
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FOXO1-PDGFRB (FusionGDB2 ID:31166) |
Fusion Gene Summary for FOXO1-PDGFRB |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FOXO1-PDGFRB | Fusion gene ID: 31166 | Hgene | Tgene | Gene symbol | FOXO1 | PDGFRB | Gene ID | 2308 | 5159 |
| Gene name | forkhead box O1 | platelet derived growth factor receptor beta | |
| Synonyms | FKH1|FKHR|FOXO1A | CD140B|IBGC4|IMF1|JTK12|KOGS|PDGFR|PDGFR-1|PDGFR1|PENTT | |
| Cytomap | 13q14.11 | 5q32 | |
| Type of gene | protein-coding | protein-coding | |
| Description | forkhead box protein O1forkhead box protein O1Aforkhead, Drosophila, homolog of, in rhabdomyosarcoma | platelet-derived growth factor receptor betaActivated tyrosine kinase PDGFRBCD140 antigen-like family member BNDEL1-PDGFRBPDGF-R-betaPDGFR-betabeta-type platelet-derived growth factor receptorplatelet-derived growth factor receptor 1platelet-deriv | |
| Modification date | 20200322 | 20200329 | |
| UniProtAcc | Q12778 | P09619 | |
| Ensembl transtripts involved in fusion gene | ENST00000379561, ENST00000473775, | ENST00000523456, ENST00000261799, | |
| Fusion gene scores | * DoF score | 13 X 12 X 9=1404 | 28 X 26 X 6=4368 |
| # samples | 16 | 15 | |
| ** MAII score | log2(16/1404*10)=-3.1333991254172 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(15/4368*10)=-4.86393845042397 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FOXO1 [Title/Abstract] AND PDGFRB [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FOXO1(41239720)-PDGFRB(149499686), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | FOXO1 | GO:0009267 | cellular response to starvation | 20543840 |
| Hgene | FOXO1 | GO:0032873 | negative regulation of stress-activated MAPK cascade | 19696738 |
| Hgene | FOXO1 | GO:0043066 | negative regulation of apoptotic process | 10871843 |
| Hgene | FOXO1 | GO:0045893 | positive regulation of transcription, DNA-templated | 7862145|10871843|12228231 |
| Hgene | FOXO1 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 10871843|12228231 |
| Hgene | FOXO1 | GO:0071455 | cellular response to hyperoxia | 20543840 |
| Tgene | PDGFRB | GO:0007165 | signal transduction | 10821867 |
| Tgene | PDGFRB | GO:0010863 | positive regulation of phospholipase C activity | 1653029 |
| Tgene | PDGFRB | GO:0018108 | peptidyl-tyrosine phosphorylation | 1653029|2536956|2850496 |
| Tgene | PDGFRB | GO:0030335 | positive regulation of cell migration | 17470632 |
| Tgene | PDGFRB | GO:0032516 | positive regulation of phosphoprotein phosphatase activity | 7691811 |
| Tgene | PDGFRB | GO:0038091 | positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway | 17470632 |
| Tgene | PDGFRB | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity | 1314164 |
| Tgene | PDGFRB | GO:0046777 | protein autophosphorylation | 1314164|2536956|2850496 |
| Tgene | PDGFRB | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 1314164|2536956 |
