGCTGGGTCAGGGGCTTCCCTAAGATCTTCACCTCTCTGCCTCCCTACTGCCCCAACATAGCCTTGAGGAGGCTTCT CAGCCACCAAAGGGTTCTGGCCCCTTCTCACTCTCCTCTCCTCTCAGATGGAACTCTGGTTATTATGGTGTTAGTTATCGAATAAAAACG >30553_30553_7_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000315144_PHYHD1_chr9_131696060_ENST00000372592_length(amino acids)=295AA_BP=68 MFPGYGPQCPVDLAGPPCLRPLFGGLGGYWRALQRGREGRTMTSRASELSPGRSVTAGIIIVGDEILKGSTDYFLSSGDKIRFFFEKGVF DEKGNFLVPPEKSINKIGHALHAHDPVFKSITHSFKVQTLARSLGLQMPVVVQSMYIFKQPHFGGEVSPHQDASFLYTEPLGRVLGVWIA VEDATLENGCLWFIPGSHTSGVSRRMVRAPVGSAPGTSFLGSEPARDNSLFVPTPVQRGALVLIHGEVVHKSKQNLSDRSRQAYTFHLME -------------------------------------------------------------- >30553_30553_8_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000315144_PHYHD1_chr9_131696060_ENST00000421063_length(transcript)=1213nt_BP=264nt GAACAAGGGAAAGAGCCGGTGAAGGGGCAGAACAGGCAGGTTCCCTCGACCCAGGACCCCCTGTTCCCAGGCTATGGCCCCCAGTGCCCT GTAGACCTGGCAGGCCCCCCGTGCTTGCGACCCCTATTTGGGGGTCTGGGTGGCTACTGGAGGGCCTTGCAGAGGGGCAGAGAAGGCAGG ACCATGACATCTAGGGCCTCTGAACTTTCTCCGGGGCGCAGCGTGACGGCTGGCATCATCATTGTTGGAGATGAGATCCTTAAGGGCAGC ACAGACTATTTCTTGAGCAGTGGTGACAAGATTCGATTCTTCTTTGAGAAAGGCGTTTTTGATGAGAAAGGAAATTTCCTGGTCCCTCCG GAGAAATCCATCAACAAAATTGGCCACGCTCTGCACGCCCACGACCCCGTCTTCAAGAGCATCACACACTCCTTCAAGGTGCAGCAACCT CACTTTGGCGGTGAAGTCTCCCCTCATCAGGACGCCTCCTTCCTGTACACGGAGCCCCTGGGCCGGGTGCTGGGCGTGTGGATCGCAGTG GAGGATGCCACGCTGGAGAACGGCTGTCTCTGGTTCATCCCTGGCTCCCACACCAGTGGTGTGTCAAGAAGGATGGTCCGGGCCCCTGTT GGCTCAGCGCCTGGTACCAGCTTCCTTGGGTCAGAGCCAGCCCGGGATAACAGCCTCTTTGTGCCCACCCCAGTGCAGAGAGGGGCCCTG GTCCTCATCCATGGAGAAGTGGTACACAAGAGCAAGCAGAACCTCTCTGACCGCTCGCGCCAGGCCTACACTTTCCACCTCATGGAGGCC TCTGGCACCACCTGGAGCCCGGAGAACTGGCTCCAGCCAACAGCTGAACTGCCCTTTCCCCAACTGTACACCTAAAGGCTCTCGCAGGGC AGGAGCCCTCGCCCCTCCCGGGTGAAGCTGTGGGCTGTAAACACCAGTGCCTTGCTCAGCCTCCTGGTTGCAACAGGGAGGTCTTGTCTC CCCTCCTGGGCTTTCCTCCTGCCCTGTGGGCAGCAGCCTAGGCTGGGTCAGGGGCTTCCCTAAGATCTTCACCTCTCTGCCTCCCTACTG CCCCAACATAGCCTTGAGGAGGCTTCTCAGCCACCAAAGGGTTCTGGCCCCTTCTCACTCTCCTCTCCTCTCAGATGGAACTCTGGTTAT >30553_30553_8_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000315144_PHYHD1_chr9_131696060_ENST00000421063_length(amino acids)=274AA_BP=68 MFPGYGPQCPVDLAGPPCLRPLFGGLGGYWRALQRGREGRTMTSRASELSPGRSVTAGIIIVGDEILKGSTDYFLSSGDKIRFFFEKGVF DEKGNFLVPPEKSINKIGHALHAHDPVFKSITHSFKVQQPHFGGEVSPHQDASFLYTEPLGRVLGVWIAVEDATLENGCLWFIPGSHTSG VSRRMVRAPVGSAPGTSFLGSEPARDNSLFVPTPVQRGALVLIHGEVVHKSKQNLSDRSRQAYTFHLMEASGTTWSPENWLQPTAELPFP -------------------------------------------------------------- >30553_30553_9_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000368432_PHYHD1_chr9_131696060_ENST00000308941_length(transcript)=1245nt_BP=258nt GGGAAAGAGCCGGTGAAGGGGCAGAACAGGCAGGTTCCCTCGACCCAGGACCCCCTGTTCCCAGGCTATGGCCCCCAGTGCCCTGTAGAC CTGGCAGGCCCCCCGTGCTTGCGACCCCTATTTGGGGGTCTGGGTGGCTACTGGAGGGCCTTGCAGAGGGGCAGAGAAGGCAGGACCATG ACATCTAGGGCCTCTGAACTTTCTCCGGGGCGCAGCGTGACGGCTGGCATCATCATTGTTGGAGATGAGATCCTTAAGGGCAGCACAGAC TATTTCTTGAGCAGTGGTGACAAGATTCGATTCTTCTTTGAGAAAGGCGTTTTTGATGAGAAAGGAAATTTCCTGGTCCCTCCGGAGAAA TCCATCAACAAAATTGGCCACGCTCTGCACGCCCACGACCCCGTCTTCAAGAGCATCACACACTCCTTCAAGGTGCAGACCTTGGCCAGA AGTCTGGGCCTCCAGATGCCCGTGGTGGTGCAGAGCATGTACATCTTTAAGTCTCCCCTCATCAGGACGCCTCCTTCCTGTACACGGAGC CCCTGGGCCGGGTGCTGGGCGTGTGGATCGCAGTGGAGGATGCCACGCTGGAGAACGGCTGTCTCTGGTTCATCCCTGGCTCCCACACCA GTGGTGTGTCAAGAAGGATGGTCCGGGCCCCTGTTGGCTCAGCGCCTGGTACCAGCTTCCTTGGGTCAGAGCCAGCCCGGGATAACAGCC TCTTTGTGCCCACCCCAGTGCAGAGAGGGGCCCTGGTCCTCATCCATGGAGAAGTGGTACACAAGAGCAAGCAGAACCTCTCTGACCGCT CGCGCCAGGCCTACACTTTCCACCTCATGGAGGCCTCTGGCACCACCTGGAGCCCGGAGAACTGGCTCCAGCCAACAGCTGAACTGCCCT TTCCCCAACTGTACACCTAAAGGCTCTCGCAGGGCAGGAGCCCTCGCCCCTCCCGGGTGAAGCTGTGGGCTGTAAACACCAGTGCCTTGC TCAGCCTCCTGGTTGCAACAGGGAGGTCTTGTCTCCCCTCCTGGGCTTTCCTCCTGCCCTGTGGGCAGCAGCCTAGGCTGGGTCAGGGGC TTCCCTAAGATCTTCACCTCTCTGCCTCCCTACTGCCCCAACATAGCCTTGAGGAGGCTTCTCAGCCACCAAAGGGTTCTGGCCCCTTCT >30553_30553_9_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000368432_PHYHD1_chr9_131696060_ENST00000308941_length(amino acids)=301AA_BP=68 MFPGYGPQCPVDLAGPPCLRPLFGGLGGYWRALQRGREGRTMTSRASELSPGRSVTAGIIIVGDEILKGSTDYFLSSGDKIRFFFEKGVF DEKGNFLVPPEKSINKIGHALHAHDPVFKSITHSFKVQTLARSLGLQMPVVVQSMYIFKSPLIRTPPSCTRSPWAGCWACGSQWRMPRWR TAVSGSSLAPTPVVCQEGWSGPLLAQRLVPASLGQSQPGITASLCPPQCREGPWSSSMEKWYTRASRTSLTARARPTLSTSWRPLAPPGA -------------------------------------------------------------- >30553_30553_10_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000368432_PHYHD1_chr9_131696060_ENST00000353176_length(transcript)=1204nt_BP=258nt GGGAAAGAGCCGGTGAAGGGGCAGAACAGGCAGGTTCCCTCGACCCAGGACCCCCTGTTCCCAGGCTATGGCCCCCAGTGCCCTGTAGAC CTGGCAGGCCCCCCGTGCTTGCGACCCCTATTTGGGGGTCTGGGTGGCTACTGGAGGGCCTTGCAGAGGGGCAGAGAAGGCAGGACCATG ACATCTAGGGCCTCTGAACTTTCTCCGGGGCGCAGCGTGACGGCTGGCATCATCATTGTTGGAGATGAGATCCTTAAGGGCAGCACAGAC TATTTCTTGAGCAGTGGTGACAAGATTCGATTCTTCTTTGAGAAAGGCGTTTTTGATGAGAAAGGAAATTTCCTGGTCCCTCCGGAGAAA