| Tgene | PDGFRB | GO:0060326 | cell chemotaxis | 2554309|17991872 |
 Fusion gene breakpoints across FOXO1 (5'-gene) Fusion gene breakpoints across FOXO1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
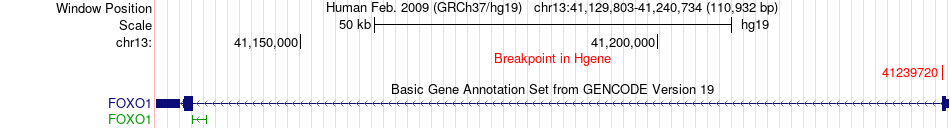 |
 Fusion gene breakpoints across PDGFRB (3'-gene) Fusion gene breakpoints across PDGFRB (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
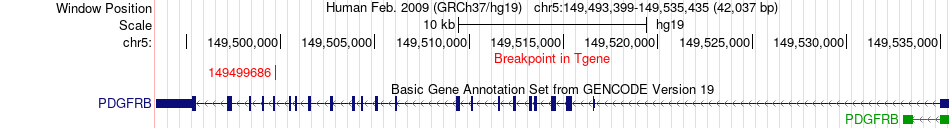 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | KICH | TCGA-KM-8443-01A | FOXO1 | chr13 | 41239720 | - | PDGFRB | chr5 | 149499686 | - |
Top |
Fusion Gene ORF analysis for FOXO1-PDGFRB |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000379561 | ENST00000523456 | FOXO1 | chr13 | 41239720 | - | PDGFRB | chr5 | 149499686 | - |
| In-frame | ENST00000379561 | ENST00000261799 | FOXO1 | chr13 | 41239720 | - | PDGFRB | chr5 | 149499686 | - |
| intron-3CDS | ENST00000473775 | ENST00000261799 | FOXO1 | chr13 | 41239720 | - | PDGFRB | chr5 | 149499686 | - |
| intron-intron | ENST00000473775 | ENST00000523456 | FOXO1 | chr13 | 41239720 | - | PDGFRB | chr5 | 149499686 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000379561 | FOXO1 | chr13 | 41239720 | - | ENST00000261799 | PDGFRB | chr5 | 149499686 | - | 3676 | 1015 | 43 | 1749 | 568 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000379561 | ENST00000261799 | FOXO1 | chr13 | 41239720 | - | PDGFRB | chr5 | 149499686 | - | 0.015304945 | 0.984695 |
Top |
Fusion Genomic Features for FOXO1-PDGFRB |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
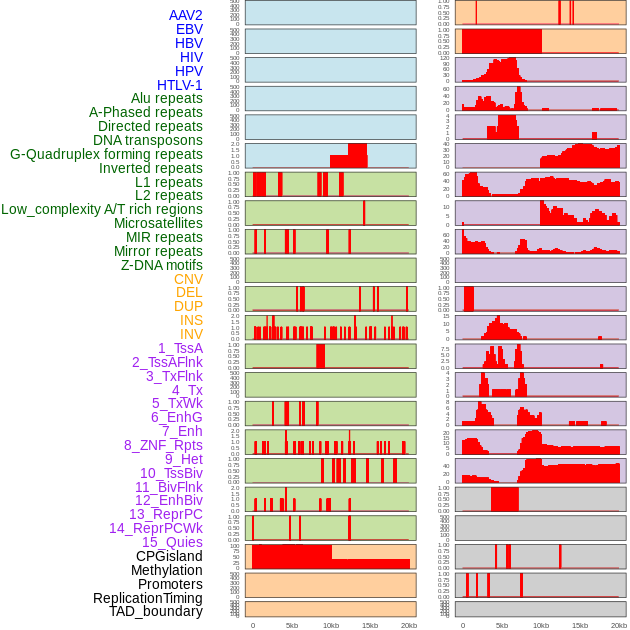 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for FOXO1-PDGFRB |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr13:41239720/chr5:149499686) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FOXO1 | PDGFRB |
| FUNCTION: Transcription factor that is the main target of insulin signaling and regulates metabolic homeostasis in response to oxidative stress (PubMed:10358076, PubMed:12228231, PubMed:15220471, PubMed:15890677, PubMed:18356527, PubMed:19221179, PubMed:20543840, PubMed:21245099). Binds to the insulin response element (IRE) with consensus sequence 5'-TT[G/A]TTTTG-3' and the related Daf-16 family binding element (DBE) with consensus sequence 5'-TT[G/A]TTTAC-3' (PubMed:10358076). Activity suppressed by insulin (PubMed:10358076). Main regulator of redox balance and osteoblast numbers and controls bone mass (By similarity). Orchestrates the endocrine function of the skeleton in regulating glucose metabolism (By similarity). Also acts as a key regulator of chondrogenic commitment of skeletal progenitor cells in response to lipid availability: when lipids levels are low, translocates to the nucleus and promotes expression of SOX9, which induces chondrogenic commitment and suppresses fatty acid oxidation (By similarity). Acts synergistically with ATF4 to suppress osteocalcin/BGLAP activity, increasing glucose levels and triggering glucose intolerance and insulin insensitivity (By similarity). Also suppresses the transcriptional activity of RUNX2, an upstream activator of osteocalcin/BGLAP (By similarity). In hepatocytes, promotes gluconeogenesis by acting together with PPARGC1A and CEBPA to activate the expression of genes such as IGFBP1, G6PC1 and PCK1 (By similarity). Important regulator of cell death acting downstream of CDK1, PKB/AKT1 and STK4/MST1 (PubMed:18356527, PubMed:19221179). Promotes neural cell death (PubMed:18356527). Mediates insulin action on adipose tissue (By similarity). Regulates the expression of adipogenic genes such as PPARG during preadipocyte differentiation and, adipocyte size and adipose tissue-specific gene expression in response to excessive calorie intake (By similarity). Regulates the transcriptional activity of GADD45A and repair of nitric oxide-damaged DNA in beta-cells (By similarity). Required for the autophagic cell death induction in response to starvation or oxidative stress in a transcription-independent manner (PubMed:20543840). Mediates the function of MLIP in cardiomyocytes hypertrophy and cardiac remodeling (By similarity). Regulates endothelial cell (EC) viability and apoptosis in a PPIA/CYPA-dependent manner via transcription of CCL2 and BCL2L11 which are involved in EC chemotaxis and apoptosis (PubMed:31063815). {ECO:0000250|UniProtKB:A4L7N3, ECO:0000250|UniProtKB:G3V7R4, ECO:0000250|UniProtKB:Q9R1E0, ECO:0000269|PubMed:10358076, ECO:0000269|PubMed:12228231, ECO:0000269|PubMed:15220471, ECO:0000269|PubMed:15890677, ECO:0000269|PubMed:18356527, ECO:0000269|PubMed:19221179, ECO:0000269|PubMed:20543840, ECO:0000269|PubMed:21245099, ECO:0000269|PubMed:31063815}. | FUNCTION: Tyrosine-protein kinase that acts as cell-surface receptor for homodimeric PDGFB and PDGFD and for heterodimers formed by PDGFA and PDGFB, and plays an essential role in the regulation of embryonic development, cell proliferation, survival, differentiation, chemotaxis and migration. Plays an essential role in blood vessel development by promoting proliferation, migration and recruitment of pericytes and smooth muscle cells to endothelial cells. Plays a role in the migration of vascular smooth muscle cells and the formation of neointima at vascular injury sites. Required for normal development of the cardiovascular system. Required for normal recruitment of pericytes (mesangial cells) in the kidney glomerulus, and for normal formation of a branched network of capillaries in kidney glomeruli. Promotes rearrangement of the actin cytoskeleton and the formation of membrane ruffles. Binding of its cognate ligands - homodimeric PDGFB, heterodimers formed by PDGFA and PDGFB or homodimeric PDGFD -leads to the activation of several signaling cascades; the response depends on the nature of the bound ligand and is modulated by