TCCATCAACAAAATTGGCCACGCTCTGCACGCCCACGACCCCGTCTTCAAGAGCATCACACACTCCTTCAAGGTGCAGCAACCTCACTTT GGCGGTGAAGTCTCCCCTCATCAGGACGCCTCCTTCCTGTACACGGAGCCCCTGGGCCGGGTGCTGGGCGTGTGGATCGCAGTGGAGGAT GCCACGCTGGAGAACGGCTGTCTCTGGTTCATCCCTGGCTCCCACACCAGTGGTGTGTCAAGAAGGATGGTCCGGGCCCCTGTTGGCTCA GCGCCTGGTACCAGCTTCCTTGGGTCAGAGCCAGCCCGGGATAACAGCCTCTTTGTGCCCACCCCAGTGCAGAGAGGGGCCCTGGTCCTC ATCCATGGAGAAGTGGTACACAAGAGCAAGCAGAACCTCTCTGACCGCTCGCGCCAGGCCTACACTTTCCACCTCATGGAGGCCTCTGGC ACCACCTGGAGCCCGGAGAACTGGCTCCAGCCAACAGCTGAACTGCCCTTTCCCCAACTGTACACCTAAAGGCTCTCGCAGGGCAGGAGC CCTCGCCCCTCCCGGGTGAAGCTGTGGGCTGTAAACACCAGTGCCTTGCTCAGCCTCCTGGTTGCAACAGGGAGGTCTTGTCTCCCCTCC TGGGCTTTCCTCCTGCCCTGTGGGCAGCAGCCTAGGCTGGGTCAGGGGCTTCCCTAAGATCTTCACCTCTCTGCCTCCCTACTGCCCCAA CATAGCCTTGAGGAGGCTTCTCAGCCACCAAAGGGTTCTGGCCCCTTCTCACTCTCCTCTCCTCTCAGATGGAACTCTGGTTATTATGGT >30553_30553_10_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000368432_PHYHD1_chr9_131696060_ENST00000353176_length(amino acids)=274AA_BP=68 MFPGYGPQCPVDLAGPPCLRPLFGGLGGYWRALQRGREGRTMTSRASELSPGRSVTAGIIIVGDEILKGSTDYFLSSGDKIRFFFEKGVF DEKGNFLVPPEKSINKIGHALHAHDPVFKSITHSFKVQQPHFGGEVSPHQDASFLYTEPLGRVLGVWIAVEDATLENGCLWFIPGSHTSG VSRRMVRAPVGSAPGTSFLGSEPARDNSLFVPTPVQRGALVLIHGEVVHKSKQNLSDRSRQAYTFHLMEASGTTWSPENWLQPTAELPFP -------------------------------------------------------------- >30553_30553_11_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000368432_PHYHD1_chr9_131696060_ENST00000372592_length(transcript)=1266nt_BP=258nt GGGAAAGAGCCGGTGAAGGGGCAGAACAGGCAGGTTCCCTCGACCCAGGACCCCCTGTTCCCAGGCTATGGCCCCCAGTGCCCTGTAGAC CTGGCAGGCCCCCCGTGCTTGCGACCCCTATTTGGGGGTCTGGGTGGCTACTGGAGGGCCTTGCAGAGGGGCAGAGAAGGCAGGACCATG ACATCTAGGGCCTCTGAACTTTCTCCGGGGCGCAGCGTGACGGCTGGCATCATCATTGTTGGAGATGAGATCCTTAAGGGCAGCACAGAC TATTTCTTGAGCAGTGGTGACAAGATTCGATTCTTCTTTGAGAAAGGCGTTTTTGATGAGAAAGGAAATTTCCTGGTCCCTCCGGAGAAA TCCATCAACAAAATTGGCCACGCTCTGCACGCCCACGACCCCGTCTTCAAGAGCATCACACACTCCTTCAAGGTGCAGACCTTGGCCAGA AGTCTGGGCCTCCAGATGCCCGTGGTGGTGCAGAGCATGTACATCTTTAAGCAACCTCACTTTGGCGGTGAAGTCTCCCCTCATCAGGAC GCCTCCTTCCTGTACACGGAGCCCCTGGGCCGGGTGCTGGGCGTGTGGATCGCAGTGGAGGATGCCACGCTGGAGAACGGCTGTCTCTGG TTCATCCCTGGCTCCCACACCAGTGGTGTGTCAAGAAGGATGGTCCGGGCCCCTGTTGGCTCAGCGCCTGGTACCAGCTTCCTTGGGTCA GAGCCAGCCCGGGATAACAGCCTCTTTGTGCCCACCCCAGTGCAGAGAGGGGCCCTGGTCCTCATCCATGGAGAAGTGGTACACAAGAGC AAGCAGAACCTCTCTGACCGCTCGCGCCAGGCCTACACTTTCCACCTCATGGAGGCCTCTGGCACCACCTGGAGCCCGGAGAACTGGCTC CAGCCAACAGCTGAACTGCCCTTTCCCCAACTGTACACCTAAAGGCTCTCGCAGGGCAGGAGCCCTCGCCCCTCCCGGGTGAAGCTGTGG GCTGTAAACACCAGTGCCTTGCTCAGCCTCCTGGTTGCAACAGGGAGGTCTTGTCTCCCCTCCTGGGCTTTCCTCCTGCCCTGTGGGCAG CAGCCTAGGCTGGGTCAGGGGCTTCCCTAAGATCTTCACCTCTCTGCCTCCCTACTGCCCCAACATAGCCTTGAGGAGGCTTCTCAGCCA CCAAAGGGTTCTGGCCCCTTCTCACTCTCCTCTCCTCTCAGATGGAACTCTGGTTATTATGGTGTTAGTTATCGAATAAAAACGACTTCA >30553_30553_11_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000368432_PHYHD1_chr9_131696060_ENST00000372592_length(amino acids)=295AA_BP=68 MFPGYGPQCPVDLAGPPCLRPLFGGLGGYWRALQRGREGRTMTSRASELSPGRSVTAGIIIVGDEILKGSTDYFLSSGDKIRFFFEKGVF DEKGNFLVPPEKSINKIGHALHAHDPVFKSITHSFKVQTLARSLGLQMPVVVQSMYIFKQPHFGGEVSPHQDASFLYTEPLGRVLGVWIA VEDATLENGCLWFIPGSHTSGVSRRMVRAPVGSAPGTSFLGSEPARDNSLFVPTPVQRGALVLIHGEVVHKSKQNLSDRSRQAYTFHLME -------------------------------------------------------------- >30553_30553_12_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000368432_PHYHD1_chr9_131696060_ENST00000421063_length(transcript)=1207nt_BP=258nt GGGAAAGAGCCGGTGAAGGGGCAGAACAGGCAGGTTCCCTCGACCCAGGACCCCCTGTTCCCAGGCTATGGCCCCCAGTGCCCTGTAGAC CTGGCAGGCCCCCCGTGCTTGCGACCCCTATTTGGGGGTCTGGGTGGCTACTGGAGGGCCTTGCAGAGGGGCAGAGAAGGCAGGACCATG ACATCTAGGGCCTCTGAACTTTCTCCGGGGCGCAGCGTGACGGCTGGCATCATCATTGTTGGAGATGAGATCCTTAAGGGCAGCACAGAC TATTTCTTGAGCAGTGGTGACAAGATTCGATTCTTCTTTGAGAAAGGCGTTTTTGATGAGAAAGGAAATTTCCTGGTCCCTCCGGAGAAA TCCATCAACAAAATTGGCCACGCTCTGCACGCCCACGACCCCGTCTTCAAGAGCATCACACACTCCTTCAAGGTGCAGCAACCTCACTTT GGCGGTGAAGTCTCCCCTCATCAGGACGCCTCCTTCCTGTACACGGAGCCCCTGGGCCGGGTGCTGGGCGTGTGGATCGCAGTGGAGGAT GCCACGCTGGAGAACGGCTGTCTCTGGTTCATCCCTGGCTCCCACACCAGTGGTGTGTCAAGAAGGATGGTCCGGGCCCCTGTTGGCTCA GCGCCTGGTACCAGCTTCCTTGGGTCAGAGCCAGCCCGGGATAACAGCCTCTTTGTGCCCACCCCAGTGCAGAGAGGGGCCCTGGTCCTC ATCCATGGAGAAGTGGTACACAAGAGCAAGCAGAACCTCTCTGACCGCTCGCGCCAGGCCTACACTTTCCACCTCATGGAGGCCTCTGGC ACCACCTGGAGCCCGGAGAACTGGCTCCAGCCAACAGCTGAACTGCCCTTTCCCCAACTGTACACCTAAAGGCTCTCGCAGGGCAGGAGC CCTCGCCCCTCCCGGGTGAAGCTGTGGGCTGTAAACACCAGTGCCTTGCTCAGCCTCCTGGTTGCAACAGGGAGGTCTTGTCTCCCCTCC TGGGCTTTCCTCCTGCCCTGTGGGCAGCAGCCTAGGCTGGGTCAGGGGCTTCCCTAAGATCTTCACCTCTCTGCCTCCCTACTGCCCCAA CATAGCCTTGAGGAGGCTTCTCAGCCACCAAAGGGTTCTGGCCCCTTCTCACTCTCCTCTCCTCTCAGATGGAACTCTGGTTATTATGGT >30553_30553_12_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000368432_PHYHD1_chr9_131696060_ENST00000421063_length(amino acids)=274AA_BP=68 MFPGYGPQCPVDLAGPPCLRPLFGGLGGYWRALQRGREGRTMTSRASELSPGRSVTAGIIIVGDEILKGSTDYFLSSGDKIRFFFEKGVF