the formation of heterodimers between PDGFRA and PDGFRB. Phosphorylates PLCG1, PIK3R1, PTPN11, RASA1/GAP, CBL, SHC1 and NCK1. Activation of PLCG1 leads to the production of the cellular signaling molecules diacylglycerol and inositol 1,4,5-trisphosphate, mobilization of cytosolic Ca(2+) and the activation of protein kinase C. Phosphorylation of PIK3R1, the regulatory subunit of phosphatidylinositol 3-kinase, leads to the activation of the AKT1 signaling pathway. Phosphorylation of SHC1, or of the C-terminus of PTPN11, creates a binding site for GRB2, resulting in the activation of HRAS, RAF1 and down-stream MAP kinases, including MAPK1/ERK2 and/or MAPK3/ERK1. Promotes phosphorylation and activation of SRC family kinases. Promotes phosphorylation of PDCD6IP/ALIX and STAM. Receptor signaling is down-regulated by protein phosphatases that dephosphorylate the receptor and its down-stream effectors, and by rapid internalization of the activated receptor. {ECO:0000269|PubMed:11297552, ECO:0000269|PubMed:11331881, ECO:0000269|PubMed:1314164, ECO:0000269|PubMed:1396585, ECO:0000269|PubMed:1653029, ECO:0000269|PubMed:1709159, ECO:0000269|PubMed:1846866, ECO:0000269|PubMed:20494825, ECO:0000269|PubMed:20529858, ECO:0000269|PubMed:21098708, ECO:0000269|PubMed:21679854, ECO:0000269|PubMed:21733313, ECO:0000269|PubMed:2554309, ECO:0000269|PubMed:26599395, ECO:0000269|PubMed:2835772, ECO:0000269|PubMed:2850496, ECO:0000269|PubMed:7685273, ECO:0000269|PubMed:7691811, ECO:0000269|PubMed:7692233, ECO:0000269|PubMed:8195171}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FOXO1 | chr13:41239720 | chr5:149499686 | ENST00000379561 | - | 1 | 3 | 120_130 | 210 | 1440.3333333333333 | Compositional bias | Note=Poly-Pro |
| Hgene | FOXO1 | chr13:41239720 | chr5:149499686 | ENST00000379561 | - | 1 | 3 | 152_155 | 210 | 1440.3333333333333 | Compositional bias | Note=Poly-Ser |
| Hgene | FOXO1 | chr13:41239720 | chr5:149499686 | ENST00000379561 | - | 1 | 3 | 91_102 | 210 | 1440.3333333333333 | Compositional bias | Note=Poly-Ala |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FOXO1 | chr13:41239720 | chr5:149499686 | ENST00000379561 | - | 1 | 3 | 159_235 | 210 | 1440.3333333333333 | DNA binding | Fork-head |
| Hgene | FOXO1 | chr13:41239720 | chr5:149499686 | ENST00000379561 | - | 1 | 3 | 211_218 | 210 | 1440.3333333333333 | Region | Note=DNA-binding |
| Hgene | FOXO1 | chr13:41239720 | chr5:149499686 | ENST00000379561 | - | 1 | 3 | 234_237 | 210 | 1440.3333333333333 | Region | Note=DNA-binding |
| Tgene | PDGFRB | chr13:41239720 | chr5:149499686 | ENST00000261799 | 17 | 23 | 129_210 | 862 | 1107.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | PDGFRB | chr13:41239720 | chr5:149499686 | ENST00000261799 | 17 | 23 | 214_309 | 862 | 1107.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | PDGFRB | chr13:41239720 | chr5:149499686 | ENST00000261799 | 17 | 23 | 331_403 | 862 | 1107.0 | Domain | Note=Ig-like C2-type 4 | |
| Tgene | PDGFRB | chr13:41239720 | chr5:149499686 | ENST00000261799 | 17 | 23 | 33_120 | 862 | 1107.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | PDGFRB | chr13:41239720 | chr5:149499686 | ENST00000261799 | 17 | 23 | 416_524 | 862 | 1107.0 | Domain | Note=Ig-like C2-type 5 | |
| Tgene | PDGFRB | chr13:41239720 | chr5:149499686 | ENST00000261799 | 17 | 23 | 600_962 | 862 | 1107.0 | Domain | Protein kinase | |
| Tgene | PDGFRB | chr13:41239720 | chr5:149499686 | ENST00000261799 | 17 | 23 | 606_614 | 862 | 1107.0 | Nucleotide binding | ATP | |
| Tgene | PDGFRB | chr13:41239720 | chr5:149499686 | ENST00000261799 | 17 | 23 | 33_532 | 862 | 1107.0 | Topological domain | Extracellular | |
| Tgene | PDGFRB | chr13:41239720 | chr5:149499686 | ENST00000261799 | 17 | 23 | 554_1106 | 862 | 1107.0 | Topological domain | Cytoplasmic | |
| Tgene | PDGFRB | chr13:41239720 | chr5:149499686 | ENST00000261799 | 17 | 23 | 533_553 | 862 | 1107.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for FOXO1-PDGFRB |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >31166_31166_1_FOXO1-PDGFRB_FOXO1_chr13_41239720_ENST00000379561_PDGFRB_chr5_149499686_ENST00000261799_length(transcript)=3676nt_BP=1015nt GCAGCCGCCACATTCAACAGGCAGCAGCGCAGCGGGCGCGCCGCTGGGGAGAGCAAGCGGCCCGCGGCGTCCGTCCGTCCTTCCGTCCGC GGCCCTGTCAGCTGGAGCGCGGCGCAGGCTCTGCCCCGGCCCGGCGGCTCTGGCCGGCCGTCCAGTCCGTGCGGCGGACCCCGAGGAGCC TCGATGTGGATGGCCCCGCGAAGTTAAGTTCTGGGCTCGCGCTTCCACTCCGCCGCGCCTTCCTCCCAGTTTCCGTCCGCTCGCCGCACC GGCTTCGTTCCCCCAAATCTCGGACCGTCCCTTCGCGCCCCCTCCCCGTCCGCCCCCAGTGCTGCGTTCTCCCCCTCTTGGCTCTCCTGC GGCTGGGGGAGGGGCGGGGGTCACCATGGCCGAGGCGCCTCAGGTGGTGGAGATCGACCCGGACTTCGAGCCGCTGCCCCGGCCGCGCTC GTGCACCTGGCCGCTGCCCAGGCCGGAGTTTAGCCAGTCCAACTCGGCCACCTCCAGCCCGGCGCCGTCGGGCAGCGCGGCTGCCAACCC CGACGCCGCGGCGGGCCTGCCCTCGGCCTCGGCTGCCGCTGTCAGCGCCGACTTCATGAGCAACCTGAGCTTGCTGGAGGAGAGCGAGGA CTTCCCGCAGGCGCCCGGCTCCGTGGCGGCGGCGGTGGCGGCGGCGGCCGCCGCGGCCGCCACCGGGGGGCTGTGCGGGGACTTCCAGGG CCCGGAGGCGGGCTGCCTGCACCCAGCGCCACCGCAGCCCCCGCCGCCCGGGCCGCTGTCGCAGCACCCGCCGGTGCCCCCCGCCGCCGC TGGGCCGCTCGCGGGGCAGCCGCGCAAGAGCAGCTCGTCCCGCCGCAACGCGTGGGGCAACCTGTCCTACGCCGACCTCATCACCAAGGC CATCGAGAGCTCGGCGGAGAAGCGGCTCACGCTGTCGCAGATCTACGAGTGGATGGTCAAGAGCGTGCCCTACTTCAAGGATAAGGGTGA CAGCAACAGCTCGGCGGGCTGGAAGACCTTTTTGCCTTTAAAGTGGATGGCTCCGGAGAGCATCTTCAACAGCCTCTACACCACCCTGAG CGACGTGTGGTCCTTCGGGATCCTGCTCTGGGAGATCTTCACCTTGGGTGGCACCCCTTACCCAGAGCTGCCCATGAACGAGCAGTTCTA CAATGCCATCAAACGGGGTTACCGCATGGCCCAGCCTGCCCATGCCTCCGACGAGATCTATGAGATCATGCAGAAGTGCTGGGAAGAGAA GTTTGAGATTCGGCCCCCCTTCTCCCAGCTGGTGCTGCTTCTCGAGAGACTGTTGGGCGAAGGTTACAAAAAGAAGTACCAGCAGGTGGA TGAGGAGTTTCTGAGGAGTGACCACCCAGCCATCCTTCGGTCCCAGGCCCGCTTGCCTGGGTTCCATGGCCTCCGATCTCCCCTGGACAC CAGCTCCGTCCTCTATACTGCCGTGCAGCCCAATGAGGGTGACAACGACTATATCATCCCCCTGCCTGACCCCAAACCCGAGGTTGCTGA CGAGGGCCCACTGGAGGGTTCCCCCAGCCTAGCCAGCTCCACCCTGAATGAAGTCAACACCTCCTCAACCATCTCCTGTGACAGCCCCCT