DEKGNFLVPPEKSINKIGHALHAHDPVFKSITHSFKVQQPHFGGEVSPHQDASFLYTEPLGRVLGVWIAVEDATLENGCLWFIPGSHTSG VSRRMVRAPVGSAPGTSFLGSEPARDNSLFVPTPVQRGALVLIHGEVVHKSKQNLSDRSRQAYTFHLMEASGTTWSPENWLQPTAELPFP -------------------------------------------------------------- >30553_30553_13_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000368433_PHYHD1_chr9_131696060_ENST00000308941_length(transcript)=1702nt_BP=715nt AGGAAAGGAACAAGGGAAAGAGCCGGTGAAGGGGCAGAACAGGCAGGTGAGAGTCTAAGAGGGCTCAGTAATCTGAAGCTTGGTGGGAAG AAGGGATTCTGGGCTAGAAAGGGTGCAGAAGCCTGAAGTAGAAAGAGACGGGATTTTGGTCCGGGGTGGAGAGCGAATGCATTGAAAAGG GCCAAGGCCCAGGATAAGGTAGACATTTAAAGGGGTACGGATGCCCAAGGTAGAGCAGACACTTGAGGAGACCAGCTCAGCAAACGGAAG ACACTTAAAGTGGTAGGTTCTCAAGAGAGAAGAAGTTTTTAAGACTAGAGCTAAGCAAGACATTTAAAAGGACATGGGTTGGGATTTGGG AACACGTTTATTCCAGAGGCAGGAACAAAGGAGTCGCTTGTCAAGGATCTGGTTAGAGAAGACTAGGGTCTTCCTCGAAGGAAGCACGCG CACGCCTGCTCTCCCCCATTGTCTTTTCTGGCTTCTCCAGGTTCCCTCGACCCAGGACCCCCTGTTCCCAGGCTATGGCCCCCAGTGCCC TGTAGACCTGGCAGGCCCCCCGTGCTTGCGACCCCTATTTGGGGGTCTGGGTGGCTACTGGAGGGCCTTGCAGAGGGGCAGAGAAGGCAG GACCATGACATCTAGGGCCTCTGAACTTTCTCCGGGGCGCAGCGTGACGGCTGGCATCATCATTGTTGGAGATGAGATCCTTAAGGGCAG CACAGACTATTTCTTGAGCAGTGGTGACAAGATTCGATTCTTCTTTGAGAAAGGCGTTTTTGATGAGAAAGGAAATTTCCTGGTCCCTCC GGAGAAATCCATCAACAAAATTGGCCACGCTCTGCACGCCCACGACCCCGTCTTCAAGAGCATCACACACTCCTTCAAGGTGCAGACCTT GGCCAGAAGTCTGGGCCTCCAGATGCCCGTGGTGGTGCAGAGCATGTACATCTTTAAGTCTCCCCTCATCAGGACGCCTCCTTCCTGTAC ACGGAGCCCCTGGGCCGGGTGCTGGGCGTGTGGATCGCAGTGGAGGATGCCACGCTGGAGAACGGCTGTCTCTGGTTCATCCCTGGCTCC CACACCAGTGGTGTGTCAAGAAGGATGGTCCGGGCCCCTGTTGGCTCAGCGCCTGGTACCAGCTTCCTTGGGTCAGAGCCAGCCCGGGAT AACAGCCTCTTTGTGCCCACCCCAGTGCAGAGAGGGGCCCTGGTCCTCATCCATGGAGAAGTGGTACACAAGAGCAAGCAGAACCTCTCT GACCGCTCGCGCCAGGCCTACACTTTCCACCTCATGGAGGCCTCTGGCACCACCTGGAGCCCGGAGAACTGGCTCCAGCCAACAGCTGAA CTGCCCTTTCCCCAACTGTACACCTAAAGGCTCTCGCAGGGCAGGAGCCCTCGCCCCTCCCGGGTGAAGCTGTGGGCTGTAAACACCAGT GCCTTGCTCAGCCTCCTGGTTGCAACAGGGAGGTCTTGTCTCCCCTCCTGGGCTTTCCTCCTGCCCTGTGGGCAGCAGCCTAGGCTGGGT CAGGGGCTTCCCTAAGATCTTCACCTCTCTGCCTCCCTACTGCCCCAACATAGCCTTGAGGAGGCTTCTCAGCCACCAAAGGGTTCTGGC >30553_30553_13_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000368433_PHYHD1_chr9_131696060_ENST00000308941_length(amino acids)=357AA_BP=124 MGWDLGTRLFQRQEQRSRLSRIWLEKTRVFLEGSTRTPALPHCLFWLLQVPSTQDPLFPGYGPQCPVDLAGPPCLRPLFGGLGGYWRALQ RGREGRTMTSRASELSPGRSVTAGIIIVGDEILKGSTDYFLSSGDKIRFFFEKGVFDEKGNFLVPPEKSINKIGHALHAHDPVFKSITHS