GGAGCCCCAGGACGAACCAGAGCCAGAGCCCCAGCTTGAGCTCCAGGTGGAGCCGGAGCCAGAGCTGGAACAGTTGCCGGATTCGGGGTG CCCTGCGCCTCGGGCGGAAGCAGAGGATAGCTTCCTGTAGGGGGCTGGCCCCTACCCTGCCCTGCCTGAAGCTCCCCCCCTGCCAGCACC CAGCATCTCCTGGCCTGGCCTGACCGGGCTTCCTGTCAGCCAGGCTGCCCTTATCAGCTGTCCCCTTCTGGAAGCTTTCTGCTCCTGACG TGTTGTGCCCCAAACCCTGGGGCTGGCTTAGGAGGCAAGAAAACTGCAGGGGCCGTGACCAGCCCTCTGCCTCCAGGGAGGCCAACTGAC TCTGAGCCAGGGTTCCCCCAGGGAACTCAGTTTTCCCATATGTAAAATGGGAAAGTTAGGCTTGATGACCCAGAATCTAGGATTCTCTCC CTGGCTGACAGGTGGGGAGACCGAATCCCTCCCTGGGAAGATTCTTGGAGTTACTGAGGTGGTAAATTAACTTTTTTCTGTTCAGCCAGC TACCCCTCAAGGAATCATAGCTCTCTCCTCGCACTTTTATCCACCCAGGAGCTAGGGAAGAGACCCTAGCCTCCCTGGCTGCTGGCTGAG CTAGGGCCTAGCCTTGAGCAGTGTTGCCTCATCCAGAAGAAAGCCAGTCTCCTCCCTATGATGCCAGTCCCTGCGTTCCCTGGCCCGAGC TGGTCTGGGGCCATTAGGCAGCCTAATTAATGCTGGAGGCTGAGCCAAGTACAGGACACCCCCAGCCTGCAGCCCTTGCCCAGGGCACTT GGAGCACACGCAGCCATAGCAAGTGCCTGTGTCCCTGTCCTTCAGGCCCATCAGTCCTGGGGCTTTTTCTTTATCACCCTCAGTCTTAAT CCATCCACCAGAGTCTAGAAGGCCAGACGGGCCCCGCATCTGTGATGAGAATGTAAATGTGCCAGTGTGGAGTGGCCACGTGTGTGTGCC AGTATATGGCCCTGGCTCTGCATTGGACCTGCTATGAGGCTTTGGAGGAATCCCTCACCCTCTCTGGGCCTCAGTTTCCCCTTCAAAAAA TGAATAAGTCGGACTTATTAACTCTGAGTGCCTTGCCAGCACTAACATTCTAGAGTATTCCAGGTGGTTGCACATTTGTCCAGATGAAGC AAGGCCATATACCCTAAACTTCCATCCTGGGGGTCAGCTGGGCTCCTGGGAGATTCCAGATCACACATCACACTCTGGGGACTCAGGAAC CATGCCCCTTCCCCAGGCCCCCAGCAAGTCTCAAGAACACAGCTGCACAGGCCTTGACTTAGAGTGACAGCCGGTGTCCTGGAAAGCCCC CAGCAGCTGCCCCAGGGACATGGGAAGACCACGGGACCTCTTTCACTACCCACGATGACCTCCGGGGGTATCCTGGGCAAAAGGGACAAA GAGGGCAAATGAGATCACCTCCTGCAGCCCACCACTCCAGCACCTGTGCCGAGGTCTGCGTCGAAGACAGAATGGACAGTGAGGACAGTT ATGTCTTGTAAAAGACAAGAAGCTTCAGATGGGTACCCCAAGAAGGATGTGAGAGGTGGGCGCTTTGGAGGTTTGCCCCTCACCCACCAG CTGCCCCATCCCTGAGGCAGCGCTCCATGGGGGTATGGTTTTGTCACTGCCCAGACCTAGCAGTGACATCTCATTGTCCCCAGCCCAGTG GGCATTGGAGGTGCCAGGGGAGTCAGGGTTGTAGCCAAGACGCCCCCGCACGGGGAGGGTTGGGAAGGGGGTGCAGGAAGCTCAACCCCT CTGGGCACCAACCCTGCATTGCAGGTTGGCACCTTACTTCCCTGGGATCCCCAGAGTTGGTCCAAGGAGGGAGAGTGGGTTCTCAATACG GTACCAAAGATATAATCACCTAGGTTTACAAATATTTTTAGGACTCACGTTAACTCACATTTATACAGCAGAAATGCTATTTTGTATGCT >31166_31166_1_FOXO1-PDGFRB_FOXO1_chr13_41239720_ENST00000379561_PDGFRB_chr5_149499686_ENST00000261799_length(amino acids)=568AA_BP=324 MGRASGPRRPSVLPSAALSAGARRRLCPGPAALAGRPVRAADPEEPRCGWPREVKFWARASTPPRLPPSFRPLAAPASFPQISDRPFAPP PRPPPVLRSPPLGSPAAGGGAGVTMAEAPQVVEIDPDFEPLPRPRSCTWPLPRPEFSQSNSATSSPAPSGSAAANPDAAAGLPSASAAAV SADFMSNLSLLEESEDFPQAPGSVAAAVAAAAAAAATGGLCGDFQGPEAGCLHPAPPQPPPPGPLSQHPPVPPAAAGPLAGQPRKSSSSR RNAWGNLSYADLITKAIESSAEKRLTLSQIYEWMVKSVPYFKDKGDSNSSAGWKTFLPLKWMAPESIFNSLYTTLSDVWSFGILLWEIFT LGGTPYPELPMNEQFYNAIKRGYRMAQPAHASDEIYEIMQKCWEEKFEIRPPFSQLVLLLERLLGEGYKKKYQQVDEEFLRSDHPAILRS QARLPGFHGLRSPLDTSSVLYTAVQPNEGDNDYIIPLPDPKPEVADEGPLEGSPSLASSTLNEVNTSSTISCDSPLEPQDEPEPEPQLEL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FOXO1-PDGFRB |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FOXO1-PDGFRB |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FOXO1-PDGFRB |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