FKVQTLARSLGLQMPVVVQSMYIFKSPLIRTPPSCTRSPWAGCWACGSQWRMPRWRTAVSGSSLAPTPVVCQEGWSGPLLAQRLVPASLG -------------------------------------------------------------- >30553_30553_14_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000368433_PHYHD1_chr9_131696060_ENST00000353176_length(transcript)=1661nt_BP=715nt AGGAAAGGAACAAGGGAAAGAGCCGGTGAAGGGGCAGAACAGGCAGGTGAGAGTCTAAGAGGGCTCAGTAATCTGAAGCTTGGTGGGAAG AAGGGATTCTGGGCTAGAAAGGGTGCAGAAGCCTGAAGTAGAAAGAGACGGGATTTTGGTCCGGGGTGGAGAGCGAATGCATTGAAAAGG GCCAAGGCCCAGGATAAGGTAGACATTTAAAGGGGTACGGATGCCCAAGGTAGAGCAGACACTTGAGGAGACCAGCTCAGCAAACGGAAG ACACTTAAAGTGGTAGGTTCTCAAGAGAGAAGAAGTTTTTAAGACTAGAGCTAAGCAAGACATTTAAAAGGACATGGGTTGGGATTTGGG AACACGTTTATTCCAGAGGCAGGAACAAAGGAGTCGCTTGTCAAGGATCTGGTTAGAGAAGACTAGGGTCTTCCTCGAAGGAAGCACGCG CACGCCTGCTCTCCCCCATTGTCTTTTCTGGCTTCTCCAGGTTCCCTCGACCCAGGACCCCCTGTTCCCAGGCTATGGCCCCCAGTGCCC TGTAGACCTGGCAGGCCCCCCGTGCTTGCGACCCCTATTTGGGGGTCTGGGTGGCTACTGGAGGGCCTTGCAGAGGGGCAGAGAAGGCAG GACCATGACATCTAGGGCCTCTGAACTTTCTCCGGGGCGCAGCGTGACGGCTGGCATCATCATTGTTGGAGATGAGATCCTTAAGGGCAG CACAGACTATTTCTTGAGCAGTGGTGACAAGATTCGATTCTTCTTTGAGAAAGGCGTTTTTGATGAGAAAGGAAATTTCCTGGTCCCTCC GGAGAAATCCATCAACAAAATTGGCCACGCTCTGCACGCCCACGACCCCGTCTTCAAGAGCATCACACACTCCTTCAAGGTGCAGCAACC TCACTTTGGCGGTGAAGTCTCCCCTCATCAGGACGCCTCCTTCCTGTACACGGAGCCCCTGGGCCGGGTGCTGGGCGTGTGGATCGCAGT GGAGGATGCCACGCTGGAGAACGGCTGTCTCTGGTTCATCCCTGGCTCCCACACCAGTGGTGTGTCAAGAAGGATGGTCCGGGCCCCTGT TGGCTCAGCGCCTGGTACCAGCTTCCTTGGGTCAGAGCCAGCCCGGGATAACAGCCTCTTTGTGCCCACCCCAGTGCAGAGAGGGGCCCT GGTCCTCATCCATGGAGAAGTGGTACACAAGAGCAAGCAGAACCTCTCTGACCGCTCGCGCCAGGCCTACACTTTCCACCTCATGGAGGC CTCTGGCACCACCTGGAGCCCGGAGAACTGGCTCCAGCCAACAGCTGAACTGCCCTTTCCCCAACTGTACACCTAAAGGCTCTCGCAGGG CAGGAGCCCTCGCCCCTCCCGGGTGAAGCTGTGGGCTGTAAACACCAGTGCCTTGCTCAGCCTCCTGGTTGCAACAGGGAGGTCTTGTCT CCCCTCCTGGGCTTTCCTCCTGCCCTGTGGGCAGCAGCCTAGGCTGGGTCAGGGGCTTCCCTAAGATCTTCACCTCTCTGCCTCCCTACT GCCCCAACATAGCCTTGAGGAGGCTTCTCAGCCACCAAAGGGTTCTGGCCCCTTCTCACTCTCCTCTCCTCTCAGATGGAACTCTGGTTA >30553_30553_14_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000368433_PHYHD1_chr9_131696060_ENST00000353176_length(amino acids)=330AA_BP=124 MGWDLGTRLFQRQEQRSRLSRIWLEKTRVFLEGSTRTPALPHCLFWLLQVPSTQDPLFPGYGPQCPVDLAGPPCLRPLFGGLGGYWRALQ RGREGRTMTSRASELSPGRSVTAGIIIVGDEILKGSTDYFLSSGDKIRFFFEKGVFDEKGNFLVPPEKSINKIGHALHAHDPVFKSITHS FKVQQPHFGGEVSPHQDASFLYTEPLGRVLGVWIAVEDATLENGCLWFIPGSHTSGVSRRMVRAPVGSAPGTSFLGSEPARDNSLFVPTP -------------------------------------------------------------- >30553_30553_15_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000368433_PHYHD1_chr9_131696060_ENST00000372592_length(transcript)=1723nt_BP=715nt AGGAAAGGAACAAGGGAAAGAGCCGGTGAAGGGGCAGAACAGGCAGGTGAGAGTCTAAGAGGGCTCAGTAATCTGAAGCTTGGTGGGAAG AAGGGATTCTGGGCTAGAAAGGGTGCAGAAGCCTGAAGTAGAAAGAGACGGGATTTTGGTCCGGGGTGGAGAGCGAATGCATTGAAAAGG GCCAAGGCCCAGGATAAGGTAGACATTTAAAGGGGTACGGATGCCCAAGGTAGAGCAGACACTTGAGGAGACCAGCTCAGCAAACGGAAG ACACTTAAAGTGGTAGGTTCTCAAGAGAGAAGAAGTTTTTAAGACTAGAGCTAAGCAAGACATTTAAAAGGACATGGGTTGGGATTTGGG AACACGTTTATTCCAGAGGCAGGAACAAAGGAGTCGCTTGTCAAGGATCTGGTTAGAGAAGACTAGGGTCTTCCTCGAAGGAAGCACGCG CACGCCTGCTCTCCCCCATTGTCTTTTCTGGCTTCTCCAGGTTCCCTCGACCCAGGACCCCCTGTTCCCAGGCTATGGCCCCCAGTGCCC TGTAGACCTGGCAGGCCCCCCGTGCTTGCGACCCCTATTTGGGGGTCTGGGTGGCTACTGGAGGGCCTTGCAGAGGGGCAGAGAAGGCAG GACCATGACATCTAGGGCCTCTGAACTTTCTCCGGGGCGCAGCGTGACGGCTGGCATCATCATTGTTGGAGATGAGATCCTTAAGGGCAG CACAGACTATTTCTTGAGCAGTGGTGACAAGATTCGATTCTTCTTTGAGAAAGGCGTTTTTGATGAGAAAGGAAATTTCCTGGTCCCTCC GGAGAAATCCATCAACAAAATTGGCCACGCTCTGCACGCCCACGACCCCGTCTTCAAGAGCATCACACACTCCTTCAAGGTGCAGACCTT GGCCAGAAGTCTGGGCCTCCAGATGCCCGTGGTGGTGCAGAGCATGTACATCTTTAAGCAACCTCACTTTGGCGGTGAAGTCTCCCCTCA TCAGGACGCCTCCTTCCTGTACACGGAGCCCCTGGGCCGGGTGCTGGGCGTGTGGATCGCAGTGGAGGATGCCACGCTGGAGAACGGCTG TCTCTGGTTCATCCCTGGCTCCCACACCAGTGGTGTGTCAAGAAGGATGGTCCGGGCCCCTGTTGGCTCAGCGCCTGGTACCAGCTTCCT TGGGTCAGAGCCAGCCCGGGATAACAGCCTCTTTGTGCCCACCCCAGTGCAGAGAGGGGCCCTGGTCCTCATCCATGGAGAAGTGGTACA CAAGAGCAAGCAGAACCTCTCTGACCGCTCGCGCCAGGCCTACACTTTCCACCTCATGGAGGCCTCTGGCACCACCTGGAGCCCGGAGAA CTGGCTCCAGCCAACAGCTGAACTGCCCTTTCCCCAACTGTACACCTAAAGGCTCTCGCAGGGCAGGAGCCCTCGCCCCTCCCGGGTGAA GCTGTGGGCTGTAAACACCAGTGCCTTGCTCAGCCTCCTGGTTGCAACAGGGAGGTCTTGTCTCCCCTCCTGGGCTTTCCTCCTGCCCTG TGGGCAGCAGCCTAGGCTGGGTCAGGGGCTTCCCTAAGATCTTCACCTCTCTGCCTCCCTACTGCCCCAACATAGCCTTGAGGAGGCTTC TCAGCCACCAAAGGGTTCTGGCCCCTTCTCACTCTCCTCTCCTCTCAGATGGAACTCTGGTTATTATGGTGTTAGTTATCGAATAAAAAC >30553_30553_15_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000368433_PHYHD1_chr9_131696060_ENST00000372592_length(amino acids)=351AA_BP=124 MGWDLGTRLFQRQEQRSRLSRIWLEKTRVFLEGSTRTPALPHCLFWLLQVPSTQDPLFPGYGPQCPVDLAGPPCLRPLFGGLGGYWRALQ RGREGRTMTSRASELSPGRSVTAGIIIVGDEILKGSTDYFLSSGDKIRFFFEKGVFDEKGNFLVPPEKSINKIGHALHAHDPVFKSITHS FKVQTLARSLGLQMPVVVQSMYIFKQPHFGGEVSPHQDASFLYTEPLGRVLGVWIAVEDATLENGCLWFIPGSHTSGVSRRMVRAPVGSA -------------------------------------------------------------- >30553_30553_16_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000368433_PHYHD1_chr9_131696060_ENST00000421063_length(transcript)=1664nt_BP=715nt AGGAAAGGAACAAGGGAAAGAGCCGGTGAAGGGGCAGAACAGGCAGGTGAGAGTCTAAGAGGGCTCAGTAATCTGAAGCTTGGTGGGAAG AAGGGATTCTGGGCTAGAAAGGGTGCAGAAGCCTGAAGTAGAAAGAGACGGGATTTTGGTCCGGGGTGGAGAGCGAATGCATTGAAAAGG GCCAAGGCCCAGGATAAGGTAGACATTTAAAGGGGTACGGATGCCCAAGGTAGAGCAGACACTTGAGGAGACCAGCTCAGCAAACGGAAG ACACTTAAAGTGGTAGGTTCTCAAGAGAGAAGAAGTTTTTAAGACTAGAGCTAAGCAAGACATTTAAAAGGACATGGGTTGGGATTTGGG AACACGTTTATTCCAGAGGCAGGAACAAAGGAGTCGCTTGTCAAGGATCTGGTTAGAGAAGACTAGGGTCTTCCTCGAAGGAAGCACGCG CACGCCTGCTCTCCCCCATTGTCTTTTCTGGCTTCTCCAGGTTCCCTCGACCCAGGACCCCCTGTTCCCAGGCTATGGCCCCCAGTGCCC TGTAGACCTGGCAGGCCCCCCGTGCTTGCGACCCCTATTTGGGGGTCTGGGTGGCTACTGGAGGGCCTTGCAGAGGGGCAGAGAAGGCAG GACCATGACATCTAGGGCCTCTGAACTTTCTCCGGGGCGCAGCGTGACGGCTGGCATCATCATTGTTGGAGATGAGATCCTTAAGGGCAG CACAGACTATTTCTTGAGCAGTGGTGACAAGATTCGATTCTTCTTTGAGAAAGGCGTTTTTGATGAGAAAGGAAATTTCCTGGTCCCTCC GGAGAAATCCATCAACAAAATTGGCCACGCTCTGCACGCCCACGACCCCGTCTTCAAGAGCATCACACACTCCTTCAAGGTGCAGCAACC TCACTTTGGCGGTGAAGTCTCCCCTCATCAGGACGCCTCCTTCCTGTACACGGAGCCCCTGGGCCGGGTGCTGGGCGTGTGGATCGCAGT GGAGGATGCCACGCTGGAGAACGGCTGTCTCTGGTTCATCCCTGGCTCCCACACCAGTGGTGTGTCAAGAAGGATGGTCCGGGCCCCTGT TGGCTCAGCGCCTGGTACCAGCTTCCTTGGGTCAGAGCCAGCCCGGGATAACAGCCTCTTTGTGCCCACCCCAGTGCAGAGAGGGGCCCT GGTCCTCATCCATGGAGAAGTGGTACACAAGAGCAAGCAGAACCTCTCTGACCGCTCGCGCCAGGCCTACACTTTCCACCTCATGGAGGC CTCTGGCACCACCTGGAGCCCGGAGAACTGGCTCCAGCCAACAGCTGAACTGCCCTTTCCCCAACTGTACACCTAAAGGCTCTCGCAGGG CAGGAGCCCTCGCCCCTCCCGGGTGAAGCTGTGGGCTGTAAACACCAGTGCCTTGCTCAGCCTCCTGGTTGCAACAGGGAGGTCTTGTCT CCCCTCCTGGGCTTTCCTCCTGCCCTGTGGGCAGCAGCCTAGGCTGGGTCAGGGGCTTCCCTAAGATCTTCACCTCTCTGCCTCCCTACT GCCCCAACATAGCCTTGAGGAGGCTTCTCAGCCACCAAAGGGTTCTGGCCCCTTCTCACTCTCCTCTCCTCTCAGATGGAACTCTGGTTA >30553_30553_16_FLAD1-PHYHD1_FLAD1_chr1_154956542_ENST00000368433_PHYHD1_chr9_131696060_ENST00000421063_length(amino acids)=330AA_BP=124 MGWDLGTRLFQRQEQRSRLSRIWLEKTRVFLEGSTRTPALPHCLFWLLQVPSTQDPLFPGYGPQCPVDLAGPPCLRPLFGGLGGYWRALQ RGREGRTMTSRASELSPGRSVTAGIIIVGDEILKGSTDYFLSSGDKIRFFFEKGVFDEKGNFLVPPEKSINKIGHALHAHDPVFKSITHS FKVQQPHFGGEVSPHQDASFLYTEPLGRVLGVWIAVEDATLENGCLWFIPGSHTSGVSRRMVRAPVGSAPGTSFLGSEPARDNSLFVPTP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FLAD1-PHYHD1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FLAD1-PHYHD1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FLAD1-PHYHD1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
